Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s
Abstract
1. Introduction
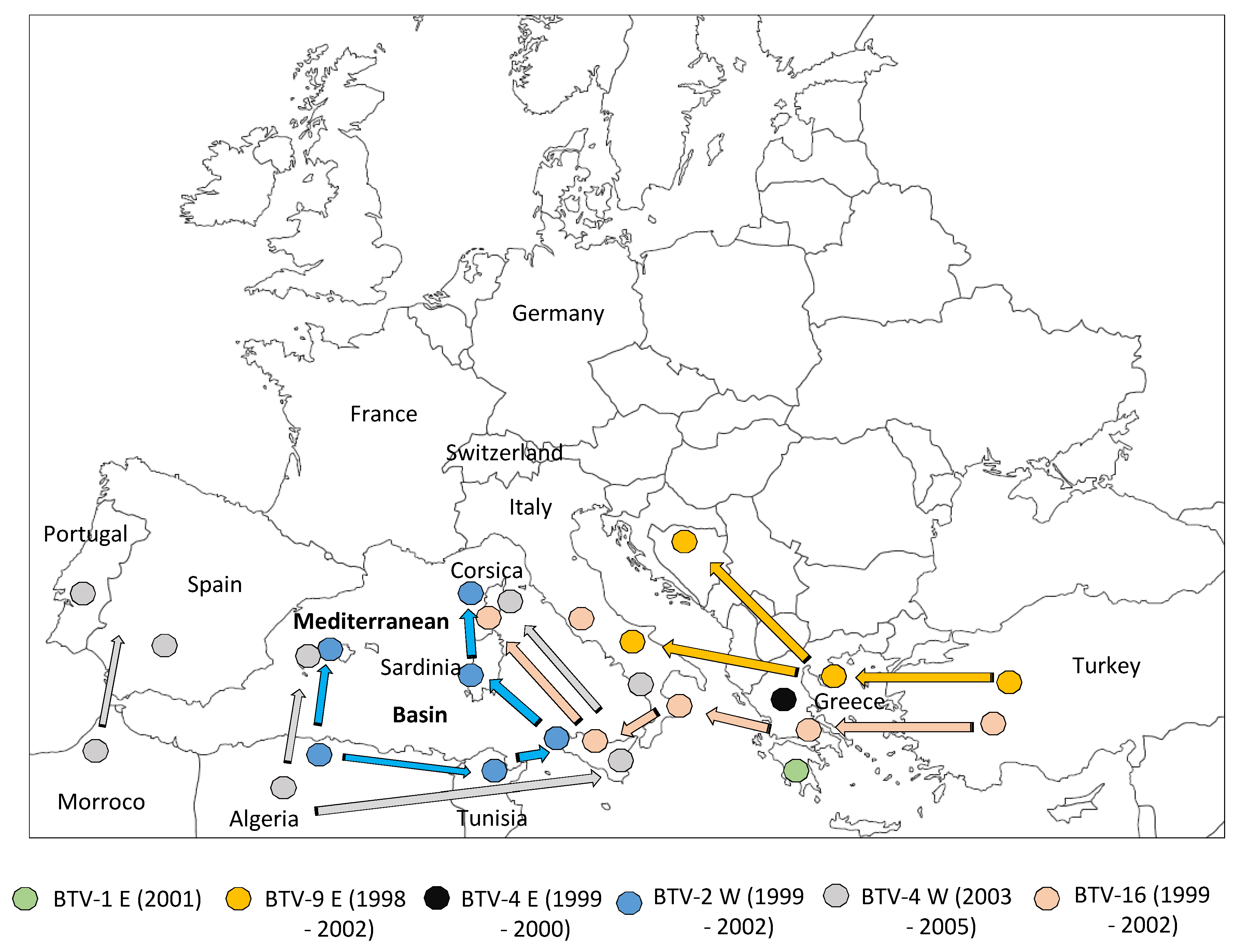
2. First Period: Bluetongue Virus (BTV) Incursions in Countries Bordering the Mediterranean Basin
3. The Second Period: A New Area of Emergence in Northern Europe Changed the BTV Introduction Rules in 2006
4. First Eastern BTV Strain Introduction in Mainland France
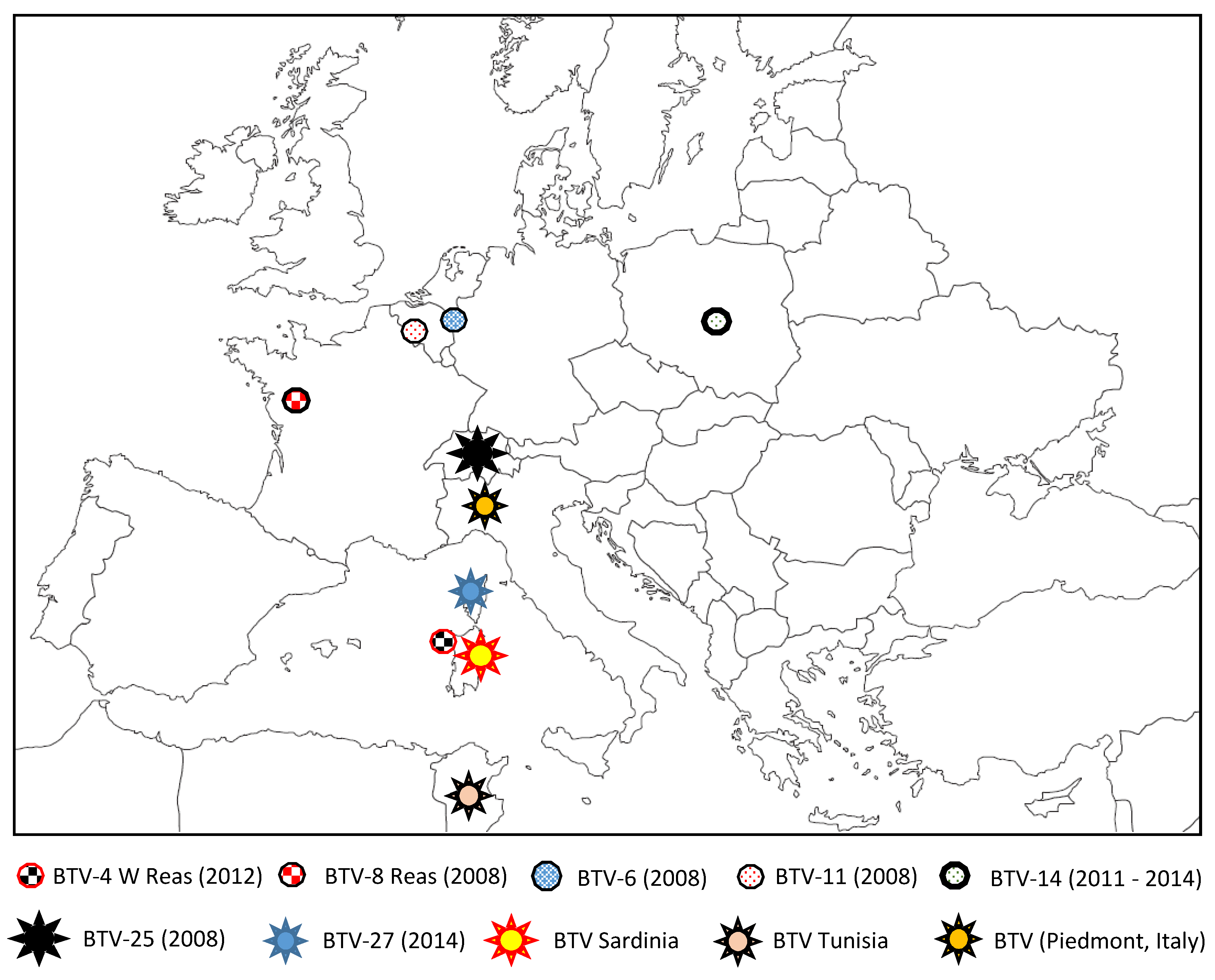
5. Discovery of New BTV Serotypes
6. BTV Vectors
7. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Maclachlan, N.J.; Drew, C.P.; Darpel, K.E.; Worwa, G. The pathology and pathogenesis of bluetongue. J. Comp. Pathol. 2009, 141, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nolan, D.V.; Dallas, J.F.; Piertney, S.B.; Mordue Luntz, A.J. Incursion and range expansion in the bluetongue vector Culicoides imicola in the Mediterranean basin: a phylogeographic analysis. Med. Vet. Entomol. 2008, 22, 340–351. [Google Scholar] [CrossRef] [PubMed]
- MacLachlan, N.J.; Crafford, J.E.; Vernau, W.; Gardner, I.A.; Goddard, A.; Guthrie, A.J.; Venter, E.H. Experimental reproduction of severe bluetongue in sheep. Vet. Pathol. 2008, 45, 310–315. [Google Scholar] [CrossRef] [PubMed]
- Savini, G.; Puggioni, G.; Meloni, G.; Marcacci, M.; Di Domenico, M.; Rocchigiani, A.M.; Spedicato, M.; Oggiano, A.; Manunta, D.; Teodori, L.; et al. Novel putative Bluetongue virus in healthy goats from Sardinia, Italy. Infect. Genet. Evol. 2017, 51, 108–117. [Google Scholar] [CrossRef] [PubMed]
- MacLachlan, N.J. Bluetongue: pathogenesis and duration of viraemia. Vet. Ital. 2004, 40, 462–467. [Google Scholar] [PubMed]
- Bonneau, K.R.; DeMaula, C.D.; Mullens, B.A.; MacLachlan, N.J. Duration of viraemia infectious to Culicoides sonorensis in bluetongue virus-infected cattle and sheep. Vet. Microbiol. 2002, 88, 115–125. [Google Scholar] [CrossRef]
- Zanella, G.; Martinelle, L.; Guyot, H.; Mauroy, A.; De Clercq, K.; Saegerman, C. Clinical pattern characterization of cattle naturally infected by BTV-8. Transbound. Emerg. Dis. 2013, 60, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Ratinier, M.; Caporale, M.; Golder, M.; Franzoni, G.; Allan, K.; Nunes, S.F.; Armezzani, A.; Bayoumy, A.; Rixon, F.; Shaw, A.; et al. Identification and characterization of a novel non-structural protein of bluetongue virus. PLoS Pathog. 2011, 7, e1002477. [Google Scholar] [CrossRef]
- Stewart, M.; Hardy, A.; Barry, G.; Pinto, R.M.; Caporale, M.; Melzi, E.; Hughes, J.; Taggart, A.; Janowicz, A.; Varela, M.; et al. Characterization of a second open reading frame in genome segment 10 of bluetongue virus. J. Gen. Virol. 2015, 96, 3280–3293. [Google Scholar] [CrossRef]
