Upstream Stimulatory Factors Regulate HIV-1 Latency and Are Required for Robust T Cell Activation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell and Virus Culture
2.2. Immunoblotting
2.3. Luciferase Expression Assays
2.4. Transfection Assays
2.5. Flow Cytometry
2.6. Generation of USF1 and USF2 Knockout Clonal Lines
2.7. USF1 and USF2 shRNA Knockdown
2.8. ChIP-qPCR
2.9. RT-PCR
2.10. RNA-Seq
2.11. Statistics and Reproducibility
3. Results
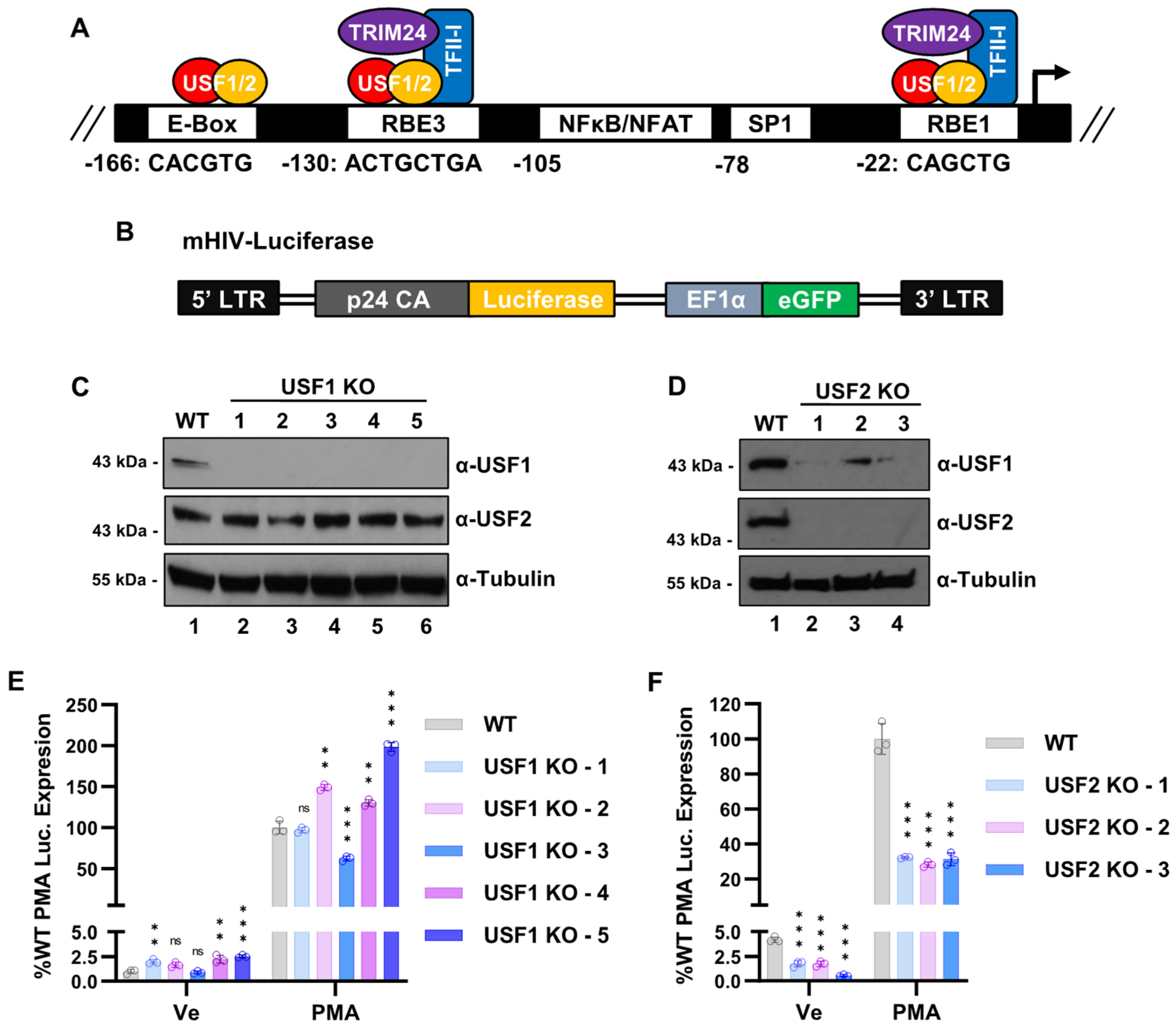
3.1. Regulation of the HIV-1 Expression by USF1 and USF2
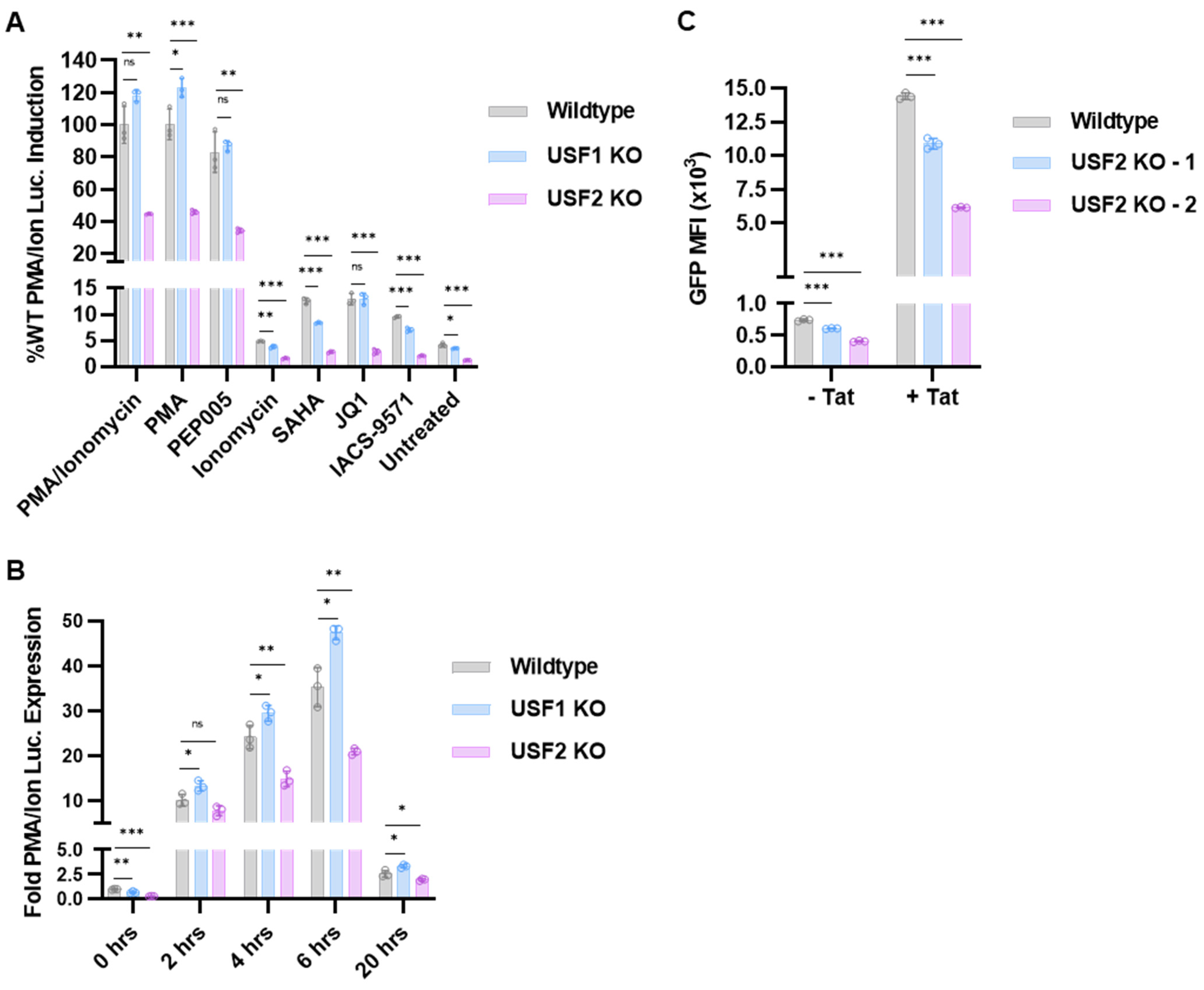
3.2. Effect of USF1 and USF2 on Response of HIV-1 to Latency Reversing Agents and Tat
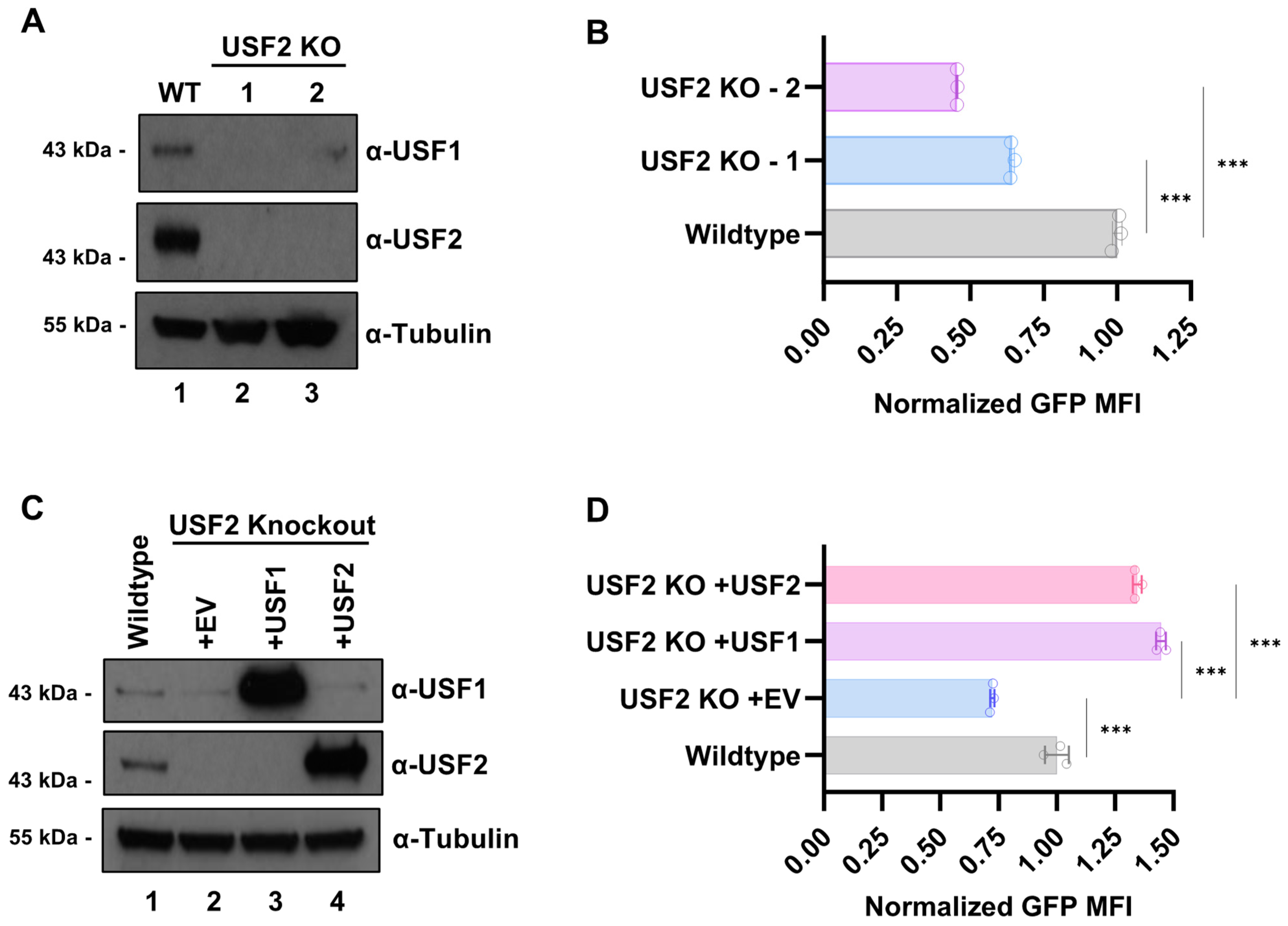
