All the Same? The Secret Life of Prion Strains within Their Target Cells
Abstract
1. Introduction
2. Restricted Susceptibility of Cell Lines to Different Prion Strains
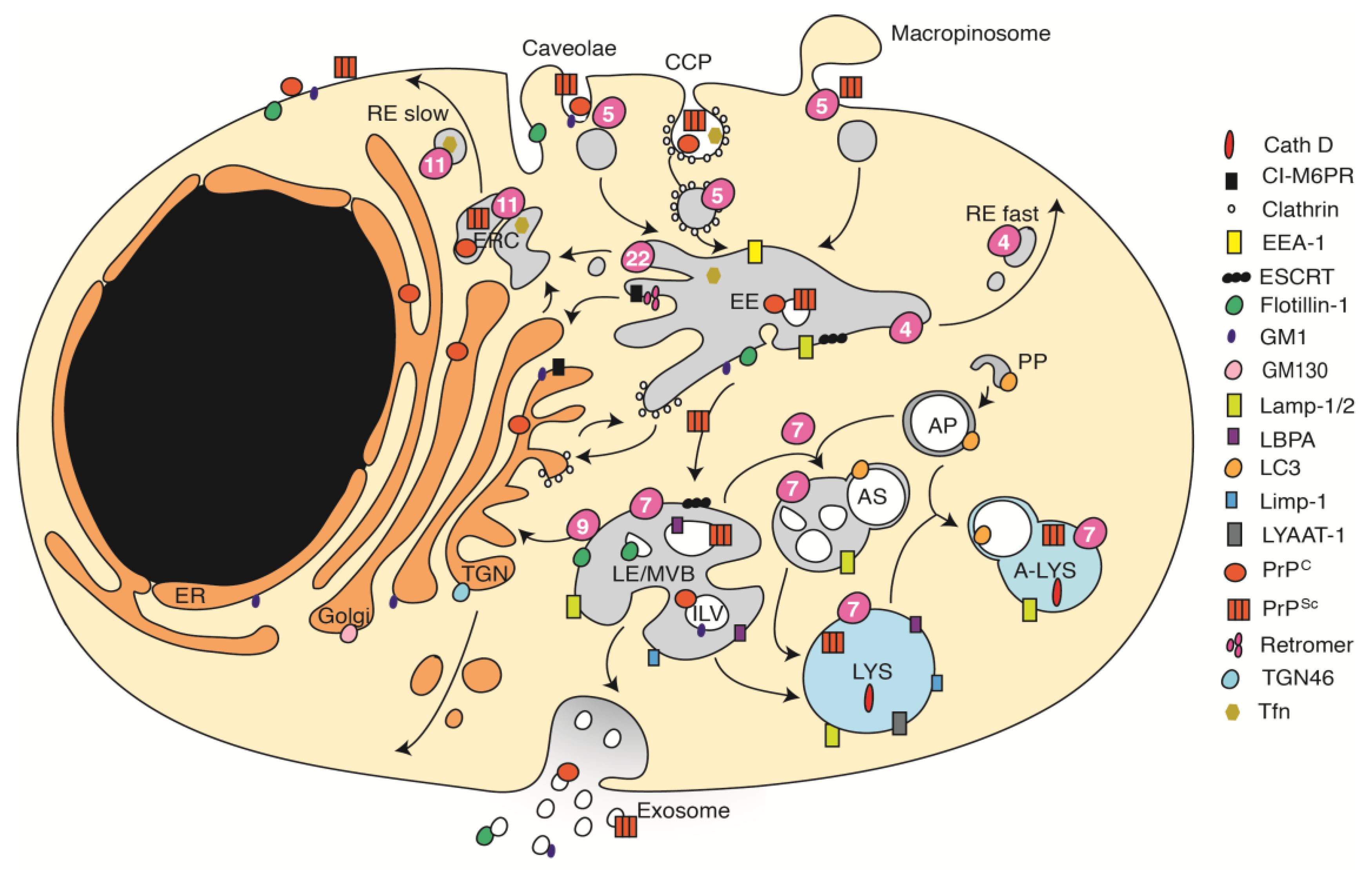
3. The Complex Organization of the Endocytic Pathway
4. Involvement of the Endocytic Trafficking Pathway in Prion Biogenesis
5. Subcellular Distribution of PrPSc in Cells Infected with Different Prion Strains
6. Cellular Factors Involved in Prion Attachment and/or Uptake
7. Establishment of Productive Infections
8. First Sites of PrPSc Formation during Acute Infection
9. Acute and Chronic Prion Infections Depend on Different Intracellular Trafficking Processes
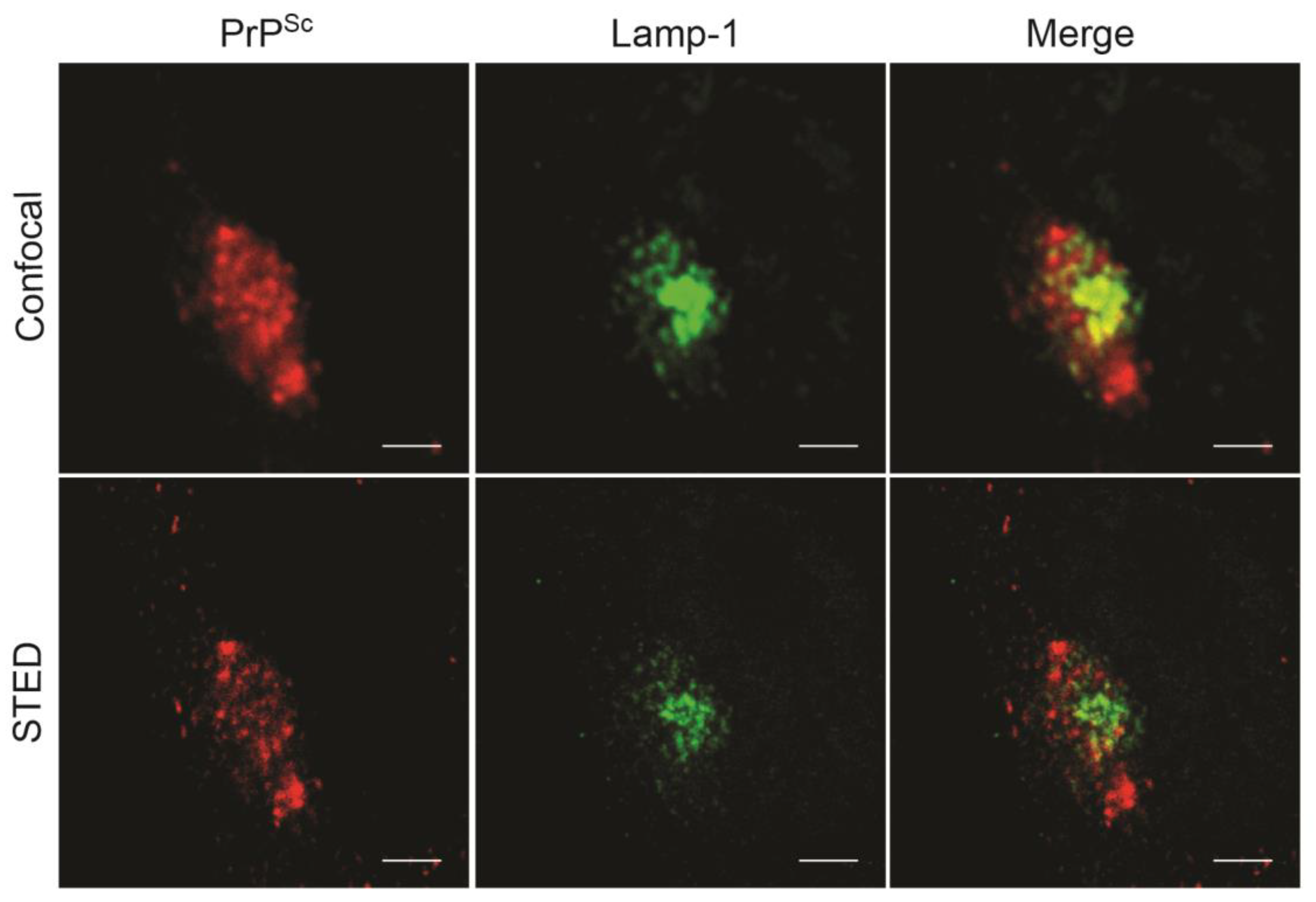
10. Subcellular Sites of PrPSc Formation in Chronically Infected Cells
11. Considerations for Future Research on Prion Cell Biology
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jeffrey, M.; McGovern, G.; Siso, S.; Gonzalez, L. Cellular and sub-cellular pathology of animal prion diseases: Relationship between morphological changes, accumulation of abnormal prion protein and clinical disease. Acta. Neuropathol. 2011, 121, 113–134. [Google Scholar] [CrossRef]
- Mabbott, N.A.; Mackay, F.; Minns, F.; Bruce, M.E. Temporary inactivation of follicular dendritic cells delays neuroinvasion of scrapie. Nat. Med. 2000, 6, 719–720. [Google Scholar] [CrossRef] [PubMed]
- Andreoletti, O.; Simon, S.; Lacroux, C.; Morel, N.; Tabouret, G.; Chabert, A.; Lugan, S.; Corbiere, F.; Ferre, P.; Foucras, G.; et al. Prpsc accumulation in myocytes from sheep incubating natural scrapie. Nat. Med. 2004, 10, 591–593. [Google Scholar] [CrossRef] [PubMed]
- Van Keulen, L.J.; Schreuder, B.E.; Vromans, M.E.; Langeveld, J.P.; Smits, M.A. Scrapie-associated prion protein in the gastrointestinal tract of sheep with natural scrapie. J. Comp. Pathol. 1999, 121, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Tuo, W.; O’Rourke, K.I.; Zhuang, D.; Cheevers, W.P.; Spraker, T.R.; Knowles, D.P. Pregnancy status and fetal prion genetics determine prpsc accumulation in placentomes of scrapie-infected sheep. Proc. Natl. Acad. Sci. USA 2002, 99, 6310–6315. [Google Scholar] [CrossRef] [PubMed]
- Heikenwalder, M.; Zeller, N.; Seeger, H.; Prinz, M.; Klohn, P.C.; Schwarz, P.; Ruddle, N.H.; Weissmann, C.; Aguzzi, A. Chronic lymphocytic inflammation specifies the organ tropism of prions. Science 2005, 307, 1107–1110. [Google Scholar] [CrossRef]
- Prusiner, S.B. Prions. Proc. Natl. Acad. Sci. USA 1998, 95, 13363–13383. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.; Waudby, C.A.; Devlin, G.L.; Cohen, S.I.; Aguzzi, A.; Vendruscolo, M.; Terentjev, E.M.; Welland, M.E.; Dobson, C.M. An analytical solution to the kinetics of breakable filament assembly. Science 2009, 326, 1533–1537. [Google Scholar] [CrossRef]
- Cobb, N.J.; Sonnichsen, F.D.; McHaourab, H.; Surewicz, W.K. Molecular architecture of human prion protein amyloid: A parallel, in-register beta-structure. Proc. Natl. Acad. Sci. USA 2007, 104, 18946–18951. [Google Scholar] [CrossRef]
- Groveman, B.R.; Dolan, M.A.; Taubner, L.M.; Kraus, A.; Wickner, R.B.; Caughey, B. Parallel in-register intermolecular beta-sheet architectures for prion-seeded prion protein (prp) amyloids. J. Biol. Chem. 2014, 289, 24129–24142. [Google Scholar] [CrossRef]
