Immunogenicity in Rabbits of Virus-Like Particles from a Contemporary Rabbit Haemorrhagic Disease Virus Type 2 (GI.2/RHDV2/b) Isolated in The Netherlands
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. RHDV2 Virus Sequence Analysis and Phylogenetic Analysis
2.3. RHDV2 VP60 Cloning and Expression Vector Construction
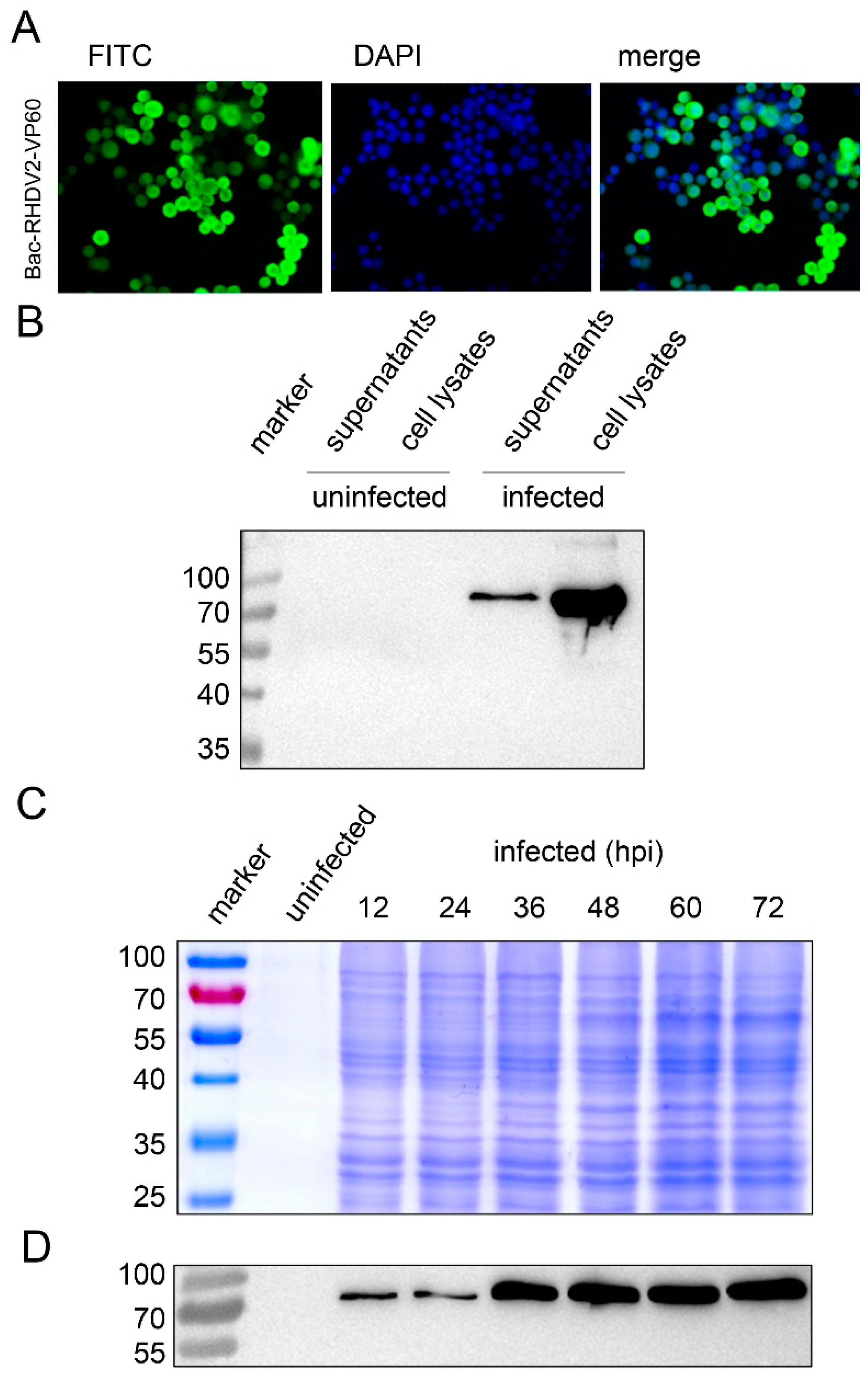
2.4. Cells and Recombinant Baculovirus Expressing RHDV2 VLPs
2.5. Immunofluorescence Assay and Western Blot Analysis
2.6. Transmission Electron Microscopy
2.7. Preparation of RHDV2 VLPs and Control Antigen
2.8. Vaccination of Rabbits with RHDV2 VLPs and ELISA Assays
2.9. Cytokine Assays
2.10. Statistical Analysis
3. Results
3.1. Pathology of RHDV2 Infection in a European Rabbit
3.2. Complete Genome Sequence and Phylogeny of RHDV2 Isolate NL2016
3.3. Baculovirus Expression of RHDV2 VP60 in Sf9 Insect Cells
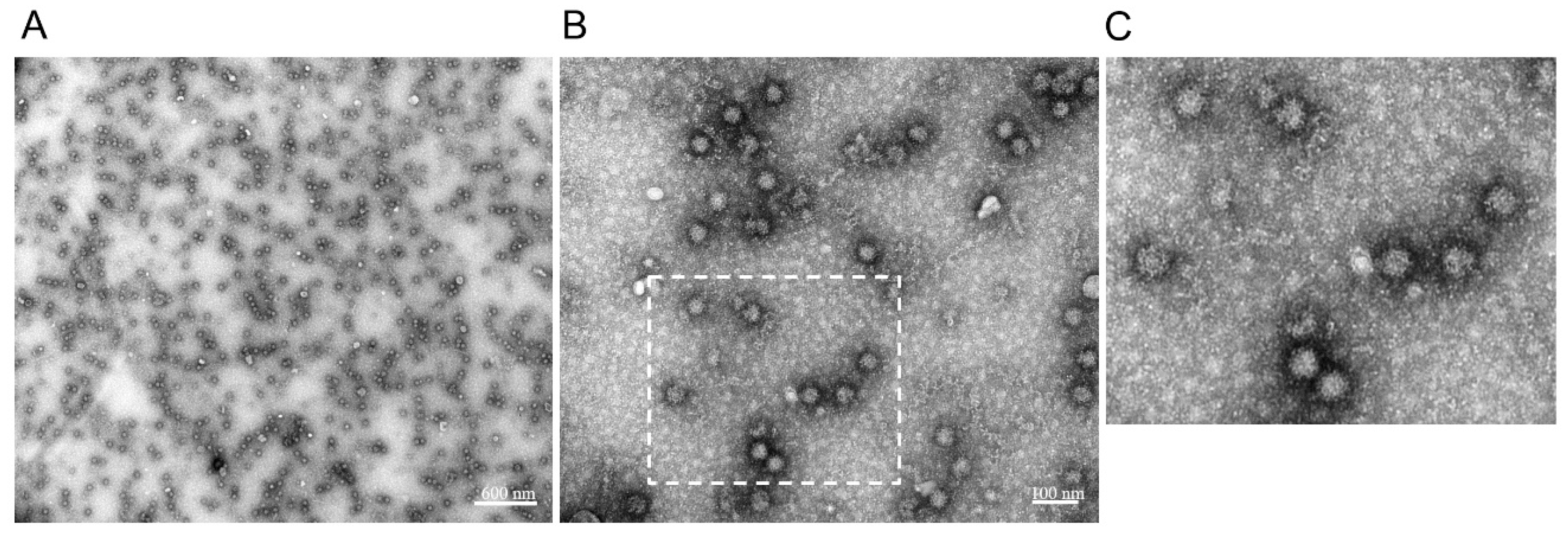
3.4. Characterization of RHDV2 VLPs by Electron Microscopy
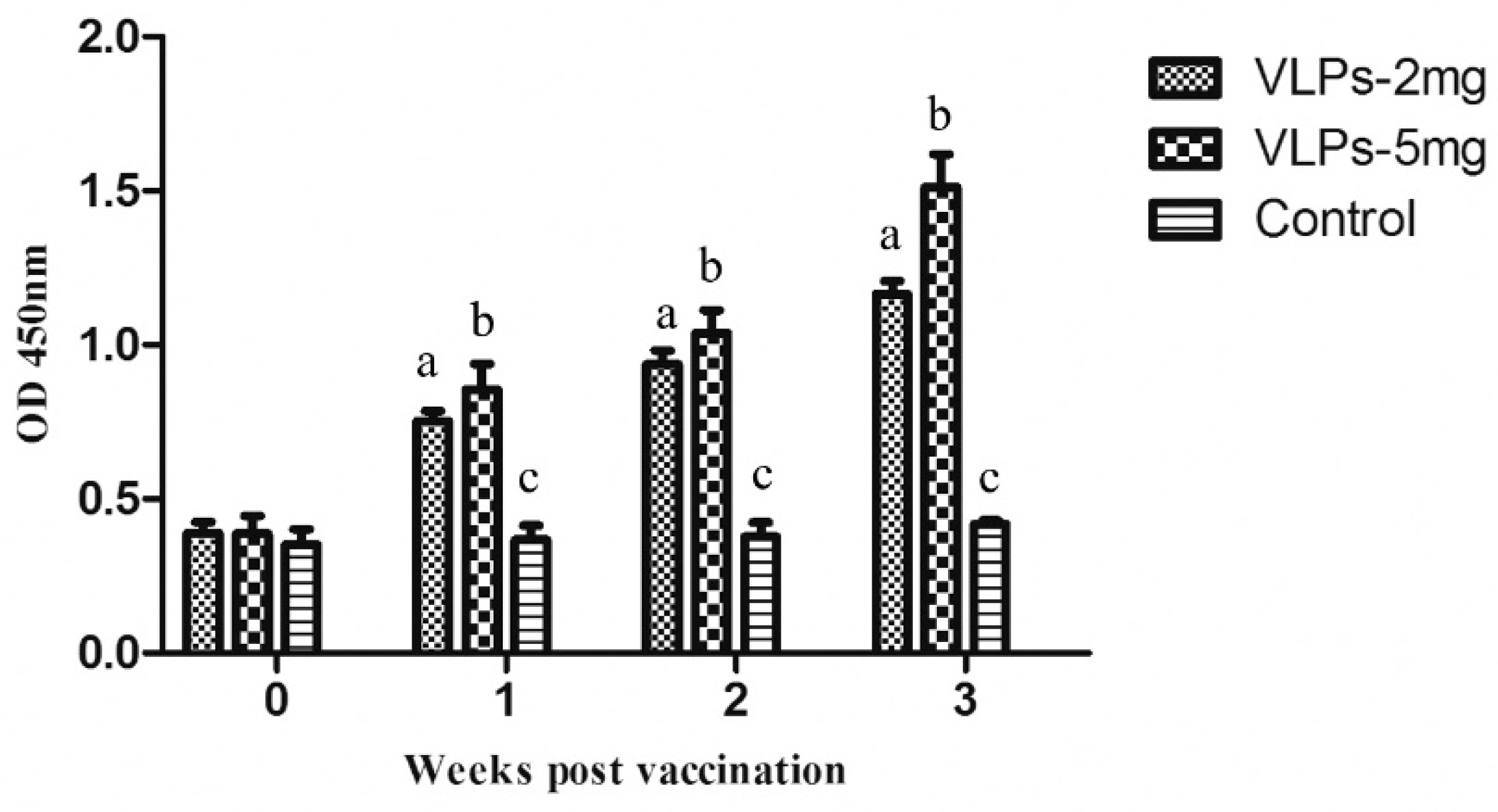
3.5. Immunogenicity of RHDV2 VLPs in Rabbits
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Abrantes, J.; van der Loo, W.; Le Pendu, J.; Esteves, P.J. Rabbit haemorrhagic disease (RHD) and rabbit haemorrhagic disease virus (RHDV): A review. Vet. Res. 2012, 43, 12. [Google Scholar] [CrossRef] [PubMed]
- Ohlinger, V.F.; Haas, B.; Meyers, G.; Weiland, F.; Thiel, H.J. Identification and characterization of the virus causing rabbit hemorrhagic disease. J. Virol. 1990, 64, 3331–3336. [Google Scholar] [PubMed]
- Le Gall-Recule, G.; Zwingelstein, F.; Boucher, S.; Le Normand, B.; Plassiart, G.; Portejoie, Y.; Decors, A.; Bertagnoli, S.; Guerin, J.L.; Marchandeau, S. Detection of a new variant of rabbit haemorrhagic disease virus in France. Vet. Rec. 2011, 168, 137–138. [Google Scholar] [CrossRef] [PubMed]
- Office International des Epizooties. Rabbit Haemorrhagic Disease, Immediate Notification Report; Office International des Epizooties: Udine, Italy, 2011. [Google Scholar]
- Dalton, K.P.; Nicieza, I.; Balseiro, A.; Muguerza, M.A.; Rosell, J.M.; Casais, R.; Alvarez, A.L.; Parra, F. Variant rabbit hemorrhagic disease virus in young rabbits, Spain. Emerg. Infect. Dis. 2012, 18, 2009–2012. [Google Scholar] [CrossRef] [PubMed]
- Le Pendu, J.; Abrantes, J.; Bertagnoli, S.; Guitton, J.S.; Le Gall-Recule, G.; Lopes, A.M.; Marchandeau, S.; Alda, F.; Almeida, T.; Celio, A.P.; et al. Proposal for a unified classification system and nomenclature of lagoviruses. J. Gen. Virol. 2017, 98, 1658–1666. [Google Scholar] [CrossRef] [PubMed]