3.3. USF2 Stabilizes USF1 Protein
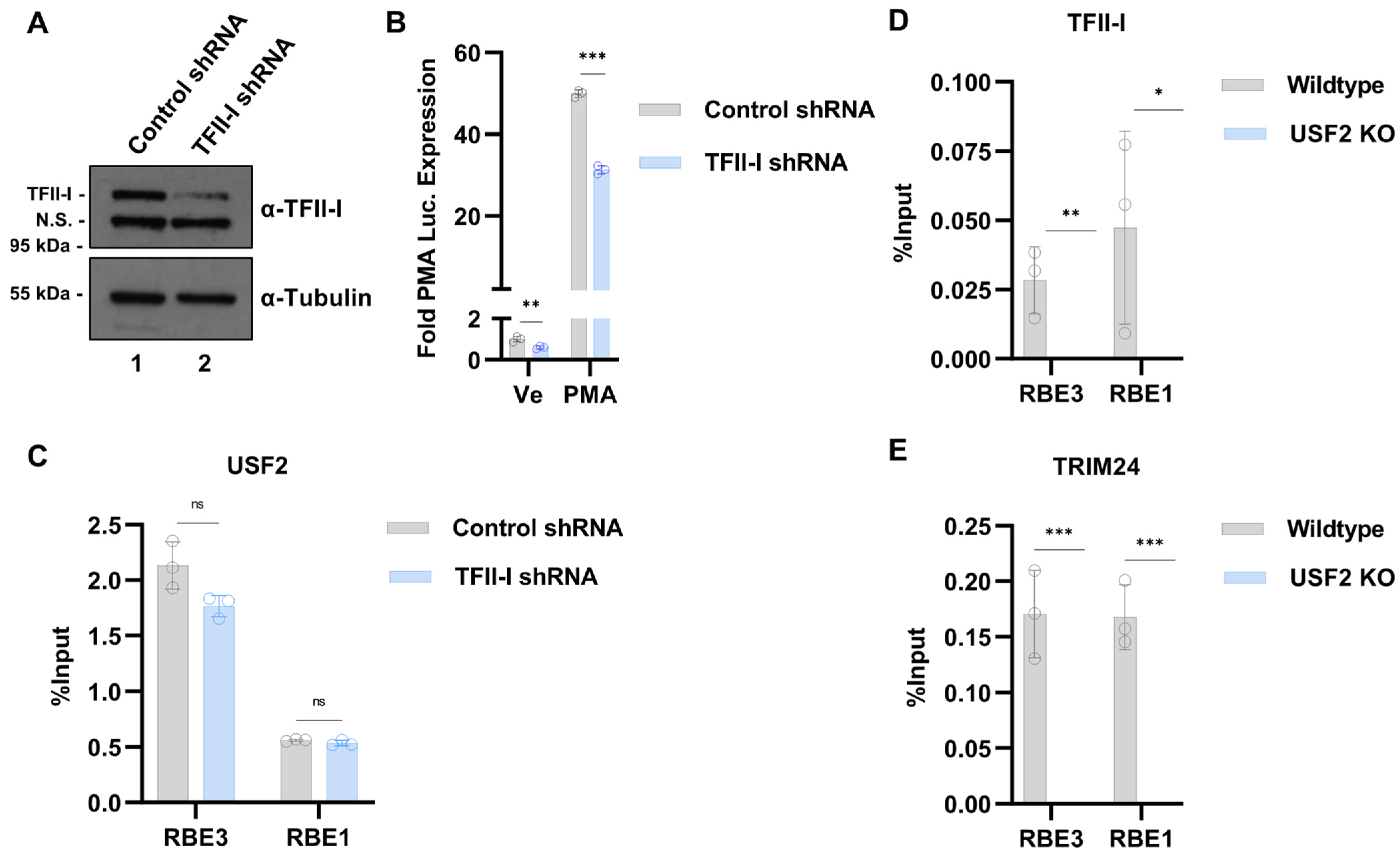
3.4. Effect of USF Knockouts on the Interaction of RBF-2 Proteins with the LTR
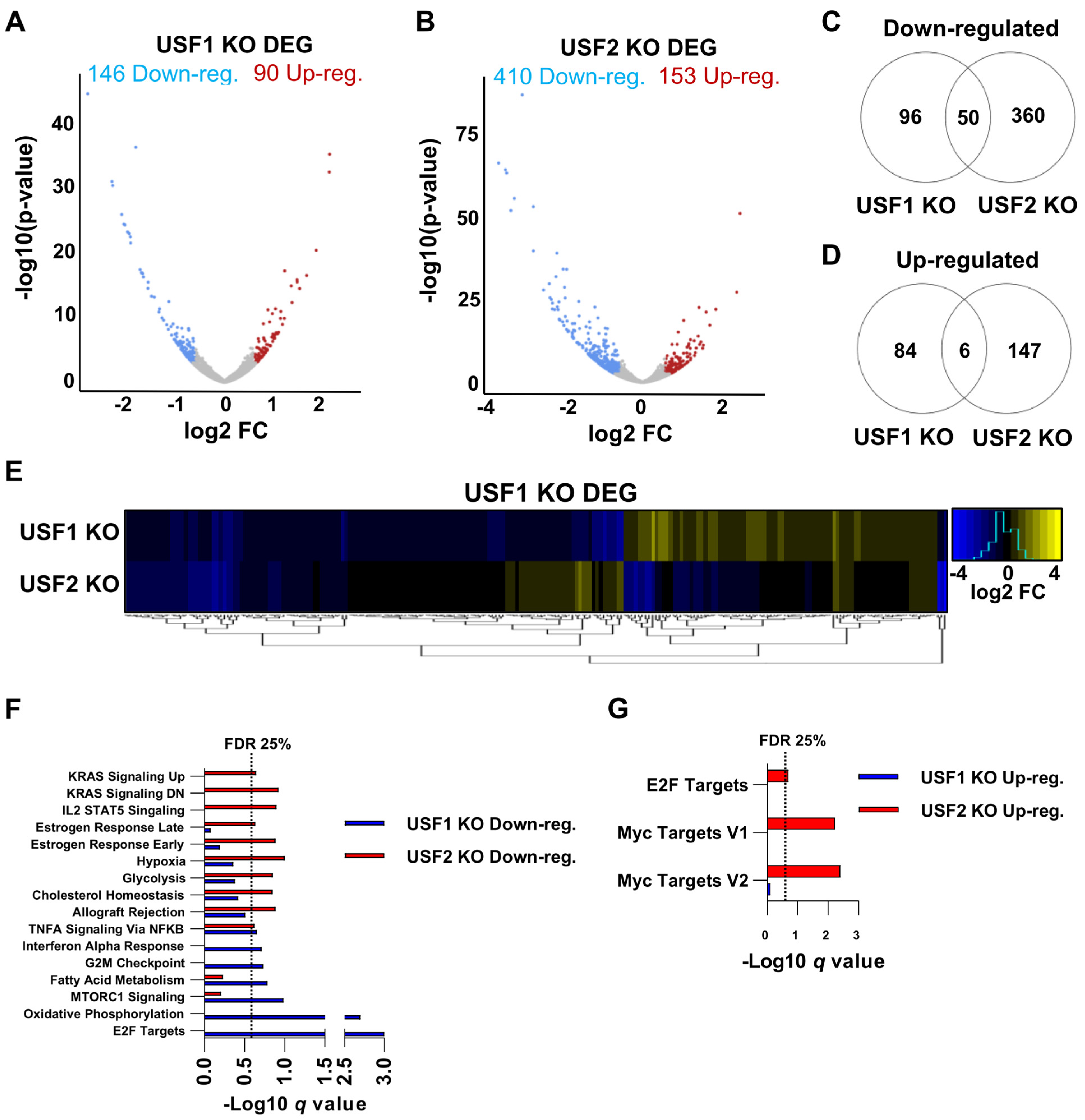
3.5. Global Regulation of Transcription by USF1 and USF2 in Unstimulated T Cells
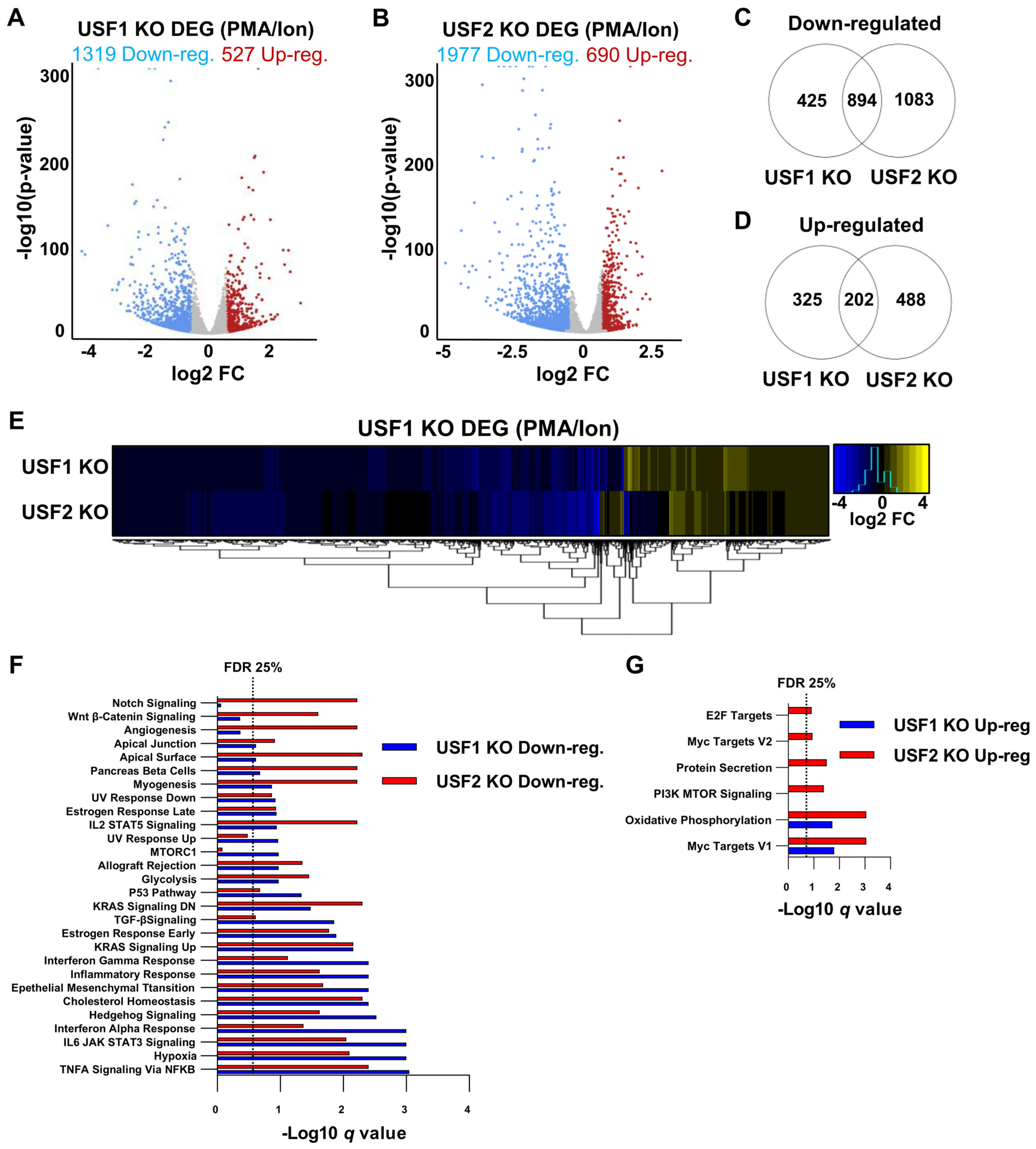
3.6. USF1 and USF2 Cooperatively Regulate Global Gene Expression in Activated T Cells
3.7. USF1 and USF2 Facilitated T Cell Activation
3.8. USF2 Deficient Cells Are Refractory for HIV-1 Infection
3.9. USF2 Promoted Productive HIV-1 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- UNAIDS In Danger—UNAIDS Global AIDS Update 2022; Joint United Nations Programme on HIV/AIDS: Geneva, Switzerland, 2022.