- Vazquez-Fernandez, E.; Vos, M.R.; Afanasyev, P.; Cebey, L.; Sevillano, A.M.; Vidal, E.; Rosa, I.; Renault, L.; Ramos, A.; Peters, P.J.; et al. The structural architecture of an infectious mammalian prion using electron cryomicroscopy. PLoS Pathog. 2016, 12, e1005835. [Google Scholar] [CrossRef]
- Dickinson, A.G. Scrapie in sheep and goats. Front. Biol. 1976, 44, 209–241. [Google Scholar]
- Bruce, M.E.; McConnell, I.; Fraser, H.; Dickinson, A.G. The disease characteristics of different strains of scrapie in sinc congenic mouse lines: Implications for the nature of the agent and host control of pathogenesis. J. Gen. Virol. 1991, 72 Pt 3, 595–603. [Google Scholar] [CrossRef]
- Kimberlin, R.H.; Walker, C.A.; Fraser, H. The genomic identity of different strains of mouse scrapie is expressed in hamsters and preserved on reisolation in mice. J. Gen. Virol. 1989, 70 Pt 8, 2017–2025. [Google Scholar] [CrossRef]
- Kimberlin, R.H.; Walker, C.A. Evidence that the transmission of one source of scrapie agent to hamsters involves separation of agent strains from a mixture. J. Gen. Virol. 1978, 39, 487–496. [Google Scholar] [CrossRef]
- Striebel, J.F.; Race, B.; Meade-White, K.D.; LaCasse, R.; Chesebro, B. Strain specific resistance to murine scrapie associated with a naturally occurring human prion protein polymorphism at residue 171. PLoS Pathog. 2011, 7, e1002275. [Google Scholar] [CrossRef]
- Zlotnik, I.; Rennie, J.C. Further observations on the experimental transmission of scrapie from sheep and goats to laboratory mice. J. Comp. Pathol. 1963, 73, 150–162. [Google Scholar] [CrossRef]
- Tateishi, J.; Ohta, M.; Koga, M.; Sato, Y.; Kuroiwa, Y. Transmission of chronic spongiform encephalopathy with kuru plaques from humans to small rodents. Ann. Neurol. 1979, 5, 581–584. [Google Scholar] [CrossRef]
- Bruce, M.E.; Fraser, H. Scrapie strain variation and its implications. Curr. Top. Microbiol. Immunol. 1991, 172, 125–138. [Google Scholar]
- Gonzalez, L.; Martin, S.; Jeffrey, M. Distinct profiles of prp(d) immunoreactivity in the brain of scrapie- and bse-infected sheep: Implications for differential cell targeting and prp processing. J. Gen. Virol. 2003, 84, 1339–1350. [Google Scholar] [CrossRef]
- Carroll, J.A.; Striebel, J.F.; Rangel, A.; Woods, T.; Phillips, K.; Peterson, K.E.; Race, B.; Chesebro, B. Prion strain differences in accumulation of prpsc on neurons and glia are associated with similar expression profiles of neuroinflammatory genes: Comparison of three prion strains. PLoS Pathog. 2016, 12, e1005551. [Google Scholar] [CrossRef]
- Dearmond, S.J.; Bajsarowicz, K. Prpsc accumulation in neuronal plasma membranes links notch-1 activation to dendritic degeneration in prion diseases. Mol. Neurodegener 2010, 5, 6. [Google Scholar] [CrossRef]
- Arnold, J.E.; Tipler, C.; Laszlo, L.; Hope, J.; Landon, M.; Mayer, R.J. The abnormal isoform of the prion protein accumulates in late-endosome-like organelles in scrapie-infected mouse brain. J. Pathol. 1995, 176, 403–411. [Google Scholar] [CrossRef]
- Laszlo, L.; Lowe, J.; Self, T.; Kenward, N.; Landon, M.; McBride, T.; Farquhar, C.; McConnell, I.; Brown, J.; Hope, J.; et al. Lysosomes as key organelles in the pathogenesis of prion encephalopathies. J. Pathol. 1992, 166, 333–341. [Google Scholar] [CrossRef]