- Huismans, H.; Erasmus, B.J. Identification of the serotype-specific and group-specific antigens of bluetongue virus. Onderstepoort J. Vet. Res. 1981, 48, 51–58. [Google Scholar]
- Kahlon, J.; Sugiyama, K.; Roy, P. Molecular basis of bluetongue virus neutralization. J. Virol. 1983, 48, 627–632. [Google Scholar] [PubMed]
- Shaw, A.E.; Ratinier, M.; Nunes, S.F.; Nomikou, K.; Caporale, M.; Golder, M.; Allan, K.; Hamers, C.; Hudelet, P.; Zientara, S.; et al. Reassortment between two serologically unrelated bluetongue virus strains is flexible and can involve any genome segment. J. Virol. 2013, 87, 543–557. [Google Scholar] [CrossRef] [PubMed]
- Zientara, S.; Sailleau, C.; Viarouge, C.; Hoper, D.; Beer, M.; Jenckel, M.; Hoffmann, B.; Romey, A.; Bakkali-Kassimi, L.; Fablet, A.; et al. Novel bluetongue virus in goats, Corsica, France, 2014. Emerg. Infect. Dis. 2014, 20, 2123–2125. [Google Scholar] [CrossRef] [PubMed]
- Bumbarov, V.; Golender, N.; Erster, O.; Khinich, Y. Detection and isolation of Bluetongue virus from commercial vaccine batches. Vaccine 2016, 34, 3317–3323. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.C.; Huang, L.P.; Xu, Q.Y.; Wang, H.X.; Xue, X.M.; Lu, P.; Li, W.J.; Liu, W.; Bu, Z.G.; Wu, D.L. Emergence of a Novel Bluetongue Virus Serotype, China 2014. Transbound. Emerg. Dis. 2016, 63, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Marcacci, M.; Sant, S.; Mangone, I.; Goria, M.; Dondo, A.; Zoppi, S.; van Gennip, R.G.P.; Radaelli, M.C.; Camma, C.; van Rijn, P.A.; et al. One after the other: A novel Bluetongue virus strain related to Toggenburg virus detected in the Piedmont region (North-western Italy), extends the panel of novel atypical BTV strains. Transbound. Emerg. Dis. 2018, 65, 370–374. [Google Scholar] [CrossRef]
- Lorusso, A.; Sghaier, S.; Di Domenico, M.; Barbria, M.E.; Zaccaria, G.; Megdich, A.; Portanti, O.; Seliman, I.B.; Spedicato, M.; Pizzurro, F.; et al. Analysis of bluetongue serotype 3 spread in Tunisia and discovery of a novel strain related to the bluetongue virus isolated from a commercial sheep pox vaccine. Infect. Genet. Evol. 2018, 59, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Chaignat, V.; Worwa, G.; Scherrer, N.; Hilbe, M.; Ehrensperger, F.; Batten, C.; Cortyen, M.; Hofmann, M.; Thuer, B. Toggenburg Orbivirus, a new bluetongue virus: initial detection, first observations in field and experimental infection of goats and sheep. Vet. Microbiol. 2009, 138, 11–19. [Google Scholar] [CrossRef]
- Maan, S.; Maan, N.S.; Ross-smith, N.; Batten, C.A.; Shaw, A.E.; Anthony, S.J.; Samuel, A.R.; Darpel, K.E.; Veronesi, E.; Oura, C.A.; et al. Sequence analysis of bluetongue virus serotype 8 from the Netherlands 2006 and comparison to other European strains. Virology 2008, 377, 308–318. [Google Scholar] [CrossRef]
