Detection of a Novel Alphaherpesvirus and Avihepadnavirus in a Plantar Papilloma from a Rainbow Lorikeet (Trichoglosis moluccanus)
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Extraction and Sequencing
2.2. Sequence Data Analysis
2.3. Genome Annotation and Bioinformatics
2.4. Phylogenetic Analyses
3. Results
3.1. Evidence of a Novel Psittacid Alphaherpesvirus 6
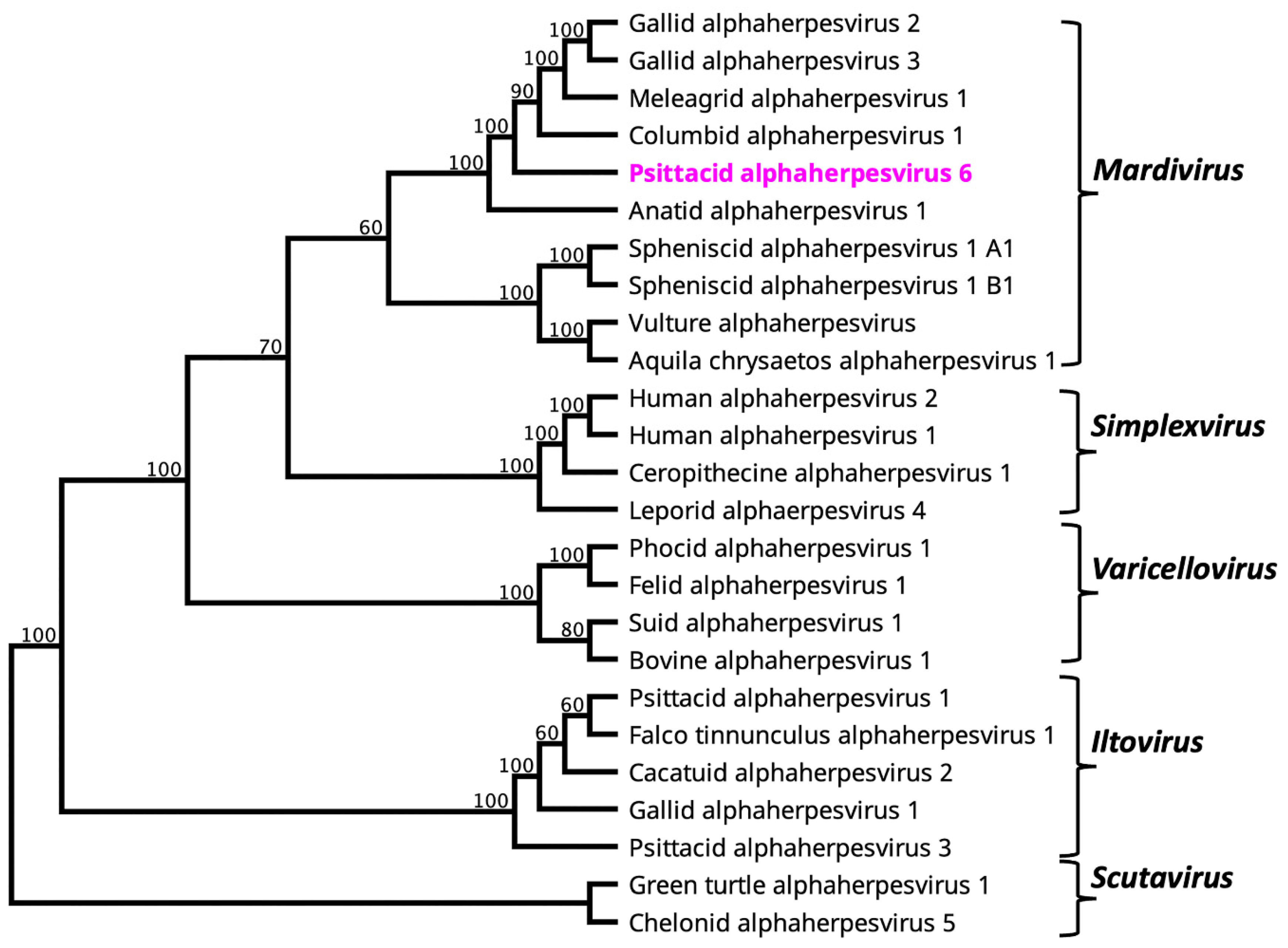
3.2. Evolutionary Relationship of PsAHV6
3.3. Evidence of a Novel Avihepadnavirus
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gatherer, D.; Depledge, D.P.; Hartley, C.A.; Szpara, M.L.; Vaz, P.K.; Benkő, M.; Brandt, C.R.; Bryant, N.A.; Dastjerdi, A.; Doszpoly, A.; et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J. Gen. Virol. 2021, 102, 001673. [Google Scholar] [CrossRef] [PubMed]
- Legler, M.; Kothe, R.; Wohlsein, P.; Hewicker-Trautwein, M.; Kummerfeld, N.; Rautenschlein, S. First detection of psittacid herpesvirus 2 in Congo African grey parrots (Psittacus erithacus erithacus) associated with pharyngeal papillomas and cloacal inflammation in Germany. Berl. Munch. Tierarztl. Wochenschr. 2014, 127, 222–226. [Google Scholar] [PubMed]
- Shivaprasad, H.L.; Phalen, D.N. A novel herpesvirus associated with respiratory disease in Bourke’s parrots (Neopsephotus bourkii). Avian Pathol. 2012, 41, 531–539. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Styles, D.K.; Tomaszewski, E.K.; Phalen, D.N. A novel psittacid herpesvirus found in African grey parrots (Psittacus erithacus erithacus). Avian Pathol. 2005, 34, 150–154. [Google Scholar] [CrossRef]
- Sutherland, M.; Sarker, S.; Raidal, S.R. Molecular and microscopic characterisation of a novel pathogenic herpesvirus from Indian ringneck parrots (Psittacula krameri). Vet. Microbiol. 2019, 239, 108428. [Google Scholar] [CrossRef]
- Sutherland, M.; Sarker, S.; Vaz, P.K.; Legione, A.R.; Devlin, J.M.; Macwhirter, P.L.; Whiteley, P.L.; Raidal, S.R. Disease surveillance in wild Victorian cacatuids reveals co-infection with multiple agents and detection of novel avian viruses. Vet. Microbiol. 2019, 235, 257–264. [Google Scholar] [CrossRef]
- Tomaszewski, E.K.; Kaleta, E.F.; Phalen, D.N. Molecular phylogeny of the psittacid herpesviruses causing Pacheco’s disease: Correlation of genotype with phenotypic expression. J. Virol. 2003, 77, 11260–11267. [Google Scholar] [CrossRef]
- Styles, D.K.; Tomaszewski, E.K.; Jaeger, L.A.; Phalen, D.N. Psittacid herpesviruses associated with mucosal papillomas in neotropical parrots. Virology 2004, 325, 24–35. [Google Scholar] [CrossRef]