- Riesner, D. Biochemistry and structure of prp(c) and prp(sc). Br. Med. Bull. 2003, 66, 21–33. [Google Scholar] [CrossRef]
- Bessen, R.A.; Marsh, R.F. Distinct prp properties suggest the molecular basis of strain variation in transmissible mink encephalopathy. J. Virol. 1994, 68, 7859–7868. [Google Scholar]
- Deleault, N.R.; Lucassen, R.W.; Supattapone, S. Rna molecules stimulate prion protein conversion. Nature 2003, 425, 717–720. [Google Scholar] [CrossRef]
- Deleault, N.R.; Harris, B.T.; Rees, J.R.; Supattapone, S. Formation of native prions from minimal components in vitro. Proc. Natl. Acad. Sci. USA 2007, 104, 9741–9746. [Google Scholar] [CrossRef]
- Kim, C.; Xiao, X.; Chen, S.; Haldiman, T.; Smirnovas, V.; Kofskey, D.; Warren, M.; Surewicz, K.; Maurer, N.R.; Kong, Q.; et al. Artificial strain of human prions created in vitro. Nat. Commun. 2018, 9, 2166. [Google Scholar] [CrossRef]
- Ayers, J.I.; Kincaid, A.E.; Bartz, J.C. Prion strain targeting independent of strain-specific neuronal tropism. J. Virol. 2009, 83, 81–87. [Google Scholar] [CrossRef]
- Priola, S.A. Cell biology of prion infection. Handb. Clin. Neurol. 2018, 153, 45–68. [Google Scholar]
- Grassmann, A.; Wolf, H.; Hofmann, J.; Graham, J.; Vorberg, I. Cellular aspects of prion replication in vitro. Viruses 2013, 5, 374–405. [Google Scholar] [CrossRef]
- Oelschlegel, A.M.; Geissen, M.; Lenk, M.; Riebe, R.; Angermann, M.; Schatzl, H.; Groschup, M.H. A bovine cell line that can be infected by natural sheep scrapie prions. PLoS ONE 2015, 10, e0117154. [Google Scholar] [CrossRef]
- Butler, D.A.; Scott, M.R.; Bockman, J.M.; Borchelt, D.R.; Taraboulos, A.; Hsiao, K.K.; Kingsbury, D.T.; Prusiner, S.B. Scrapie-infected murine neuroblastoma cells produce protease-resistant prion proteins. J. Virol. 1988, 62, 1558–1564. [Google Scholar]
- Borchelt, D.R.; Taraboulos, A.; Prusiner, S.B. Evidence for synthesis of scrapie prion proteins in the endocytic pathway. J. Biol. Chem. 1992, 267, 16188–16199. [Google Scholar]
- Taraboulos, A.; Raeber, A.J.; Borchelt, D.R.; Serban, D.; Prusiner, S.B. Synthesis and trafficking of prion proteins in cultured cells. Mol. Biol. Cell 1992, 3, 851–863. [Google Scholar] [CrossRef]
- McKinley, M.P.; Taraboulos, A.; Kenaga, L.; Serban, D.; Stieber, A.; DeArmond, S.J.; Prusiner, S.B.; Gonatas, N. Ultrastructural localization of scrapie prion proteins in cytoplasmic vesicles of infected cultured cells. Lab. Investig. 1991, 65, 622–630. [Google Scholar]
- Borchelt, D.R.; Scott, M.; Taraboulos, A.; Stahl, N.; Prusiner, S.B. Scrapie and cellular prion proteins differ in their kinetics of synthesis and topology in cultured cells. J. Cell Biol. 1990, 110, 743–752. [Google Scholar] [CrossRef]
- Birkett, C.R.; Hennion, R.M.; Bembridge, D.A.; Clarke, M.C.; Chree, A.; Bruce, M.E.; Bostock, C.J. Scrapie strains maintain biological phenotypes on propagation in a cell line in culture. EMBO J. 2001, 20, 3351–3358. [Google Scholar] [CrossRef]
- Schatzl, H.M.; Laszlo, L.; Holtzman, D.M.; Tatzelt, J.; DeArmond, S.J.; Weiner, R.I.; Mobley, W.C.; Prusiner, S.B. A hypothalamic neuronal cell line persistently infected with scrapie prions exhibits apoptosis. J. Virol. 1997, 71, 8821–8831. [Google Scholar]