- Gibbs, E.P.; Greiner, E.C. The epidemiology of bluetongue. Comp. Immunol. Microbiol. Infect. Dis. 1994, 17, 207–220. [Google Scholar] [CrossRef]
- Saegerman, C.; Berkvens, D.; Mellor, P.S. Bluetongue epidemiology in the European Union. Emerg. Infect. Dis. 2008, 14, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Guis, H.; Caminade, C.; Calvete, C.; Morse, A.P.; Tran, A.; Baylis, M. Modelling the effects of past and future climate on the risk of bluetongue emergence in Europe. J. R. Soc. Interface 2012, 9, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.J.; Mellor, P.S. Bluetongue in Europe: past, present and future. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2009, 364, 2669–2681. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, A.; Sghaier, S.; Ancora, M.; Marcacci, M.; Di Gennaro, A.; Portanti, O.; Mangone, I.; Teodori, L.; Leone, A.; Camma, C.; et al. Molecular epidemiology of bluetongue virus serotype 1 circulating in Italy and its connection with northern Africa. Infect. Genet. Evol. 2014, 28, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Savini, G.; Potgieter, A.C.; Monaco, F.; Mangana-Vougiouka, O.; Nomikou, K.; Yadin, H.; Caporale, V. VP2 gene sequence analysis of some isolates of bluetongue virus recovered in the Mediterranean Basin during the 1998-2002 outbreak. Vet. Ital. 2004, 40, 473–478. [Google Scholar]
- Baylis, M.; Mellor, P.S. Bluetongue around the Mediterranean in 2001. Vet. Rec. 2001, 149, 659. [Google Scholar]
- Mellor, P.S.; Wittmann, E.J. Bluetongue virus in the Mediterranean Basin 1998–2001. Vet. J. 2002, 164, 20–37. [Google Scholar] [CrossRef]
- Breard, E.; Sailleau, C.; Coupier, H.; Mure-Ravaud, K.; Hammoumi, S.; Gicquel, B.; Hamblin, C.; Dubourget, P.; Zientara, S. Comparison of genome segments 2, 7 and 10 of bluetongue viruses serotype 2 for differentiation between field isolates and the vaccine strain. Vet. Res. 2003, 34, 777–789. [Google Scholar] [CrossRef]
- Breard, E.; Hamblin, C.; Hammoumi, S.; Sailleau, C.; Dauphin, G.; Zientara, S. The epidemiology and diagnosis of bluetongue with particular reference to Corsica. Res. Vet. Sci. 2004, 77, 1–8. [Google Scholar] [CrossRef]
- Breard, E.; Sailleau, C.; Nomikou, K.; Hamblin, C.; Mertens, P.P.; Mellor, P.S.; El Harrak, M.; Zientara, S. Molecular epidemiology of bluetongue virus serotype 4 isolated in the Mediterranean Basin between 1979 and 2004. Virus Res. 2007, 125, 191–197. [Google Scholar] [CrossRef]