- Wyss, F.; Schumacher, V.; Wenker, C.; Hoby, S.; Gobeli, S.; Arnaud, A.; Engels, M.; Friess, M.; Lange, C.E.; Stoffel, M.H.; et al. Pododermatitis in Captive and Free-Ranging Greater Flamingos (Phoenicopterus roseus). Vet. Pathol. 2015, 52, 1235–1242. [Google Scholar] [CrossRef]
- De Thoisy, B.; Lavergne, A.; Semelin, J.; Pouliquen, J.F.; Blanchard, F.; Hansen, E.; Lacoste, V. Outbreaks of disease possibly due to a natural avian herpesvirus infection in a colony of young Magnificent Frigatebirds (Fregata magnificens) in French Guiana. J. Wildl. Dis. 2009, 45, 802–807. [Google Scholar] [CrossRef][Green Version]
- Sebastiano, M.; Canestrelli, D.; Bisconti, R.; Lavergne, A.; Pineau, K.; Chastel, O.; Lacoste, V.; Costantini, D. Detection and Phylogenetic Characterization of a Novel Herpesvirus in Sooty Terns Onychoprion fuscatus. Front. Vet. Sci. 2020, 7, 563309. [Google Scholar] [CrossRef]
- Magnius, L.; Mason, W.S.; Taylor, J.; Kann, M.; Glebe, D.; Dény, P.; Sureau, C.; Norder, H.; Ictv Report, C. ICTV Virus Taxonomy Profile: Hepadnaviridae. J. Gen. Virol. 2020, 101, 571–572. [Google Scholar] [CrossRef]
- Jo, W.K.; Pfankuche, V.M.; Petersen, H.; Frei, S.; Kummrow, M.; Lorenzen, S.; Ludlow, M.; Metzger, J.; Baumgärtner, W.; Osterhaus, A.; et al. New Avian Hepadnavirus in Palaeognathous Bird, Germany. Emerg. Infect. Dis. 2017, 23, 2089–2091. [Google Scholar] [CrossRef]
- Funk, A.; Mhamdi, M.; Will, H.; Sirma, H. Avian hepatitis B viruses: Molecular and cellular biology, phylogenesis, and host tropism. World J. Gastroenterol. 2007, 13, 91–103. [Google Scholar] [CrossRef]
- Piasecki, T.; Harkins, G.W.; Chrząstek, K.; Julian, L.; Martin, D.P.; Varsani, A. Avihepadnavirus diversity in parrots is comparable to that found amongst all other avian species. Virology 2013, 438, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Pan, S.; Yang, H.; Bai, W.; Shen, Z.; Liu, J.; Xie, Y. The first full-length endogenous hepadnaviruses: Identification and analysis. J. Virol. 2012, 86, 9510–9513. [Google Scholar] [CrossRef] [PubMed]
- Athukorala, A.; Phalen, D.N.; Das, A.; Helbig, K.J.; Forwood, J.K.; Sarker, S. Genomic Characterisation of a Highly Divergent Siadenovirus (Psittacine Siadenovirus F) from the Critically Endangered Orange-Bellied Parrot (Neophema chrysogaster). Viruses 2021, 13, 1714. [Google Scholar] [CrossRef] [PubMed]