- Mahal, S.P.; Baker, C.A.; Demczyk, C.A.; Smith, E.W.; Julius, C.; Weissmann, C. Prion strain discrimination in cell culture: The cell panel assay. Proc. Natl. Acad. Sci. USA 2007, 104, 20908–20913. [Google Scholar] [CrossRef]
- Courageot, M.P.; Daude, N.; Nonno, R.; Paquet, S.; Di Bari, M.A.; Le Dur, A.; Chapuis, J.; Hill, A.F.; Agrimi, U.; Laude, H.; et al. A cell line infectible by prion strains from different species. J. Gen. Virol. 2008, 89, 341–347. [Google Scholar] [CrossRef]
- Vorberg, I.; Raines, A.; Story, B.; Priola, S.A. Susceptibility of common fibroblast cell lines to transmissible spongiform encephalopathy agents. J. Infect. Dis. 2004, 189, 431–439. [Google Scholar] [CrossRef]
- Arjona, A.; Simarro, L.; Islinger, F.; Nishida, N.; Manuelidis, L. Two creutzfeldt-jakob disease agents reproduce prion protein-independent identities in cell cultures. Proc. Natl. Acad. Sci. USA 2004, 101, 8768–8773. [Google Scholar] [CrossRef]
- Vilette, D.; Andreoletti, O.; Archer, F.; Madelaine, M.F.; Vilotte, J.L.; Lehmann, S.; Laude, H. Ex vivo propagation of infectious sheep scrapie agent in heterologous epithelial cells expressing ovine prion protein. Proc. Natl. Acad. Sci. USA 2001, 98, 4055–4059. [Google Scholar] [CrossRef]
- Bian, J.; Napier, D.; Khaychuck, V.; Angers, R.; Graham, C.; Telling, G. Cell-based quantification of chronic wasting disease prions. J. Virol. 2010, 84, 8322–8326. [Google Scholar] [CrossRef]
- Tark, D.; Kim, H.; Neale, M.H.; Kim, M.; Sohn, H.; Lee, Y.; Cho, I.; Joo, Y.; Windl, O. Generation of a persistently infected mdbk cell line with natural bovine spongiform encephalopathy (bse). PLoS ONE 2015, 10, e0115939. [Google Scholar] [CrossRef]
- Lawson, V.A.; Vella, L.J.; Stewart, J.D.; Sharples, R.A.; Klemm, H.; Machalek, D.M.; Masters, C.L.; Cappai, R.; Collins, S.J.; Hill, A.F. Mouse-adapted sporadic human creutzfeldt-jakob disease prions propagate in cell culture. Int. J. Biochem. Cell Biol. 2008, 40, 2793–2801. [Google Scholar] [CrossRef]
- Hannaoui, S.; Gougerot, A.; Privat, N.; Levavasseur, E.; Bizat, N.; Hauw, J.J.; Brandel, J.P.; Haik, S. Cycline efficacy on the propagation of human prions in primary cultured neurons is strain-specific. J. Infect. Dis. 2014, 209, 1144–1148. [Google Scholar] [CrossRef]
- Krejciova, Z.; Alibhai, J.; Zhao, C.; Krencik, R.; Rzechorzek, N.M.; Ullian, E.M.; Manson, J.; Ironside, J.W.; Head, M.W.; Chandran, S. Human stem cell-derived astrocytes replicate human prions in a prnp genotype-dependent manner. J. Exp. Med. 2017, 214, 3481–3495. [Google Scholar] [CrossRef]
- Vorberg, I.; Priola, S.A. Molecular basis of scrapie strain glycoform variation. J. Biol. Chem. 2002, 277, 36775–36781. [Google Scholar] [CrossRef]
- Choi, Y.P.; Priola, S.A. A specific population of abnormal prion protein aggregates is preferentially taken up by cells and disaggregated in a strain-dependent manner. J. Virol. 2013, 87, 11552–11561. [Google Scholar] [CrossRef]
- Supattapone, S. Synthesis of high titer infectious prions with cofactor molecules. J. Biol. Chem. 2014, 289, 19850–19854. [Google Scholar] [CrossRef]
- Klohn, P.C.; Stoltze, L.; Flechsig, E.; Enari, M.; Weissmann, C. A quantitative, highly sensitive cell-based infectivity assay for mouse scrapie prions. Proc. Natl. Acad. Sci. USA 2003, 100, 11666–11671. [Google Scholar] [CrossRef]