- Monaco, F.; Camma, C.; Serini, S.; Savini, G. Differentiation between field and vaccine strain of bluetongue virus serotype 16. Vet. Microbiol. 2006, 116, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Batten, C.A.; Maan, S.; Shaw, A.E.; Maan, N.S.; Mertens, P.P. A European field strain of bluetongue virus derived from two parental vaccine strains by genome segment reassortment. Virus Res 2008, 137, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Mellor, P.S.; Carpenter, S.; Harrup, L.; Baylis, M.; Mertens, P.P. Bluetongue in Europe and the Mediterranean Basin: history of occurrence prior to 2006. Prev. Vet. Med. 2008, 87, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Nomikou, K.; Hughes, J.; Wash, R.; Kellam, P.; Breard, E.; Zientara, S.; Palmarini, M.; Biek, R.; Mertens, P. Widespread Reassortment Shapes the Evolution and Epidemiology of Bluetongue Virus following European Invasion. PLoS Pathog. 2015, 11, e1005056. [Google Scholar] [CrossRef] [PubMed]
- Tago, D.; Hammitt, J.K.; Thomas, A.; Raboisson, D. Cost assessment of the movement restriction policy in France during the 2006 bluetongue virus episode (BTV-8). Prev. Vet. Med. 2014, 117, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Bournez, L.; Cavalerie, L.; Sailleau, C.; Breard, E.; Zanella, G.; Servan de Almeida, R.; Pedarrieu, A.; Garin, E.; Tourette, I.; Dion, F.; et al. Estimation of French cattle herd immunity against bluetongue serotype 8 at the time of its re-emergence in 2015. BMC Vet. Res. 2018, 14, 65. [Google Scholar] [CrossRef]
- Lorusso, A.; Sghaier, S.; Carvelli, A.; Di Gennaro, A.; Leone, A.; Marini, V.; Pelini, S.; Marcacci, M.; Rocchigiani, A.M.; Puggioni, G.; et al. Bluetongue virus serotypes 1 and 4 in Sardinia during autumn 2012: new incursions or re-infection with old strains? Infect. Genet. Evol. 2013, 19, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Viarouge, C.; Breard, E.; Perrin, J.B.; Doceul, V.; Vitour, D.; Zientara, S. Emergence of Bluetongue Virus Serotype 1 in French Corsica Island in September 2013. Transbound. Emerg. Dis. 2015, 62, e89–e91. [Google Scholar] [CrossRef]
- Cappai, S.; Rolesu, S.; Loi, F.; Liciardi, M.; Leone, A.; Marcacci, M.; Teodori, L.; Mangone, I.; Sghaier, S.; Portanti, O.; et al. Western Bluetongue virus serotype 3 in Sardinia, diagnosis and characterization. Transbound. Emerg. Dis. 2019. [Google Scholar] [CrossRef]
- Elbers, A.R.; Backx, A.; Mintiens, K.; Gerbier, G.; Staubach, C.; Hendrickx, G.; van der Spek, A. Field observations during the Bluetongue serotype 8 epidemic in 2006. II. Morbidity and mortality rate, case fatality and clinical recovery in sheep and cattle in the Netherlands. Prev. Vet. Med. 2008, 87, 31–40. [Google Scholar] [CrossRef]