- Le Gall-Recule, G.; Lavazza, A.; Marchandeau, S.; Bertagnoli, S.; Zwingelstein, F.; Cavadini, P.; Martinelli, N.; Lombardi, G.; Guerin, J.L.; Lemaitre, E.; et al. Emergence of a new lagovirus related to rabbit haemorrhagic disease virus. Vet. Res. 2013, 44, 81. [Google Scholar] [CrossRef]
- Dalton, K.P.; Nicieza, I.; Abrantes, J.; Esteves, P.J.; Parra, F. Spread of new variant rhdv in domestic rabbits on the Iberian Peninsula. Vet. Microbiol. 2014, 169, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Abrantes, J.; Lopes, A.M.; Dalton, K.P.; Melo, P.; Correia, J.J.; Ramada, M.; Alves, P.C.; Parra, F.; Esteves, P.J. New variant of rabbit hemorrhagic disease virus, Portugal, 2012–2013. Emerg. Infect. Dis. 2013, 19, 1900–1902. [Google Scholar] [CrossRef] [PubMed]
- Puggioni, G.; Cavadini, P.; Maestrale, C.; Scivoli, R.; Botti, G.; Ligios, C.; Le Gall-Recule, G.; Lavazza, A.; Capucci, L. The new french 2010 rabbit hemorrhagic disease virus causes an rhd-like disease in the sardinian cape hare (Lepus capensis mediterraneus). Vet. Res. 2013, 44, 96. [Google Scholar] [CrossRef]
- Westcott, D.G.; Frossard, J.P.; Everest, D.; Dastjerdi, A.; Duff, J.P.; Steinbach, F.; Choudhury, B. Incursion of rhdv2-like variant in great britain. Vet. Rec. 2014, 174, 333. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.L.; Silva, S.; Gouveia, P.; Costa, M.; Duarte, E.L.; Henriques, A.M.; Barros, S.S.; Luis, T.; Ramos, F.; Fagulha, T.; et al. Emergence of rabbit haemorrhagic disease virus 2 in the Archipelago of Madeira, Portugal (2016–2017). Virus Genes 2017, 53, 922–926. [Google Scholar] [CrossRef] [PubMed]
- Duarte, M.; Carvalho, C.; Bernardo, S.; Barros, S.V.; Benevides, S.; Flor, L.; Monteiro, M.; Marques, I.; Henriques, M.; Barros, S.C.; et al. Rabbit haemorrhagic disease virus 2 (RHDV2) outbreak in azores: Disclosure of common genetic markers and phylogenetic segregation within the european strains. Infect. Genet. Evol. 2015, 35, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.N.; Mahar, J.E.; Haboury, S.; Stevens, V.; Holmes, E.C.; Strive, T. Emerging rabbit hemorrhagic disease virus 2 (RHDVB), Australia. Emerg. Infect. Dis. 2015, 21, 2276–2278. [Google Scholar] [CrossRef] [PubMed]