- Sarker, S.; Das, S.; Lavers, J.L.; Hutton, I.; Helbig, K.; Imbery, J.; Upton, C.; Raidal, S.R. Genomic characterization of two novel pathogenic avipoxviruses isolated from pacific shearwaters (Ardenna spp.). BMC Genom. 2017, 18, 298. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sarker, S.; Isberg, R.S.; Moran, L.J.; Araujo, D.R.; Elliott, N.; Melville, L.; Beddoe, T.; Helbig, J.K. Crocodilepox Virus Evolutionary Genomics Supports Observed Poxvirus Infection Dynamics on Saltwater Crocodile (Crocodylus porosus). Viruses 2019, 11, 1116. [Google Scholar] [CrossRef]
- Benson, D.A.; Cavanaugh, M.; Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2013, 41, D36–D42. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, Y.; Sun, G.; Bao, K.; Gao, Y.; Qi, X.; Cui, H.; Wang, Y.; Li, K.; Gao, L.; et al. Genetic evolution of Gallid herpesvirus 2 isolated in China. Infect. Genet. Evol. 2017, 51, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Tusnády, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Stoffel, W. Tmbase—A database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 1993, 374, 166. [Google Scholar]
- Zimmermann, L.; Stephens, A.; Nam, S.Z.; Rau, D.; Kubler, J.; Lozajic, M.; Gabler, F.; Soding, J.; Lupas, A.N.; Alva, V. A Completely Reimplemented MPI Bioinformatics Toolkit with a New HHpred Server at its Core. J. Mol. Biol. 2018, 430, 2237–2243. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Mason, W.S.; Aldrich, C.E.; Saputelli, J.R.; Miller, D.S.; Jilbert, A.R.; Newbold, J.E. Identification and characterization of avihepadnaviruses isolated from exotic anseriformes maintained in captivity. J. Virol. 2005, 79, 2729–2742. [Google Scholar] [CrossRef] [PubMed]

| PsAHV6 Synteny | PsAHV6 Gene Coordinate | Nt Length | AA Length | Best Hit Gene Product | Best Hit (%Identity/Query Coverage/e-Value/PI/Organism) |
|---|---|---|---|---|---|
| PsAHV6-001 | 1128-301 | 828 | 275 | envelope protein UL20 | 38.43/85/6.00E-36/YP_009352926.1/CoAHV1 |
| PsAHV6-002 | 1628-1885 | 258 | 85 | hypothetical protein | no significant BLAST hits |
| PsAHV6-003 | 1959-3761 | 1803 | 600 | tegument protein UL21 | 33.28/92/7.00E-89/YP_009046517.1/FaAHV1 |
| PsAHV6-004 | 3847-4176 | 330 | 109 | hypothetical protein | no significant BLAST hits |
| PsAHV6-005 | 6679-4010 | 2670 | 889 | envelope glycoprotein H UL22 | 33.87/88/1.00E-120/YP_009046519.1/FaAHV1 |
| PsAHV6-006 | 8196-7066 | 1131 | 376 | thymidine kinase UL23 | 36.47/85/7.00E-59/YP_009046521.1/FaAHV1 |
| PsAHV6-007 | 8112-9194 | 1083 | 360 | nuclear protein UL24 | 35.27/66/4.00E-26/ABK55352.1/AnAHV1 |