- Elbers, A.R.; Backx, A.; Meroc, E.; Gerbier, G.; Staubach, C.; Hendrickx, G.; van der Spek, A.; Mintiens, K. Field observations during the bluetongue serotype 8 epidemic in 2006. I. Detection of first outbreaks and clinical signs in sheep and cattle in Belgium, France and the Netherlands. Prev. Vet. Med. 2008, 87, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Darpel, K.E.; Batten, C.A.; Veronesi, E.; Shaw, A.E.; Anthony, S.; Bachanek-Bankowska, K.; Kgosana, L.; bin-Tarif, A.; Carpenter, S.; Muller-Doblies, U.U.; et al. Clinical signs and pathology shown by British sheep and cattle infected with bluetongue virus serotype 8 derived from the 2006 outbreak in northern Europe. Vet. Rec. 2007, 161, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Backx, A.; Heutink, R.; van Rooij, E.; van Rijn, P. Transplacental and oral transmission of wild-type bluetongue virus serotype 8 in cattle after experimental infection. Vet. Microbiol. 2009, 138, 235–243. [Google Scholar] [CrossRef] [PubMed]
- Backx, A.; Heutink, C.G.; van Rooij, E.M.; van Rijn, P.A. Clinical signs of bluetongue virus serotype 8 infection in sheep and goats. Vet. Rec. 2007, 161, 591–592. [Google Scholar] [CrossRef] [PubMed]
- Rossi, S.; Pioz, M.; Beard, E.; Durand, B.; Gibert, P.; Gauthier, D.; Klein, F.; Maillard, D.; Saint-Andrieux, C.; Saubusse, T.; et al. Bluetongue dynamics in French wildlife: exploring the driving forces. Transbound. Emerg. Dis. 2014, 61, e12–e24. [Google Scholar] [CrossRef]
- Linden, A.; Gregoire, F.; Nahayo, A.; Hanrez, D.; Mousset, B.; Massart, A.L.; De Leeuw, I.; Vandemeulebroucke, E.; Vandenbussche, F.; De Clercq, K. Bluetongue virus in wild deer, Belgium, 2005–2008. Emerg. Infect. Dis. 2010, 16, 833–836. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, K.; Vandenbussche, F.; Vandemeulebroucke, E.; Vanbinst, T.; De Leeuw, I.; Verheyden, B.; Goris, N.; Mintiens, K.; Meroc, E.; Herr, C.; et al. Transplacental bluetongue infection in cattle. Vet. Rec. 2008, 162, 564. [Google Scholar] [CrossRef]
- Darpel, K.E.; Batten, C.A.; Veronesi, E.; Williamson, S.; Anderson, P.; Dennison, M.; Clifford, S.; Smith, C.; Philips, L.; Bidewell, C.; et al. Transplacental transmission of bluetongue virus 8 in cattle, UK. Emerg. Infect. Dis. 2009, 15, 2025–2028. [Google Scholar] [CrossRef]
- Zanella, G.; Durand, B.; Sellal, E.; Breard, E.; Sailleau, C.; Zientara, S.; Batten, C.A.; Mathevet, P.; Audeval, C. Bluetongue virus serotype 8: abortion and transplacental transmission in cattle in the Burgundy region, France, 2008–2009. Theriogenology 2012, 77, 65–72. [Google Scholar] [CrossRef]
- MacLachlan, N.J.; Conley, A.J.; Kennedy, P.C. Bluetongue and equine viral arteritis viruses as models of virus-induced fetal injury and abortion. Anim. Reprod. Sci. 2000, 60-61, 643–651. [Google Scholar] [CrossRef]