- Neimanis, A.S.; Ahola, H.; Zohari, S.; Larsson Pettersson, U.; Brojer, C.; Capucci, L.; Gavier-Widen, D. Arrival of rabbit haemorrhagic disease virus 2 to northern europe: Emergence and outbreaks in wild and domestic rabbits (Oryctolagus cuniculus) in Sweden. Transbound. Emerg. Dis. 2018, 65, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Lopes, A.M.; Rouco, C.; Esteves, P.J.; Abrantes, J. Gi.1b/gi.1b/gi.2 recombinant rabbit hemorrhagic disease virus 2 (lagovirus europaeus/gi.2) in Morocco, Africa. Arch. Virol. 2019, 164, 279–283. [Google Scholar] [CrossRef] [PubMed]
- Rouco, C.; Aguayo-Adán, J.A.; Santoro, S.; Abrantes, J.; Delibes-Mateos, M. Worldwide rapid spread of the novel rabbit haemorrhagic disease virus (GI.2/RHDV 2/b). Transbound. Emerg. Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Mahar, J.E.; Hall, R.N.; Peacock, D.; Kovaliski, J.; Piper, M.; Mourant, R.; Huang, N.; Campbell, S.; Gu, X.; Read, A.; et al. Rabbit hemorrhagic disease virus 2 (RHDV2; GI.2) is replacing endemic strains of RHDV in the australian landscape within 18 months of its arrival. J. Virol. 2018, 92, e01374-17. [Google Scholar] [CrossRef]
- Lopes, A.M.; Correia, J.; Abrantes, J.; Melo, P.; Ramada, M.; Magalhaes, M.J.; Alves, P.C.; Esteves, P.J. Is the new variant rhdv replacing genogroup 1 in portuguese wild rabbit populations? Viruses 2014, 7, 27–36. [Google Scholar] [CrossRef]
- Peacock, D.; Kovaliski, J.; Sinclair, R.; Mutze, G.; Iannella, A.; Capucci, L. Rhdv2 overcoming rhdv immunity in wild rabbits (Oryctolagus cuniculus) in Australia. Vet. Rec. 2017, 180, 280. [Google Scholar] [CrossRef]
- Capucci, L.; Cavadini, P.; Schiavitto, M.; Lombardi, G.; Lavazza, A. Increased pathogenicity in rabbit haemorrhagic disease virus type 2 (RHDV2). Vet. Rec. 2017, 180, 426. [Google Scholar] [CrossRef]
- Kushnir, N.; Streatfield, S.J.; Yusibov, V. Virus-like particles as a highly efficient vaccine platform: Diversity of targets and production systems and advances in clinical development. Vaccine 2012, 31, 58–83. [Google Scholar] [CrossRef] [PubMed]
- Wagner, R.; Deml, L.; Schirmbeck, R.; Reimann, J.; Wolf, H. Induction of a mhc class I-restricted, CD8 positive cytolytic T-cell response by chimeric HIV-1 virus-like particles in vivo: Implications on HIV vaccine development. Behring Inst. Mitt. 1994, 95, 23–34. [Google Scholar]