| PsAHV6-008 | 9374-11428 | 2055 | 684 | DNA packaging tegument protein UL25 | 54.55/82/5.00E-155/YP_009352933.1/CoAHV1 |
| PsAHV6-009 | 11370-11570 | 201 | 66 | hypothetical protein | no significant BLAST hits |
| PsAHV6-010 | 11752-14400 | 2649 | 882 | capsid maturation protease UL26 | 56.57/28/2.00E-73/UJO49828.1/AnAHV1 |
| PsAHV6-011 | 17814-14731 | 3084 | 1027 | envelope glycoprotein B UL2 | 60.91/80/0/CAA63039.1/GaAHV2 |
| PsAHV6-012 | 20681-17586 | 3096 | 1031 | DNA packaging terminase subunit 2 UL28 | 51.93/71/1.00E-151/NP_073322.1/MeAHV1 |
| PsAHV6-013 | 20801-21118 | 318 | 105 | hypothetical protein | no significant BLAST hits |
| PsAHV6-014 | 25126-21098 | 4029 | 1342 | single-stranded DNA-binding protein UL29 | 51.90/99/0/YP_009352937.1/CoAHV1 |
| PsAHV6-015 | 25389-25129 | 261 | 86 | hypothetical protein | no significant BLAST hits |
| PsAHV6-016 | 25677-29612 | 3936 | 1311 | DNA polymerase UL30 | 54.06/98/0/UOW62139.1/GaAHV2 |
| PsAHV6-017 | 30888-29536 | 1353 | 450 | nuclear egress lamina protein UL31 | 58.70/67/6.00E-111/YP_009046529.1/FaAHV1 |
| PsAHV6-018 | 33797-31323 | 2475 | 824 | DNA packaging protein UL32 | 51.12/84/2.00E-126/YP_009352940.1/CoAHV1 |
| PsAHV6-019 | 33808-34290 | 483 | 160 | DNA packaging protein UL33 | 56.49/81/2.00E-34/YP_009046531.1/FaAHV1 |
| PsAHV6-020 | 34360-34557 | 198 | 65 | hypothetical protein | no significant BLAST hits |
| PsAHV6-021 | 34643-35719 | 1077 | 358 | nuclear egress membrane protein UL34 | 58.62/48/5.00E-66/YP_009352942.1/CoAHV1 |
| PsAHV6-022 | 35815-36162 | 348 | 115 | small capsid protein UL35 | 47.92/81/9.00E-12/YP_009046533.1/FaAHV1 |
| PsAHV6-023 | 46419-36400 | 10020 | 3339 | large tegument protein UL36 | 37.71/71/0/YP_009046534.1/FaAHV1 |
| PsAHV6-024 | 46742-46416 | 327 | 108 | hypothetical protein | no significant BLAST hits |
| PsAHV6-025 | 50376-46966 | 3411 | 1136 | tegument protein UL37 | 36.23/98/0/YP_009046535.1/FaAHV1 |
| PsAHV6-026 | 50892-52397 | 1506 | 501 | capsid triplex subunit 1 UL38 | 54.68/93/1.00E-144/YP_009046536.1/FaAHV1 |
| PsAHV6-027 | 52766-55537 | 2772 | 923 | ribonucleotide reductase subunit 1 UL39 | 56.43/86/0/NP_073333.1/MeAHV1 |
| PsAHV6-028 | 56022-57398 | 1377 | 458 | ribonucleotide reductase subunit 2 UL40 | 67.17/71/1.00E-156/YP_009046538.1/FaAHV1 |
| PsAHV6-029 | 59232-57688 | 1545 | 514 | tegument host shutoff protein UL41 | 44.74/73/3.00E-79/YP_009352949.1/CoAHV1 |
| PsAHV6-030 | 59365-59556 | 192 | 63 | hypothetical protein | no significant BLAST hits |