- Flanagan, M.; Johnson, S.J. The effects of vaccination of Merino ewes with an attenuated Australian bluetongue virus serotype 23 at different stages of gestation. Aust. Vet. J. 1995, 72, 455–457. [Google Scholar] [CrossRef] [PubMed]
- Mintiens, K.; Meroc, E.; Mellor, P.S.; Staubach, C.; Gerbier, G.; Elbers, A.R.; Hendrickx, G.; De Clercq, K. Possible routes of introduction of bluetongue virus serotype 8 into the epicentre of the 2006 epidemic in north-western Europe. Prev. Vet. Med. 2008, 87, 131–144. [Google Scholar] [CrossRef] [PubMed]
- Mintiens, K.; Meroc, E.; Faes, C.; Abrahantes, J.C.; Hendrickx, G.; Staubach, C.; Gerbier, G.; Elbers, A.R.; Aerts, M.; De Clercq, K. Impact of human interventions on the spread of bluetongue virus serotype 8 during the 2006 epidemic in north-western Europe. Prev. Vet. Med. 2008, 87, 145–161. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, S.; McArthur, C.; Selby, R.; Ward, R.; Nolan, D.V.; Luntz, A.J.; Dallas, J.F.; Tripet, F.; Mellor, P.S. Experimental infection studies of UK Culicoides species midges with bluetongue virus serotypes 8 and 9. Vet. Rec. 2008, 163, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, E.; van der Ven, I.J.; Meiswinkel, R.; Holzel, D.R.; Van Rijn, P.A.; Meiswinkel, R. Culicoides chiopterus as a potential vector of bluetongue virus in Europe. Vet. Rec. 2008, 162, 422. [Google Scholar] [CrossRef] [PubMed]
- Martinelle, L.; Dal Pozzo, F.; Sarradin, P.; Van Campe, W.; De Leeuw, I.; De Clercq, K.; Thys, C.; Thiry, E.; Saegerman, C. Experimental bluetongue virus superinfection in calves previously immunized with bluetongue virus serotype 8. Vet. Res. 2016, 47, 73. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rushton, J.; Lyons, N. Economic impact of Bluetongue: a review of the effects on production. Vet. Ital. 2015, 51, 401–406. [Google Scholar]
- De Clercq, K.; Mertens, P.; De Leeuw, I.; Oura, C.; Houdart, P.; Potgieter, A.C.; Maan, S.; Hooyberghs, J.; Batten, C.; Vandemeulebroucke, E.; et al. Emergence of bluetongue serotypes in Europe, part 2: the occurrence of a BTV-11 strain in Belgium. Transbound. Emerg. Dis. 2009, 56, 355–361. [Google Scholar] [CrossRef]
- Orlowska, A.; Trebas, P.; Smreczak, M.; Marzec, A.; Zmudzinski, J.F. First detection of bluetongue virus serotype 14 in Poland. Arch. Virol. 2016, 161, 1969–1972. [Google Scholar] [CrossRef]
- Eschbaumer, M.; Hoffmann, B.; Moss, A.; Savini, G.; Leone, A.; Konig, P.; Zemke, J.; Conraths, F.; Beer, M. Emergence of bluetongue virus serotype 6 in Europe--German field data and experimental infection of cattle. Vet. Microbiol. 2010, 143, 189–195. [Google Scholar] [CrossRef]