- Schirmbeck, R.; Bohm, W.; Reimann, J. Virus-like particles induce mhc class I-restricted T-cell responses. Lessons learned from the hepatitis B small surface antigen. Intervirology 1996, 39, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.; Frazer, I.H.; Fernando, G.J.; Zhou, J. Papillomavirus virus-like particles can deliver defined CTL epitopes to the mhc class I pathway. Virology 1998, 240, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Win, S.J.; Ward, V.K.; Dunbar, P.R.; Young, S.L.; Baird, M.A. Cross-presentation of epitopes on virus-like particles via the MHC I receptor recycling pathway. Immunol. Cell Biol. 2011, 89, 681–688. [Google Scholar] [CrossRef]
- Monie, A.; Hung, C.F.; Roden, R.; Wu, T.C. Cervarix: A vaccine for the prevention of HPV 16, 18-associated cervical cancer. Biologics 2008, 2, 97–105. [Google Scholar]
- Ji, M.; Xie, X.X.; Liu, D.Q.; Yu, X.L.; Zhang, Y.; Zhang, L.X.; Wang, S.W.; Huang, Y.R.; Liu, R.T. Hepatitis B core VLP-based mis-disordered Tau vaccine elicits strong immune response and alleviates cognitive deficits and neuropathology progression in tau.P301s mouse model of alzheimer’s disease and frontotemporal dementia. Alzheimers Res. Ther. 2018, 10, 55. [Google Scholar] [CrossRef] [PubMed]
- Fort, M.; Sibila, M.; Perez-Martin, E.; Nofrarias, M.; Mateu, E.; Segales, J. One dose of a porcine circovirus 2 (PCV2) sub-unit vaccine administered to 3-week-old conventional piglets elicits cell-mediated immunity and significantly reduces PCV2 viremia in an experimental model. Vaccine 2009, 27, 4031–4037. [Google Scholar] [CrossRef] [PubMed]
- Akahata, W.; Yang, Z.-Y.; Andersen, H.; Sun, S.; Holdaway, H.A.; Kong, W.-P.; Lewis, M.G.; Higgs, S.; Rossmann, M.G.; Rao, S.; et al. A virus-like particle vaccine for epidemic chikungunya virus protects nonhuman primates against infection. Nat. Med. 2010, 16, 334. [Google Scholar] [CrossRef]
- Boigard, H.; Alimova, A.; Martin, G.R.; Katz, A.; Gottlieb, P.; Galarza, J.M. Zika virus-like particle (VLP) based vaccine. PLoS Negl. Trop. Dis. 2017, 11, e0005608. [Google Scholar] [CrossRef]
- Nagesha, H.S.; Wang, L.F.; Hyatt, A.D.; Morrissy, C.J.; Lenghaus, C.; Westbury, H.A. Self-assembly, antigenicity, and immunogenicity of the rabbit haemorrhagic disease virus (czechoslovakian strain v-351) capsid protein expressed in baculovirus. Arch. Virol. 1995, 140, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, J.; Tan, Y.; Li, C.; Chen, Z.; Sun, S.; Liu, G. Self-assembly of virus-like particles of rabbit hemorrhagic disease virus capsid protein expressed in escherichia coli and their immunogenicity in rabbits. Antivir. Res. 2016, 131, 85–91. [Google Scholar] [CrossRef]