- Temereanca, A.; Ruta, S. Strategies to Overcome HIV Drug Resistance-Current and Future Perspectives. Front. Microbiol. 2023, 14, 1133407. [Google Scholar] [CrossRef] [PubMed]
- Sadowski, I.; Hashemi, F.B. Strategies to Eradicate HIV from Infected Patients: Elimination of Latent Provirus Reservoirs. Cell. Mol. Life Sci. 2019, 76, 3583–3600. [Google Scholar] [CrossRef] [PubMed]
- Puertas, M.C.; Ploumidis, G.; Ploumidis, M.; Fumero, E.; Clotet, B.; Walworth, C.M.; Petropoulos, C.J.; Martinez-Picado, J. Pan-Resistant HIV-1 Emergence in the Era of Integrase Strand-Transfer Inhibitors: A Case Report. Lancet Microbe 2020, 1, e130–e135. [Google Scholar] [CrossRef] [PubMed]
- Finzi, D. Identification of a Reservoir for HIV-1 in Patients on Highly Active Antiretroviral Therapy. Science 1997, 278, 1295–1300. [Google Scholar] [CrossRef] [PubMed]
- Abner, E.; Jordan, A. HIV “Shock and Kill” Therapy: In Need of Revision. Antivir. Res. 2019, 166, 19–34. [Google Scholar] [CrossRef]
- Pasquereau, S.; Herbein, G. CounterAKTing HIV: Toward a “Block and Clear” Strategy? Front. Cell. Infect. Microbiol. 2022, 12, 827717. [Google Scholar] [CrossRef]
- Petravic, J.; Rasmussen, T.A.; Lewin, S.R.; Kent, S.J.; Davenport, M.P. Relationship between Measures of HIV Reactivation and Decline of the Latent Reservoir under Latency-Reversing Agents. J. Virol. 2017, 91, e02092-16. [Google Scholar] [CrossRef]
- Walker-Sperling, V.E.; Pohlmeyer, C.W.; Tarwater, P.M.; Blankson, J.N. The Effect of Latency Reversal Agents on Primary CD8+ T Cells: Implications for Shock and Kill Strategies for Human Immunodeficiency Virus Eradication. eBioMedicine 2016, 8, 217–229. [Google Scholar] [CrossRef]
- Kim, Y.; Anderson, J.L.; Lewin, S.R. Getting the “Kill” into “Shock and Kill”: Strategies to Eliminate Latent HIV. Cell Host Microbe 2018, 23, 14–26. [Google Scholar] [CrossRef]
- Pereira, L.A. SURVEY AND SUMMARY A Compilation of Cellular Transcription Factor Interactions with the HIV-1 LTR Promoter. Nucleic Acids Res. 2000, 28, 663–668. [Google Scholar] [CrossRef]
- Brooks, D.G.; Arlen, P.A.; Gao, L.; Kitchen, C.M.R.; Zack, J.A. Identification of T Cell-Signaling Pathways That Stimulate Latent HIV in Primary Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 12955–12960. [Google Scholar] [CrossRef]
- Estable, M.C.; Bell, B.; Merzouki, A.; Montaner, J.S.G.; O’Shaughnessy, M.V.; Sadowski, I.J. Human Immunodeficiency Virus Type 1 Long Terminal Repeat Variants from 42 Patients Representing All Stages of Infection Display a Wide Range of Sequence Polymorphism and Transcription Activity. J. Virol. 1996, 70, 4053–4062. [Google Scholar] [CrossRef]
- Bell, B.; Sadowski, I. Ras-Responsiveness of the HIV-1 LTR Requires RBF-1 and RBF-2 Binding Sites. Oncogene 1996, 13, 2687–2697. [Google Scholar] [PubMed]
- Chen, J.; Malcolm, T.; Estable, M.C.; Roeder, R.G.; Sadowski, I. TFII-I Regulates Induction of Chromosomally Integrated Human Immunodeficiency Virus Type 1 Long Terminal Repeat in Cooperation with USF. J. Virol. 2005, 79, 4396–4406. [Google Scholar] [CrossRef] [PubMed]
- Bernhard, W.; Barreto, K.; Raithatha, S.; Sadowski, I. An Upstream YY1 Binding Site on the HIV-1 LTR Contributes to Latent Infection. PLoS ONE 2013, 8, e77052. [Google Scholar] [CrossRef]
- Malcolm, T.; Chen, J.; Chang, C.; Sadowski, I. Induction of Chromosomally Integrated HIV-1 LTR Requires RBF-2 (USF/TFII-I) and RAS/MAPK Signaling. Virus Genes 2007, 35, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Malcolm, T.; Kam, J.; Pour, P.S.; Sadowski, I. Specific Interaction of TFII-I with an Upstream Element on the HIV-1 LTR Regulates Induction of Latent Provirus. FEBS Lett. 2008, 582, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Ooms, M.; Malcolm, T.; Simon, V.; Sadowski, I. Identification and Functional Analysis of a Second RBF-2 Binding Site within the HIV-1 Promoter. Virology 2011, 418, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Horvath, R.M.; Dahabieh, M.; Malcolm, T.; Sadowski, I. TRIM24 Controls Induction of Latent HIV-1 by Stimulating Transcriptional Elongation. Commun. Biol. 2023, 6, 86. [Google Scholar] [CrossRef]
- Horvath, R.M.; Brumme, Z.L.; Sadowski, I. Inhibition of the TRIM24 Bromodomain Reactivates Latent HIV-1. Sci. Rep. 2023, 13, 556. [Google Scholar] [CrossRef]
- Sawadogo, M. Interaction of a Gene-Specific Transcription Factor with the Adenovirus Major Late Promoter Upstream of the TATA Box Region. Cell 1985, 43, 165–175. [Google Scholar] [CrossRef]