| PsAHV6-031 | 60266-61675 | 1410 | 469 | DNA polymerase processivity subunit UL42 | 42.02/73/5.00E-84/AUB50956.1/GaAHV2 |
| PsAHV6-032 | 62162-63520 | 1359 | 452 | hypothetical protein | no significant BLAST hits |
| PsAHV6-033 | 64113-63922 | 192 | 63 | hypothetical protein | no significant BLAST hits |
| PsAHV6-034 | 64380-66293 | 1914 | 637 | glycoprotein C UL44 | 34.00/62/3.00E-78/AAM97710.1/GaAHV2 |
| PsAHV6-035 | 67702-66737 | 966 | 321 | hypothetical protein FaHV1S18_060 | 34.68/90/5.00E-45/YP_009046544.1/FaAHV1 |
| PsAHV6-036 | 68228-68416 | 189 | 62 | hypothetical protein | no significant BLAST hits |
| PsAHV6-037 | 68425-69114 | 690 | 229 | membrane protein UL45 | 40.85/71/1.00E-40/YP_009352955.1/CoAHV1 |
| PsAHV6-038 | 72032-69492 | 2541 | 846 | tegument protein VP11/12 UL46 | 28.57/47/4.00E-40/YP_009046546.1/FaAHV1 |
| PsAHV6-039 | 71997-72278 | 282 | 93 | hypothetical protein | no significant BLAST hits |
| PsAHV6-040 | 72552-72283 | 270 | 89 | hypothetical protein | no significant BLAST hits |
| PsAHV6-041 | 72854-73048 | 195 | 64 | hypothetical protein | no significant BLAST hits |
| PsAHV6-042 | 75190-73202 | 1989 | 662 | tegument protein VP13/14 UL47 | 33.16/81/3.00E-67/YP_009046547.1/FaAHV1 |
| PsAHV6-043 | 75135-75587 | 453 | 150 | hypothetical protein | no significant BLAST hits |
| PsAHV6-044 | 75720-75475 | 246 | 81 | hypothetical protein | no significant BLAST hits |
| PsAHV6-045 | 76074-75808 | 267 | 88 | hypothetical protein | no significant BLAST hits |
| PsAHV6-046 | 76919-76722 | 198 | 65 | hypothetical protein | no significant BLAST hits |
| PsAHV6-047 | 78218-77028 | 1191 | 396 | transactivating tegument protein VP16 UL48 | 45.56/89/9.00E-95/YP_009046548.1/FaAHV1 |
| PsAHV6-048 | 78299-78481 | 183 | 60 | hypothetical protein | no significant BLAST hits |
| PsAHV6-049 | 78811-78533 | 279 | 92 | hypothetical protein | no significant BLAST hits |
| PsAHV6-050 | 79856-79092 | 765 | 254 | tegument protein VP22 UL49 | 38.36/28/7.00E-07/UOW62334.1/GaAHV2 |
| PsAHV6-051 | 79855-80055 | 201 | 66 | hypothetical protein | no significant BLAST hits |
| PsAHV6-052 | 80468-80181 | 288 | 95 | envelope glycoprotein N UL49.5 | 51.04/98/8.00E-11/YP_009046550.1/FaAHV1 |
| PsAHV6-053 | 81124-82521 | 1398 | 465 | deoxyuridine triphosphatase UL50 | 40.74/98/2.00E-103/YP_009046551.1/FaAHV1 |
| PsAHV6-054 | 82780-82541 | 240 | 79 | hypothetical protein | no significant BLAST hits |