- Peacey, M.; Wilson, S.; Perret, R.; Ronchese, F.; Ward, V.K.; Young, V.; Young, S.L.; Baird, M.A. Virus-like particles from rabbit hemorrhagic disease virus can induce an anti-tumor response. Vaccine 2008, 26, 5334–5337. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, E.; Toledo, J.R.; Mendez, L.; Gonzalez, N.; Parra, F.; Martin-Alonso, J.M.; Limonta, M.; Sanchez, K.; Cabrales, A.; Estrada, M.P.; et al. Conformational and thermal stability improvements for the large-scale production of yeast-derived rabbit hemorrhagic disease virus-like particles as multipurpose vaccine. PLoS ONE 2013, 8, e56417. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Song, Y.; Fan, Z.; Jiang, P.; Hu, B.; Xue, J.; Wei, H.; Wang, F. Immunogenicity of different recombinant rabbit hemorrhagic disease virus-like particles carrying CD8+ T cell epitope from chicken ovalbumin (OVA). Virus Res. 2014, 183, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Fan, Z.; Zuo, Y.; Wei, H.; Hu, B.; Chen, M.; Qiu, R.; Xue, J.; Wang, F. Binding of rabbit hemorrhagic disease virus-like particles to host histo-blood group antigens is blocked by antisera from experimentally vaccinated rabbits. Arch. Virol. 2017, 162, 3425–3430. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Meng, C.; Chen, Z.; Li, C.; Liu, G. Codon optimization of the rabbit hemorrhagic disease virus (RHDV) capsid gene leads to increased gene expression in spodoptera frugiperda 9 (Sf9) cells. J. Vet. Sci. 2013, 14, 441–447. [Google Scholar] [CrossRef]
- Van Oers, M.M.; Pijlman, G.P.; Vlak, J.M. Thirty years of baculovirus-insect cell protein expression: From dark horse to mainstream technology. J. Gen. Virol. 2015, 96, 6–23. [Google Scholar] [CrossRef]
- Metz, S.W.; Pijlman, G.P. Arbovirus vaccines; opportunities for the baculovirus-insect cell expression system. J. Invertebr. Pathol. 2011, 107, S16–S30. [Google Scholar] [CrossRef]
- Lopes, A.M.; Marques, S.; Silva, E.; Magalhaes, M.J.; Pinheiro, A.; Alves, P.C.; Le Pendu, J.; Esteves, P.J.; Thompson, G.; Abrantes, J. Detection of rhdv strains in the iberian hare (Lepus granatensis): Earliest evidence of rabbit lagovirus cross-species infection. Vet. Res. 2014, 45, 94. [Google Scholar]