- Herva, M.E.; Weissman, C. Cell-specific susceptibility to prion strains is a property of the intact cell. Prion 2012, 6, 371–374. [Google Scholar] [CrossRef]
- Marbiah, M.M.; Harvey, A.; West, B.T.; Louzolo, A.; Banerjee, P.; Alden, J.; Grigoriadis, A.; Hummerich, H.; Kan, H.M.; Cai, Y.; et al. Identification of a gene regulatory network associated with prion replication. EMBO J. 2014, 33, 1527–1547. [Google Scholar] [CrossRef]
- Race, R.E.; Fadness, L.H.; Chesebro, B. Characterization of scrapie infection in mouse neuroblastoma cells. J. Gen. Virol. 1987, 68 Pt 5, 1391–1399. [Google Scholar] [CrossRef]
- Bosque, P.J.; Prusiner, S.B. Cultured cell sublines highly susceptible to prion infection. J. Virol. 2000, 74, 4377–4386. [Google Scholar] [CrossRef]
- Nishida, N.; Harris, D.A.; Vilette, D.; Laude, H.; Frobert, Y.; Grassi, J.; Casanova, D.; Milhavet, O.; Lehmann, S. Successful transmission of three mouse-adapted scrapie strains to murine neuroblastoma cell lines overexpressing wild-type mouse prion protein. J. Virol. 2000, 74, 320–325. [Google Scholar] [CrossRef]
- Julius, C.; Hutter, G.; Wagner, U.; Seeger, H.; Kana, V.; Kranich, J.; Klohn, P.C.; Weissmann, C.; Miele, G.; Aguzzi, A. Transcriptional stability of cultured cells upon prion infection. J. Mol. Biol. 2008, 375, 1222–1233. [Google Scholar] [CrossRef]
- Magalhaes, A.C.; Baron, G.S.; Lee, K.S.; Steele-Mortimer, O.; Dorward, D.; Prado, M.A.; Caughey, B. Uptake and neuritic transport of scrapie prion protein coincident with infection of neuronal cells. J. Neurosci. 2005, 25, 5207–5216. [Google Scholar] [CrossRef]
- Baron, G.S.; Magalhaes, A.C.; Prado, M.A.; Caughey, B. Mouse-adapted scrapie infection of sn56 cells: Greater efficiency with microsome-associated versus purified prp-res. J. Virol. 2006, 80, 2106–2117. [Google Scholar] [CrossRef]
- Piccardo, P.; Cervenakova, L.; Vasilyeva, I.; Yakovleva, O.; Bacik, I.; Cervenak, J.; McKenzie, C.; Kurillova, L.; Gregori, L.; Pomeroy, K.; et al. Candidate cell substrates, vaccine production, and transmissible spongiform encephalopathies. Emerg. Infect. Dis. 2011, 17, 2262–2269. [Google Scholar] [CrossRef]
- Vella, L.J.; Sharples, R.A.; Lawson, V.A.; Masters, C.L.; Cappai, R.; Hill, A.F. Packaging of prions into exosomes is associated with a novel pathway of prp processing. J. Pathol. 2007, 211, 582–590. [Google Scholar] [CrossRef]
- Hannaoui, S.; Maatouk, L.; Privat, N.; Levavasseur, E.; Faucheux, B.A.; Haik, S. Prion propagation and toxicity occur in vitro with two-phase kinetics specific to strain and neuronal type. J. Virol. 2013, 87, 2535–2548. [Google Scholar] [CrossRef]
- Cronier, S.; Beringue, V.; Bellon, A.; Peyrin, J.M.; Laude, H. Prion strain- and species-dependent effects of antiprion molecules in primary neuronal cultures. J. Virol. 2007, 81, 13794–13800. [Google Scholar] [CrossRef]
- Vey, M.; Pilkuhn, S.; Wille, H.; Nixon, R.; DeArmond, S.J.; Smart, E.J.; Anderson, R.G.; Taraboulos, A.; Prusiner, S.B. Subcellular colocalization of the cellular and scrapie prion proteins in caveolae-like membranous domains. Proc. Natl. Acad. Sci. USA 1996, 93, 14945–14949. [Google Scholar] [CrossRef]
- Kaneko, K.; Vey, M.; Scott, M.; Pilkuhn, S.; Cohen, F.E.; Prusiner, S.B. Cooh-terminal sequence of the cellular prion protein directs subcellular trafficking and controls conversion into the scrapie isoform. Proc. Natl. Acad. Sci. USA 1997, 94, 2333–2338. [Google Scholar] [CrossRef]