- Sawadogo, M.; Van Dyke, M.W.; Gregor, P.D.; Roeder, R.G. Multiple Forms of the Human Gene-Specific Transcription Factor USF. I. Complete Purification and Identification of USF from HeLa Cell Nuclei. J. Biol. Chem. 1988, 263, 11985–11993. [Google Scholar] [CrossRef]
- Gregor, P.D.; Sawadogo, M.; Roeder, R.G. The Adenovirus Major Late Transcription Factor USF Is a Member of the Helix-Loop-Helix Group of Regulatory Proteins and Binds to DNA as a Dimer. Genes Dev. 1990, 4, 1730–1740. [Google Scholar] [CrossRef]
- Sirito, M.; Lin, Q.; Maity, T.; Sawadogo, M. Ubiquitous Expression of the 43- and 44-KDa Forms of Transcription Factor USF in Mammalian Cells. Nucleic Acids Res. 1994, 22, 427–433. [Google Scholar] [CrossRef]
- Viollet, B.; Lefrançois-Martinez, A.-M.; Henrion, A.; Kahn, A.; Raymondjean, M.; Martinez, A. Immunochemical Characterization and Transacting Properties of Upstream Stimulatory Factor Isoforms. J. Biol. Chem. 1996, 271, 1405–1415. [Google Scholar] [CrossRef] [PubMed]
- Sirito, M.; Lin, Q.; Deng, J.M.; Behringer, R.R.; Sawadogo, M. Overlapping Roles and Asymmetrical Cross-Regulation of the USF Proteins in Mice. Proc. Natl. Acad. Sci. USA 1998, 95, 3758–3763. [Google Scholar] [CrossRef] [PubMed]
- Qyang, Y.; Luo, X.; Lu, T.; Ismail, P.M.; Krylov, D.; Vinson, C.; Sawadogo, M. Cell-Type-Dependent Activity of the Ubiquitous Transcription Factor USF in Cellular Proliferation and Transcriptional Activation. Mol. Cell. Biol. 1999, 19, 1508–1517. [Google Scholar] [CrossRef] [PubMed]
- Tiruppathi, C.; Soni, D.; Wang, D.-M.; Xue, J.; Singh, V.; Thippegowda, P.B.; Cheppudira, B.P.; Mishra, R.K.; DebRoy, A.; Qian, Z.; et al. The Transcription Factor DREAM Represses the Deubiquitinase A20 and Mediates Inflammation. Nat. Immunol. 2014, 15, 239–247. [Google Scholar] [CrossRef]
- Hu, D.; Tjon, E.C.; Andersson, K.M.; Molica, G.M.; Pham, M.C.; Healy, B.; Murugaiyan, G.; Pochet, N.; Kuchroo, V.K.; Bokarewa, M.I.; et al. Aberrant Expression of USF2 in Refractory Rheumatoid Arthritis and Its Regulation of Proinflammatory Cytokines in Th17 Cells. Proc. Natl. Acad. Sci. USA 2020, 117, 30639–30648. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Zhou, X.; Wright, S.; Hyle, J.; Zhao, L.; An, J.; Zhao, X.; Shao, Y.; Xu, B.; et al. Functional Interrogation of HOXA9 Regulome in MLLr Leukemia via Reporter-Based CRISPR/Cas9 Screen. eLife 2020, 9, e57858. [Google Scholar] [CrossRef]
- Horbach, T.; Götz, C.; Kietzmann, T.; Dimova, E.Y. Protein Kinases as Switches for the Function of Upstream Stimulatory Factors: Implications for Tissue Injury and Cancer. Front. Pharmacol. 2015, 6, 3. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.; Helleboid-Chapman, A.; Jakel, H.; Martin, G.; Duran-Sandoval, D.; Staels, B.; Rubin, E.M.; Pennacchio, L.A.; Taskinen, M.-R.; Fruchart-Najib, J.; et al. Insulin-Mediated Down-Regulation of Apolipoprotein A5 Gene Expression through the Phosphatidylinositol 3-Kinase Pathway: Role of Upstream Stimulatory Factor. Mol. Cell. Biol. 2005, 25, 1537–1548. [Google Scholar] [CrossRef] [PubMed]
- Spohrer, S.; Groß, R.; Nalbach, L.; Schwind, L.; Stumpf, H.; Menger, M.D.; Ampofo, E.; Montenarh, M.; Götz, C. Functional Interplay between the Transcription Factors USF1 and PDX-1 and Protein Kinase CK2 in Pancreatic β-Cells. Sci. Rep. 2017, 7, 16367. [Google Scholar] [CrossRef] [PubMed]
- Chi, T.; Horbach, T.; Götz, C.; Kietzmann, T.; Dimova, E. Cyclin-Dependent Kinase 5 (CDK5)-Mediated Phosphorylation of Upstream Stimulatory Factor 2 (USF2) Contributes to Carcinogenesis. Cancers 2019, 11, 523. [Google Scholar] [CrossRef] [PubMed]
- Lupp, S.; Götz, C.; Khadouma, S.; Horbach, T.; Dimova, E.Y.; Bohrer, A.-M.; Kietzmann, T.; Montenarh, M. The Upstream Stimulatory Factor USF1 Is Regulated by Protein Kinase CK2 Phosphorylation. Cell. Signal. 2014, 26, 2809–2817. [Google Scholar] [CrossRef]
- Roy, A.L. Cloning of an Inr- and E-Box-Binding Protein, TFII-I, That Interacts Physically and Functionally with USF1. EMBO J. 1997, 16, 7091–7104. [Google Scholar] [CrossRef]
- Du, H.; Roy, A.L.; Roeder, R.G. Human Transcription Factor USF Stimulates Transcription through the Initiator Elements of the HIV-1 and the Ad-ML Promoters. EMBO J. 1993, 12, 501–511. [Google Scholar] [CrossRef]