- Camarda, A.; Pugliese, N.; Cavadini, P.; Circella, E.; Capucci, L.; Caroli, A.; Legretto, M.; Mallia, E.; Lavazza, A. Detection of the new emerging rabbit haemorrhagic disease type 2 virus (RHDV2) in sicily from rabbit (Oryctolagus cuniculus) and italian hare (Lepus corsicanus). Res. Vet. Sci. 2014, 97, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.N.; Peacock, D.E.; Kovaliski, J.; Mahar, J.E.; Mourant, R.; Piper, M.; Strive, T. Detection of RHDV2 in european brown hares (Lepus europaeus) in Australia. Vet. Rec. 2017, 180, 121. [Google Scholar] [CrossRef] [PubMed]
- Velarde, R.; Cavadini, P.; Neimanis, A.; Cabezon, O.; Chiari, M.; Gaffuri, A.; Lavin, S.; Grilli, G.; Gavier-Widen, D.; Lavazza, A.; et al. Spillover events of infection of brown hares (Lepus europaeus) with rabbit haemorrhagic disease type 2 virus (RHDV2) caused sporadic cases of an european brown hare syndrome-like disease in Italy and Spain. Transbound. Emerg. Dis. 2016, 64, 1750–1761. [Google Scholar] [CrossRef] [PubMed]
- Neimanis, A.S.; Ahola, H.; Larsson Pettersson, U.; Lopes, A.M.; Abrantes, J.; Zohari, S.; Esteves, P.J.; Gavier-Widen, D. Overcoming species barriers: An outbreak of lagovirus europaeus GI.2/RHDV2 in an isolated population of mountain hares (Lepus timidus). BMC Vet. Res. 2018, 14, 367. [Google Scholar] [CrossRef] [PubMed]
- Metz, S.W.; Martina, B.E.; van den Doel, P.; Geertsema, C.; Osterhaus, A.D.; Vlak, J.M.; Pijlman, G.P. Chikungunya virus-like particles are more immunogenic in a lethal ag129 mouse model compared to glycoprotein E1 or E2 subunits. Vaccine 2013, 31, 6092–6096. [Google Scholar] [CrossRef] [PubMed]
- Peacey, M.; Wilson, S.; Baird, M.A.; Ward, V.K. Versatile rhdv virus-like particles: Incorporation of antigens by genetic modification and chemical conjugation. Biotechnol. Bioeng. 2007, 98, 968–977. [Google Scholar] [CrossRef]
- Barcena, J.; Guerra, B.; Angulo, I.; Gonzalez, J.; Valcarcel, F.; Mata, C.P.; Caston, J.R.; Blanco, E.; Alejo, A. Comparative analysis of rabbit hemorrhagic disease virus (RHDV) and new RHDV2 virus antigenicity, using specific virus-like particles. Vet. Res. 2015, 46, 106. [Google Scholar] [CrossRef] [PubMed]
- Dalton, K.P.; Podadera, A.; Granda, V.; Nicieza, I.; Del Llano, D.; Gonzalez, R.; de Los Toyos, J.R.; Garcia Ocana, M.; Vazquez, F.; Martin Alonso, J.M.; et al. Elisa for detection of variant rabbit haemorrhagic disease virus RHDV2 antigen in liver extracts. J. Virol. Methods 2018, 251, 38–42. [Google Scholar] [CrossRef]