- Langhorst, M.F.; Reuter, A.; Jaeger, F.A.; Wippich, F.M.; Luxenhofer, G.; Plattner, H.; Stuermer, C.A. Trafficking of the microdomain scaffolding protein reggie-1/flotillin-2. Eur. J. Cell Biol. 2008, 87, 211–226. [Google Scholar] [CrossRef]
- Shyng, S.L.; Moulder, K.L.; Lesko, A.; Harris, D.A. The n-terminal domain of a glycolipid-anchored prion protein is essential for its endocytosis via clathrin-coated pits. J. Biol. Chem. 1995, 270, 14793–14800. [Google Scholar] [CrossRef]
- Sunyach, C.; Jen, A.; Deng, J.; Fitzgerald, K.T.; Frobert, Y.; Grassi, J.; McCaffrey, M.W.; Morris, R. The mechanism of internalization of glycosylphosphatidylinositol-anchored prion protein. EMBO J. 2003, 22, 3591–3601. [Google Scholar] [CrossRef]
- Sarnataro, D.; Caputo, A.; Casanova, P.; Puri, C.; Paladino, S.; Tivodar, S.S.; Campana, V.; Tacchetti, C.; Zurzolo, C. Lipid rafts and clathrin cooperate in the internalization of prp in epithelial frt cells. PLoS ONE 2009, 4, e5829. [Google Scholar] [CrossRef]
- Fehlinger, A.; Wolf, H.; Hossinger, A.; Duernberger, Y.; Pleschka, C.; Riemschoss, K.; Liu, S.; Bester, R.; Paulsen, L.; Priola, S.A.; et al. Prion strains depend on different endocytic routes for productive infection. Sci. Rep. 2017, 7, 6923. [Google Scholar] [CrossRef]
- Fivaz, M.; Vilbois, F.; Thurnheer, S.; Pasquali, C.; Abrami, L.; Bickel, P.E.; Parton, R.G.; van der Goot, F.G. Differential sorting and fate of endocytosed gpi-anchored proteins. EMBO J. 2002, 21, 3989–4000. [Google Scholar] [CrossRef]
- Prado, M.A.; Alves-Silva, J.; Magalhaes, A.C.; Prado, V.F.; Linden, R.; Martins, V.R.; Brentani, R.R. Prpc on the road: Trafficking of the cellular prion protein. J. Neurochem. 2004, 88, 769–781. [Google Scholar] [CrossRef]
- Jovic, M.; Sharma, M.; Rahajeng, J.; Caplan, S. The early endosome: A busy sorting station for proteins at the crossroads. Histol. Histopathol. 2010, 25, 99–112. [Google Scholar]
- Cullen, P.J.; Steinberg, F. To degrade or not to degrade: Mechanisms and significance of endocytic recycling. Nat. Rev. Mol. Cell Biol. 2018, 19, 679–696. [Google Scholar] [CrossRef]
- Stenmark, H. Rab gtpases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Johannes, L.; Popoff, V. Tracing the retrograde route in protein trafficking. Cell 2008, 135, 1175–1187. [Google Scholar] [CrossRef]
- Matsudaira, T.; Niki, T.; Taguchi, T.; Arai, H. Transport of the cholera toxin b-subunit from recycling endosomes to the golgi requires clathrin and ap-1. J. Cell Sci. 2015, 128, 3131–3142. [Google Scholar] [CrossRef]
- McNally, K.E.; Cullen, P.J. Endosomal retrieval of cargo: Retromer is not alone. Trends. Cell Biol. 2018, 28, 807–822. [Google Scholar] [CrossRef]
- Hurley, J.H.; Emr, S.D. The escrt complexes: Structure and mechanism of a membrane-trafficking network. Annu. Rev. Biophys. Biomol. Struct. 2006, 35, 277–298. [Google Scholar] [CrossRef]
- Taylor, D.R.; Hooper, N.M. The prion protein and lipid rafts. Mol. Membr. Biol. 2006, 23, 89–99. [Google Scholar] [CrossRef]
- Pimpinelli, F.; Lehmann, S.; Maridonneau-Parini, I. The scrapie prion protein is present in flotillin-1-positive vesicles in central- but not peripheral-derived neuronal cell lines. Eur. J. Neurosci. 2005, 21, 2063–2072. [Google Scholar] [CrossRef]