- Afgan, E.; Baker, D.; Batut, B.; van den Beek, M.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Grüning, B.A.; et al. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2018 Update. Nucleic Acids Res. 2018, 46, W537–W544. [Google Scholar] [CrossRef]
- d’Adda di Fagagna, F.; Marzio, G.; Gutierrez, M.I.; Kang, L.Y.; Falaschi, A.; Giacca, M. Molecular and Functional Interactions of Transcription Factor USF with the Long Terminal Repeat of Human Immunodeficiency Virus Type 1. J. Virol. 1995, 69, 2765–2775. [Google Scholar] [CrossRef]
- Sieweke, M.H. Cooperative Interaction of Ets-1 with USF-1 Required for HIV-1 Enhancer Activity in T Cells. EMBO J. 1998, 17, 1728–1739. [Google Scholar] [CrossRef]
- Bernhard, W.; Barreto, K.; Saunders, A.; Dahabieh, M.S.; Johnson, P.; Sadowski, I. The Suv39H1 Methyltransferase Inhibitor Chaetocin Causes Induction of Integrated HIV-1 without Producing a T Cell Response. FEBS Lett. 2011, 585, 3549–3554. [Google Scholar] [CrossRef] [PubMed]
- Mandala, W.; Harawa, V.; Munyenyembe, A.; Soko, M.; Longwe, H. Optimization of Stimulation and Staining Conditions for Intracellular Cytokine Staining (ICS) for Determination of Cytokine-Producing T Cells and Monocytes. Curr. Res. Immunol. 2021, 2, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Kedei, N.; Lundberg, D.J.; Toth, A.; Welburn, P.; Garfield, S.H.; Blumberg, P.M. Characterization of the Interaction of Ingenol 3-Angelate with Protein Kinase C. Cancer Res. 2004, 64, 3243–3255. [Google Scholar] [CrossRef] [PubMed]
- Desimio, M.G.; Giuliani, E.; Doria, M. The Histone Deacetylase Inhibitor SAHA Simultaneously Reactivates HIV-1 from Latency and up-Regulates NKG2D Ligands Sensitizing for Natural Killer Cell Cytotoxicity. Virology 2017, 510, 9–21. [Google Scholar] [CrossRef]
- Li, Z.; Guo, J.; Wu, Y.; Zhou, Q. The BET Bromodomain Inhibitor JQ1 Activates HIV Latency through Antagonizing Brd4 Inhibition of Tat-Transactivation. Nucleic Acids Res. 2013, 41, 277–287. [Google Scholar] [CrossRef]
- Galibert, M.-D. The Usf-1 Transcription Factor Is a Novel Target for the Stress-Responsive P38 Kinase and Mediates UV-Induced Tyrosinase Expression. EMBO J. 2001, 20, 5022–5031. [Google Scholar] [CrossRef]
- Corre, S.; Primot, A.; Sviderskaya, E.; Bennett, D.C.; Vaulont, S.; Goding, C.R.; Galibert, M.-D. UV-Induced Expression of Key Component of the Tanning Process, the POMC and MC1R Genes, Is Dependent on the p-38-Activated Upstream Stimulating Factor-1 (USF-1). J. Biol. Chem. 2004, 279, 51226–51233. [Google Scholar] [CrossRef]
- Bouafia, A.; Corre, S.; Gilot, D.; Mouchet, N.; Prince, S.; Galibert, M.-D. P53 Requires the Stress Sensor USF1 to Direct Appropriate Cell Fate Decision. PLoS Genet. 2014, 10, e1004309. [Google Scholar] [CrossRef]
- Tong, A.-J.; Liu, X.; Thomas, B.J.; Lissner, M.M.; Baker, M.R.; Senagolage, M.D.; Allred, A.L.; Barish, G.D.; Smale, S.T. A Stringent Systems Approach Uncovers Gene-Specific Mechanisms Regulating Inflammation. Cell 2016, 165, 165–179. [Google Scholar] [CrossRef]
- Gagne, M.; Michaels, D.; Schiralli Lester, G.M.; Gummuluru, S.; Wong, W.W.; Henderson, A.J. Strength of T Cell Signaling Regulates HIV-1 Replication and Establishment of Latency. PLoS Pathog. 2019, 15, e1007802. [Google Scholar] [CrossRef]
- Korin, Y.D.; Zack, J.A. Progression to the G 1 b Phase of the Cell Cycle Is Required for Completion of Human Immunodeficiency Virus Type 1 Reverse Transcription in T Cells. J. Virol. 1998, 72, 3161–3168. [Google Scholar] [CrossRef] [PubMed]
- Plesa, G.; Dai, J.; Baytop, C.; Riley, J.L.; June, C.H.; O’Doherty, U. Addition of Deoxynucleosides Enhances Human Immunodeficiency Virus Type 1 Integration and 2LTR Formation in Resting CD4 + T Cells. J. Virol. 2007, 81, 13938–13942. [Google Scholar] [CrossRef] [PubMed]
- Unutmaz, D.; KewalRamani, V.N.; Marmon, S.; Littman, D.R. Cytokine Signals Are Sufficient for HIV-1 Infection of Resting Human T Lymphocytes. J. Exp. Med. 1999, 189, 1735–1746. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Ooms, M.; Brumme, C.; Taylor, J.; Harrigan, P.R.; Simon, V.; Sadowski, I. Direct Non-Productive HIV-1 Infection in a T-Cell Line Is Driven by Cellular Activation State and NFκB. Retrovirology 2014, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Dahabieh, M.S.; Ooms, M.; Simon, V.; Sadowski, I. A Doubly Fluorescent HIV-1 Reporter Shows That the Majority of Integrated HIV-1 Is Latent Shortly after Infection. J. Virol. 2013, 87, 4716–4727. [Google Scholar] [CrossRef] [PubMed]