- Muller, C.; Ulrich, R.; Franzke, K.; Muller, M.; Kollner, B. Crude extracts of recombinant baculovirus expressing rabbit hemorrhagic disease virus 2 VLPs from both insect and rabbit cells protect rabbits from rabbit hemorrhagic disease caused by RHDV2. Arch. Virol. 2019, 164, 137–148. [Google Scholar] [CrossRef]
- Lopes, A.M.; Dalton, K.P.; Magalhaes, M.J.; Parra, F.; Esteves, P.J.; Holmes, E.C.; Abrantes, J. Full genomic analysis of new variant rabbit hemorrhagic disease virus revealed multiple recombination events. J. Gen. Virol. 2015, 96, 1309–1319. [Google Scholar] [CrossRef]
- Silverio, D.; Lopes, A.M.; Melo-Ferreira, J.; Magalhaes, M.J.; Monterroso, P.; Serronha, A.; Maio, E.; Alves, P.C.; Esteves, P.J.; Abrantes, J. Insights into the evolution of the new variant rabbit haemorrhagic disease virus (GI.2) and the identification of novel recombinant strains. Transbound. Emerg. Dis. 2018, 65, 983–992. [Google Scholar] [CrossRef] [PubMed]

| Primer Name | Primer Sequence |
|---|---|
| Gate-VP60F(NcoI) | GGGGACAAGTTTGTACAAAAAAGCAGGCTTACCATGGACCATGGAGGGCAAAGCCCG |
| Gate-VP60R(NsiI) | GGGGACCACTTTGTACAAGAAAGCTGGGTAATGCATTCAGACATAAGAAAAGCCATTG |
| RHDV2(1-405)-F | GTGAAAGTTATGGCGGCTATG |
| RHDV2(1-405)-R | TCGGTAAGCACAGGGGATGAC |
| RHDV2(80-1355)-F | TCCTGGACCTCAGGGACAAGA |
| RHDV2(80-1355)-R | GCCATTTTCACAACTGTCAT |
| RHDV2(1332-3101)-F | GGTTATGACAGTTGTGAAAATGGC |
| RHDV2(1332-3101)-R | GTCATGTCATGTGCGTTGACA |
| RHDV2(3083-5319)-F | TCAACGCACATGACATGACTG |
| RHDV2(3083-5319)-R | GGCTTTGCCCTCCATAACATT |
| RHDV2(6964-7378)-F | CGCCCTGTGGGACCCAGA |
| RHDV2(6964-7378)-R | TCAAGCACTGGACTCGCCAGT |
| RHDV2(5295-7047)-F | TGTGAATGTTATGGAGGGCAAAGC |
| RHDV2- 3′dTNNN | GACTGACTGCCATGGCCGGCGCTAGCTTTTTTTTTTTTTTTTTTTTTTTTT |
| Genbank Accession Number | Genotype | Strain Name | Year of Isolation |
|---|---|---|---|
| KX844830 | RHDVa | SCH04 | 2016 |
| KY171748 | RHDVa | Sch07 | 2017 |
| AB300693 | RHDVa | Hokkaido/2002/JPN | 2009 |
| EU003581 | RHDVa | NY-01 | 2007 |
| KY622129 | RHDVa | P175 | 2017 |
| MF598301 | RHDVa | K5_08Q712_BatchRelease1/2008 | 2017 |
| DQ205345 | RHDVa | JX/CHA/97 | 1997 |
| JF412629 | RHDVa | HYD | 2011 |
| HM623309 | RHDVa | NJ-2009 | 2009 |
| KF677011 | RHDVa | STR2012 | 2012 |
| KY679905 | RHDVa | STR2014 | 2014 |
| AF258618 | RHDVa | Iowa2000 | 2000 |
| EF558583 | RHDVa | Triptis | 2008 |
| EU003582 | RHDVa | UT-01 | 2001 |
| EF558586 | RHDVa | Hartmannsdorf | 2007 |