| PsAHV6-055 | 83633-82671 | 963 | 320 | tegument protein UL51 | 52.58/60/7.00E-59/YP_009046552.1/FaAHV1 |
| PsAHV6-056 | 83632-87303 | 3672 | 1223 | helicase-primase primase subunit UL52 | 42.05/94/0/YP_009352963.1/CoAHV1 |
| PsAHV6-057 | 87325-88494 | 1170 | 389 | envelope glycoprotein K UL53 | 49.29/89/2.00E-98/YP_009352964.1/CoAHV1 |
| PsAHV6-058 | 88959-90398 | 1440 | 479 | multifunctional expression regulator UL54 | 37.67/44/8.00E-30/YP_009046555.1/FaAHV1 |
| PsAHV6-059 | 91648-90716 | 933 | 310 | protein LORF4 | 36.59/87/3.00E-50/YP_009046556.1/FaAHV1 |
| PsAHV6-060 | 92305-92919 | 615 | 204 | nuclear protein UL55 | 46.43/81/6.00E-46/YP_009352967.1/FaAHV1 |
| PsAHV6-061 | 94339-93110 | 1230 | 409 | myristylated tegument protein CIRC | 33.19/55/5.00E-13/YP_009352969.1/CoAHV1 |
| PsAHV6-062 | 97772-94611 | 3162 | 1053 | hypothetical protein LORF11 | 28.14/86/4.00E-81/UOW65035.1/GaAHV2 |
| PsAHV6-063 | 99543-98449 | 1095 | 364 | hypothetical protein | no significant BLAST hits |
| PsAHV6-064 | 99943-100107 | 165 | 54 | hypothetical protein | no significant BLAST hits |
| PsAHV6-065 | 100621-100463 | 159 | 52 | hypothetical protein | no significant BLAST hits |
| PsAHV6-066 | 101828-100758 | 1071 | 356 | hypothetical protein | no significant BLAST hits |
| PsAHV6-067 | 102213-101770 | 444 | 147 | hypothetical protein | no significant BLAST hits |
| PsAHV6-068 | 102515-102832 | 318 | 105 | hypothetical protein | no significant BLAST hits |
| PsAHV6-069 | 103442-103113 | 330 | 109 | hypothetical protein | no significant BLAST hits |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarker, S.; Phalen, D.N. Detection of a Novel Alphaherpesvirus and Avihepadnavirus in a Plantar Papilloma from a Rainbow Lorikeet (Trichoglosis moluccanus). Viruses 2023, 15, 2106. https://doi.org/10.3390/v15102106
Sarker S, Phalen DN. Detection of a Novel Alphaherpesvirus and Avihepadnavirus in a Plantar Papilloma from a Rainbow Lorikeet (Trichoglosis moluccanus). Viruses. 2023; 15(10):2106. https://doi.org/10.3390/v15102106
Chicago/Turabian StyleSarker, Subir, and David N. Phalen. 2023. "Detection of a Novel Alphaherpesvirus and Avihepadnavirus in a Plantar Papilloma from a Rainbow Lorikeet (Trichoglosis moluccanus)" Viruses 15, no. 10: 2106. https://doi.org/10.3390/v15102106
APA StyleSarker, S., & Phalen, D. N. (2023). Detection of a Novel Alphaherpesvirus and Avihepadnavirus in a Plantar Papilloma from a Rainbow Lorikeet (Trichoglosis moluccanus). Viruses, 15(10), 2106. https://doi.org/10.3390/v15102106