- Sailleau, C.; Breard, E.; Viarouge, C.; Vitour, D.; Romey, A.; Garnier, A.; Fablet, A.; Lowenski, S.; Gorna, K.; Caignard, G.; et al. Re-Emergence of Bluetongue Virus Serotype 8 in France, 2015. Transbound. Emerg. Dis. 2017, 64, 998–1000. [Google Scholar] [CrossRef] [PubMed]
- Breard, E.; Sailleau, C.; Quenault, H.; Lucas, P.; Viarouge, C.; Touzain, F.; Fablet, A.; Vitour, D.; Attoui, H.; Zientara, S.; et al. Complete Genome Sequence of Bluetongue Virus Serotype 8, Which Reemerged in France in August 2015. Genom. Announc. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Courtejoie, N.; Durand, B.; Bournez, L.; Gorlier, A.; Breard, E.; Sailleau, C.; Vitour, D.; Zientara, S.; Baurier, F.; Gourmelen, C.; et al. Circulation of bluetongue virus 8 in French cattle, before and after the re-emergence in 2015. Transbound. Emerg. Dis. 2018, 65, 281–284. [Google Scholar] [CrossRef] [PubMed]
- Flannery, J.; Sanz-Bernardo, B.; Ashby, M.; Brown, H.; Carpenter, S.; Cooke, L.; Corla, A.; Frost, L.; Gubbins, S.; Hicks, H.; et al. Evidence of reduced viremia, pathogenicity and vector competence in a re-emerging European strain of bluetongue virus serotype 8 in sheep. Transbound. Emerg. Dis. 2019, 66, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Breard, E.; Viarouge, C.; Gorlier, A.; Quenault, H.; Hirchaud, E.; Touzain, F.; Blanchard, Y.; Vitour, D.; Zientara, S. Complete genome sequence of bluetongue virus serotype 4 that emerged on the French island of Corsica in December 2016. Transbound. Emerg. Dis. 2018, 65, e194–e197. [Google Scholar] [CrossRef] [PubMed]
- Hornyak, A.; Malik, P.; Marton, S.; Doro, R.; Cadar, D.; Banyai, K. Emergence of multireassortant bluetongue virus serotype 4 in Hungary. Infect. Gen. Evol. 2015, 33, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Sailleau, C.; Breard, E.; Viarouge, C.; Gorlier, A.; Leroux, A.; Hirchaud, E.; Lucas, P.; Blanchard, Y.; Vitour, D.; Grandcollot-Chabot, M.; et al. Emergence of bluetongue virus serotype 4 in mainland France in November 2017. Transbound. Emerg. Dis. 2018, 65, 1158–1162. [Google Scholar] [CrossRef]
- Schulz, C.; Breard, E.; Sailleau, C.; Jenckel, M.; Viarouge, C.; Vitour, D.; Palmarini, M.; Gallois, M.; Hoper, D.; Hoffmann, B.; et al. Bluetongue virus serotype 27: detection and characterization of two novel variants in Corsica, France. J. Gen. Virol. 2016, 97, 2073–2083. [Google Scholar] [CrossRef]
- Breard, E.; Schulz, C.; Sailleau, C.; Bernelin-Cottet, C.; Viarouge, C.; Vitour, D.; Guillaume, B.; Caignard, G.; Gorlier, A.; Attoui, H.; et al. Bluetongue virus serotype 27: Experimental infection of goats, sheep and cattle with three BTV-27 variants reveal atypical characteristics and likely direct contact transmission BTV-27 between goats. Transbound. Emerg. Dis. 2018, 65, e251–e263. [Google Scholar] [CrossRef]