| Z29514 | RHDV | SD | 2005 |
| KU882095 | RHDV | ZD0 | 2000 |
| EF558576 | RHDV | Jena | 2007 |
| KY622127 | RHDV | P158 | 1998 |
| DQ189078 | RHDV | Saudi Arabia | 2005 |
| KX357670 | RHDV | AUS/ACT/MtPt-2/2010 | 2010 |
| KT006735 | RHDV | AUS/WA/Bunbury/2000 | 2000 |
| KT006733 | RHDV | AUS/WA/Gnowangerup/1999 | 1999 |
| U54983 | RHDV | Czech strain V351 | 1997 |
| EF558579 | RHDV | NZ54 | 2007 |
| KT006725 | RHDV | NZL/Otago/Queensberry/74/2013 | 2013 |
| GQ166866 | RCV | MRCV | 2001 |
| KP129397 | RHDV2(RHDVb) | Tar06-12 | 2015 |
| MF421684 | RHDV2(RHDVb) | AUS/VIC/AC-1/2016 | 2016 |
| KF442961 | RHDV2(RHDVb) | Algarve1 | 2013 |
| KM115680 | RHDV2(RHDVb) | CBEstoi13-7 | 2013 |
| MF598302 | RHDV2(RHDVb) | AUS/NSW/CAR-3/2016 | 2016 |
| KM979445 | RHDV2(RHDVb) | CBVal16 | 2012 |
| KP129395 | RHDV2(RHDVb) | Rij06-12 | 2014 |
| KP090976 | RHDV2(RHDVb) | CBAnd1 | 2012 |
| KM878681 | RHDV2(RHDVb) | N11 | 2011 |
| KP129398 | RHDV2(RHDVb) | Zar11-11 | 2010 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, Q.; Qi, R.; Veldkamp, L.; Ijzer, J.; Kik, M.L.; Zhu, J.; Tang, A.; Dong, D.; Shi, Y.; van Oers, M.M.; et al. Immunogenicity in Rabbits of Virus-Like Particles from a Contemporary Rabbit Haemorrhagic Disease Virus Type 2 (GI.2/RHDV2/b) Isolated in The Netherlands. Viruses 2019, 11, 553. https://doi.org/10.3390/v11060553
Miao Q, Qi R, Veldkamp L, Ijzer J, Kik ML, Zhu J, Tang A, Dong D, Shi Y, van Oers MM, et al. Immunogenicity in Rabbits of Virus-Like Particles from a Contemporary Rabbit Haemorrhagic Disease Virus Type 2 (GI.2/RHDV2/b) Isolated in The Netherlands. Viruses. 2019; 11(6):553. https://doi.org/10.3390/v11060553
Chicago/Turabian StyleMiao, Qiuhong, Ruibing Qi, Luut Veldkamp, Jooske Ijzer, Marja L. Kik, Jie Zhu, Aoxing Tang, Dandan Dong, Yonghong Shi, Monique M. van Oers, and et al. 2019. "Immunogenicity in Rabbits of Virus-Like Particles from a Contemporary Rabbit Haemorrhagic Disease Virus Type 2 (GI.2/RHDV2/b) Isolated in The Netherlands" Viruses 11, no. 6: 553. https://doi.org/10.3390/v11060553
APA StyleMiao, Q., Qi, R., Veldkamp, L., Ijzer, J., Kik, M. L., Zhu, J., Tang, A., Dong, D., Shi, Y., van Oers, M. M., Liu, G., & Pijlman, G. P. (2019). Immunogenicity in Rabbits of Virus-Like Particles from a Contemporary Rabbit Haemorrhagic Disease Virus Type 2 (GI.2/RHDV2/b) Isolated in The Netherlands. Viruses, 11(6), 553. https://doi.org/10.3390/v11060553