- Marijanovic, Z.; Caputo, A.; Campana, V.; Zurzolo, C. Identification of an intracellular site of prion conversion. PLoS Pathog. 2009, 5, e1000426. [Google Scholar] [CrossRef]
- Shakya, S.; Sharma, P.; Bhatt, A.M.; Jani, R.A.; Delevoye, C.; Setty, S.R. Rab22a recruits bloc-1 and bloc-2 to promote the biogenesis of recycling endosomes. EMBO Rep. 2018, 19. [Google Scholar] [CrossRef]
- Caughey, B.; Raymond, G.J. The scrapie-associated form of prp is made from a cell surface precursor that is both protease- and phospholipase-sensitive. J. Biol. Chem. 1991, 266, 18217–18223. [Google Scholar]
- Caughey, B.; Raymond, G.J.; Ernst, D.; Race, R.E. N-terminal truncation of the scrapie-associated form of prp by lysosomal protease(s): Implications regarding the site of conversion of prp to the protease-resistant state. J. Virol. 1991, 65, 6597–6603. [Google Scholar]
- Gilch, S.; Bach, C.; Lutzny, G.; Vorberg, I.; Schatzl, H.M. Inhibition of cholesterol recycling impairs cellular prp(sc) propagation. Cell Mol. Life. Sci. 2009, 66, 3979–3991. [Google Scholar] [CrossRef]
- Taraboulos, A.; Serban, D.; Prusiner, S.B. Scrapie prion proteins accumulate in the cytoplasm of persistently infected cultured cells. J. Cell Biol. 1990, 110, 2117–2132. [Google Scholar] [CrossRef]
- Marzo, L.; Marijanovic, Z.; Browman, D.; Chamoun, Z.; Caputo, A.; Zurzolo, C. 4-hydroxytamoxifen leads to prpsc clearance by conveying both prpc and prpsc to lysosomes independently of autophagy. J. Cell Sci. 2013, 126, 1345–1354. [Google Scholar] [CrossRef]
- MacDonald, L.; Baldini, G.; Storrie, B. Does super-resolution fluorescence microscopy obsolete previous microscopic approaches to protein co-localization? Methods. Mol. Biol. 2015, 1270, 255–275. [Google Scholar]
- Jongsma, M.L.; Berlin, I.; Wijdeven, R.H.; Janssen, L.; Janssen, G.M.; Garstka, M.A.; Janssen, H.; Mensink, M.; van Veelen, P.A.; Spaapen, R.M.; et al. An er-associated pathway defines endosomal architecture for controlled cargo transport. Cell 2016, 166, 152–166. [Google Scholar] [CrossRef]
- Bonifacino, J.S.; Neefjes, J. Moving and positioning the endolysosomal system. Curr. Opin. Cell Biol. 2017, 47, 1–8. [Google Scholar] [CrossRef]
- Yim, Y.I.; Park, B.C.; Yadavalli, R.; Zhao, X.; Eisenberg, E.; Greene, L.E. The multivesicular body is the major internal site of prion conversion. J. Cell Sci. 2015, 128, 1434–1443. [Google Scholar] [CrossRef]
- Tanaka, M.; Fujiwara, A.; Suzuki, A.; Yamasaki, T.; Hasebe, R.; Masujin, K.; Horiuchi, M. Comparison of abnormal isoform of prion protein in prion-infected cell lines and primary-cultured neurons by prpsc-specific immunostaining. J. Gen. Virol. 2016, 97, 2030–2042. [Google Scholar] [CrossRef]
- Ertmer, A.; Gilch, S.; Yun, S.W.; Flechsig, E.; Klebl, B.; Stein-Gerlach, M.; Klein, M.A.; Schatzl, H.M. The tyrosine kinase inhibitor sti571 induces cellular clearance of prpsc in prion-infected cells. J. Biol. Chem. 2004, 279, 41918–41927. [Google Scholar] [CrossRef]
- Taraboulos, A.; Rogers, M.; Borchelt, D.R.; McKinley, M.P.; Scott, M.; Serban, D.; Prusiner, S.B. Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc. Natl. Acad. Sci. USA 1990, 87, 8262–8266. [Google Scholar] [CrossRef]