- Batten, C.; Darpel, K.; Henstock, M.; Fay, P.; Veronesi, E.; Gubbins, S.; Graves, S.; Frost, L.; Oura, C. Evidence for transmission of bluetongue virus serotype 26 through direct contact. PLoS ONE 2014, 9, e96049. [Google Scholar] [CrossRef]
- Foxi, C.; Delrio, G.; Falchi, G.; Marche, M.G.; Satta, G.; Ruiu, L. Role of different Culicoides vectors (Diptera: Ceratopogonidae) in bluetongue virus transmission and overwintering in Sardinia (Italy). Parasit. Vectors 2016, 9, 440. [Google Scholar] [CrossRef] [PubMed]
- Foxi, C.; Meloni, G.; Puggioni, G.; Manunta, D.; Rocchigiani, A.; Vento, L.; Cabras, P.; Satta, G. Bluetongue virus detection in new Culicoides species in Sardinia, Italy. Vet. Rec. 2019, 184, 621. [Google Scholar] [CrossRef] [PubMed]
- Goffredo, M.; Catalani, M.; Federici, V.; Portanti, O.; Marini, V.; Mancini, G.; Quaglia, M.; Santilli, A.; Teodori, L.; Savini, G. Vector species of Culicoides midges implicated in the 20122014 Bluetongue epidemics in Italy. Vet. Ital. 2015, 51, 131–138. [Google Scholar] [PubMed]
- Garigliany, M.M.; Hoffmann, B.; Dive, M.; Sartelet, A.; Bayrou, C.; Cassart, D.; Beer, M.; Desmecht, D. Schmallenberg virus in calf born at term with porencephaly, Belgium. Emerg. Infect. Dis. 2012, 18, 1005–1006. [Google Scholar] [PubMed]
- Van den Brom, R.; Luttikholt, S.J.; Lievaart-Peterson, K.; Peperkamp, N.H.; Mars, M.H.; van der Poel, W.H.; Vellema, P. Epizootic of ovine congenital malformations associated with Schmallenberg virus infection. Tijdschr. Diergeneeskd. 2012, 137, 106–111. [Google Scholar] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kundlacz, C.; Caignard, G.; Sailleau, C.; Viarouge, C.; Postic, L.; Vitour, D.; Zientara, S.; Breard, E. Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses 2019, 11, 672. https://doi.org/10.3390/v11070672
Kundlacz C, Caignard G, Sailleau C, Viarouge C, Postic L, Vitour D, Zientara S, Breard E. Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses. 2019; 11(7):672. https://doi.org/10.3390/v11070672
Chicago/Turabian StyleKundlacz, Cindy, Grégory Caignard, Corinne Sailleau, Cyril Viarouge, Lydie Postic, Damien Vitour, Stéphan Zientara, and Emmanuel Breard. 2019. "Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s" Viruses 11, no. 7: 672. https://doi.org/10.3390/v11070672
APA StyleKundlacz, C., Caignard, G., Sailleau, C., Viarouge, C., Postic, L., Vitour, D., Zientara, S., & Breard, E. (2019). Bluetongue Virus in France: An Illustration of the European and Mediterranean Context since the 2000s. Viruses, 11(7), 672. https://doi.org/10.3390/v11070672