- Battivelli, E.; Verdin, E. HIVGKO: A Tool to Assess HIV-1 Latency Reversal Agents in Human Primary CD4+ T Cells. Bio-Protocol 2018, 8, e3050. [Google Scholar] [CrossRef]
- Corre, S.; Primot, A.; Baron, Y.; Le Seyec, J.; Goding, C.; Galibert, M.-D. Target Gene Specificity of USF-1 Is Directed via P38-Mediated Phosphorylation-Dependent Acetylation. J. Biol. Chem. 2009, 284, 18851–18862. [Google Scholar] [CrossRef]
- Cheung, E.; Mayr, P.; Coda-Zabetta, F.; Woodman, P.G.; Boam, D.S.W. DNA-Binding Activity of the Transcription Factor Upstream Stimulatory Factor 1 (USF-1) Is Regulated by Cyclin-Dependent Phosphorylation. Biochem. J. 1999, 344, 145–152. [Google Scholar] [CrossRef]
- Xiao, Q.; Kenessey, A.; Ojamaa, K. Role of USF1 Phosphorylation on Cardiac α-Myosin Heavy Chain Promoter Activity. Am. J. Physiol.-Heart Circ. Physiol. 2002, 283, H213–H219. [Google Scholar] [CrossRef]
- Wong, R.H.F.; Sul, H.S. DNA-PK: Relaying the Insulin Signal to USF in Lipogenesis. Cell Cycle 2009, 8, 1973–1978. [Google Scholar] [CrossRef]
- Terragni, J.; Nayak, G.; Banerjee, S.; Medrano, J.-L.; Graham, J.R.; Brennan, J.F.; Sepulveda, S.; Cooper, G.M. The E-Box Binding Factors Max/Mnt, MITF, and USF1 Act Coordinately with FoxO to Regulate Expression of Proapoptotic and Cell Cycle Control Genes by Phosphatidylinositol 3-Kinase/Akt/Glycogen Synthase Kinase 3 Signaling. J. Biol. Chem. 2011, 286, 36215–36227. [Google Scholar] [CrossRef] [PubMed]
- Maekawa, T.; Sudo, T.; Kurimoto, M.; Ishii, S. USF-Related Transcription Factor, HIV-TF1, Stimulates Transcription of Human Immunodeficiency Virus-1. Nucleic Acids Res. 1991, 19, 4689–4694. [Google Scholar] [CrossRef] [PubMed]
- Estable, M.C.; Hirst, M.; Bell, B.; O’Shaughnessy, M.V.; Sadowski, I. Purification of RBF-2, a Transcription Factor with Specificity for the Most Conserved Cis-Element of Naturally Occurring HIV-1 LTRs. J. Biomed. Sci. 1999, 6, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.-W.; Cochran, B.H. Extracellular Signal-Regulated Kinase Binds to TFII-I and Regulates Its Activation of the c-Fos Promoter. Mol. Cell. Biol. 2000, 20, 1140–1148. [Google Scholar] [CrossRef]
- Kim, D.-W.; Cheriyath, V.; Roy, A.L.; Cochran, B.H. TFII-I Enhances Activation of the c- Fos Promoter through Interactions with Upstream Elements. Mol. Cell. Biol. 1998, 18, 3310–3320. [Google Scholar] [CrossRef]
- Olave, N.C.; Grenett, M.H.; Cadeiras, M.; Grenett, H.E.; Higgins, P.J. Upstream Stimulatory Factor-2 Mediates Quercetin-Induced Suppression of PAI-1 Gene Expression in Human Endothelial Cells. J. Cell. Biochem. 2010, 111, 720–726. [Google Scholar] [CrossRef]
- Qi, L.; Higgins, C.E.; Higgins, S.P.; Law, B.K.; Simone, T.M.; Higgins, P.J. The Basic Helix-Loop-Helix/Leucine Zipper Transcription Factor USF2 Integrates Serum-Induced PAI-1 Expression and Keratinocyte Growth: THE BHLH-LZ TRANSCRIPTION FACTOR USF2. J. Cell. Biochem. 2014, 115, 1840–1847. [Google Scholar] [CrossRef]
- Johnson, P.R.; Swanson, R.; Rakhilina, L.; Hochstrasser, M. Degradation Signal Masking by Heterodimerization of MATalpha2 and MATa1 Blocks Their Mutual Destruction by the Ubiquitin-Proteasome Pathway. Cell 1998, 94, 217–227. [Google Scholar] [CrossRef]
- Huang, S.; Li, X.; Yusufzai, T.M.; Qiu, Y.; Felsenfeld, G. USF1 Recruits Histone Modification Complexes and Is Critical for Maintenance of a Chromatin Barrier. Mol. Cell. Biol. 2007, 27, 7991–8002. [Google Scholar] [CrossRef]
- West, A.G.; Huang, S.; Gaszner, M.; Litt, M.D.; Felsenfeld, G. Recruitment of Histone Modifications by USF Proteins at a Vertebrate Barrier Element. Mol. Cell 2004, 16, 453–463. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Horvath, R.M.; Sadowski, I. Upstream Stimulatory Factors Regulate HIV-1 Latency and Are Required for Robust T Cell Activation. Viruses 2023, 15, 1470. https://doi.org/10.3390/v15071470
Horvath RM, Sadowski I. Upstream Stimulatory Factors Regulate HIV-1 Latency and Are Required for Robust T Cell Activation. Viruses. 2023; 15(7):1470. https://doi.org/10.3390/v15071470
Chicago/Turabian StyleHorvath, Riley M., and Ivan Sadowski. 2023. "Upstream Stimulatory Factors Regulate HIV-1 Latency and Are Required for Robust T Cell Activation" Viruses 15, no. 7: 1470. https://doi.org/10.3390/v15071470
APA StyleHorvath, R. M., & Sadowski, I. (2023). Upstream Stimulatory Factors Regulate HIV-1 Latency and Are Required for Robust T Cell Activation. Viruses, 15(7), 1470. https://doi.org/10.3390/v15071470