- Hagiwara, K.; Nakamura, Y.; Nishijima, M.; Yamakawa, Y. Prevention of prion propagation by dehydrocholesterol reductase inhibitors in cultured cells and a therapeutic trial in mice. Biol. Pharm. Bull. 2007, 30, 835–838. [Google Scholar] [CrossRef]
- Okemoto-Nakamura, Y.; Yamakawa, Y.; Hanada, K.; Tanaka, K.; Miura, M.; Tanida, I.; Nishijima, M.; Hagiwara, K. Synthetic fibril peptide promotes clearance of scrapie prion protein by lysosomal degradation. Microbiol. Immunol. 2008, 52, 357–365. [Google Scholar] [CrossRef]
- Rouvinski, A.; Karniely, S.; Kounin, M.; Moussa, S.; Goldberg, M.D.; Warburg, G.; Lyakhovetsky, R.; Papy-Garcia, D.; Kutzsche, J.; Korth, C.; et al. Live imaging of prions reveals nascent prpsc in cell-surface, raft-associated amyloid strings and webs. J. Cell Biol. 2014, 204, 423–441. [Google Scholar] [CrossRef]
- Uchiyama, K.; Muramatsu, N.; Yano, M.; Usui, T.; Miyata, H.; Sakaguchi, S. Prions disturb post-golgi trafficking of membrane proteins. Nat. Commun. 2013, 4, 1846. [Google Scholar] [CrossRef] [PubMed]
- Veith, N.M.; Plattner, H.; Stuermer, C.A.; Schulz-Schaeffer, W.J.; Burkle, A. Immunolocalisation of prpsc in scrapie-infected n2a mouse neuroblastoma cells by light and electron microscopy. Eur. J. Cell Biol. 2009, 88, 45–63. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, T.; Suzuki, A.; Shimizu, T.; Watarai, M.; Hasebe, R.; Horiuchi, M. Characterization of intracellular localization of prp(sc) in prion-infected cells using a mab that recognizes the region consisting of aa 119–127 of mouse prp. J. Gen. Virol. 2012, 93, 668–680. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, T.; Suzuki, A.; Hasebe, R.; Horiuchi, M. Retrograde transport by clathrin-coated vesicles is involved in intracellular transport of prp(sc) in persistently prion-infected cells. Sci. Rep. 2018, 8, 12241. [Google Scholar] [CrossRef]
- Yamasaki, T.; Suzuki, A.; Hasebe, R.; Horiuchi, M. Comparison of the anti-prion mechanism of four different anti-prion compounds, anti-prp monoclonal antibody 44b1, pentosan polysulfate, chlorpromazine, and u18666a, in prion-infected mouse neuroblastoma cells. PLoS ONE 2014, 9, e106516. [Google Scholar] [CrossRef]
- Greil, C.S.; Vorberg, I.M.; Ward, A.E.; Meade-White, K.D.; Harris, D.A.; Priola, S.A. Acute cellular uptake of abnormal prion protein is cell type and scrapie-strain independent. Virology 2008, 379, 284–293. [Google Scholar] [CrossRef]
- Goold, R.; Rabbanian, S.; Sutton, L.; Andre, R.; Arora, P.; Moonga, J.; Clarke, A.R.; Schiavo, G.; Jat, P.; Collinge, J.; et al. Rapid cell-surface prion protein conversion revealed using a novel cell system. Nat. Commun. 2011, 2, 281. [Google Scholar] [CrossRef]
- Leucht, C.; Simoneau, S.; Rey, C.; Vana, K.; Rieger, R.; Lasmezas, C.I.; Weiss, S. The 37 kda/67 kda laminin receptor is required for prp(sc) propagation in scrapie-infected neuronal cells. EMBO Rep. 2003, 4, 290–295. [Google Scholar] [CrossRef]
- Horonchik, L.; Tzaban, S.; Ben-Zaken, O.; Yedidia, Y.; Rouvinski, A.; Papy-Garcia, D.; Barritault, D.; Vlodavsky, I.; Taraboulos, A. Heparan sulfate is a cellular receptor for purified infectious prions. J. Biol. Chem. 2005, 280, 17062–17067. [Google Scholar] [CrossRef] [PubMed]
- Jen, A.; Parkyn, C.J.; Mootoosamy, R.C.; Ford, M.J.; Warley, A.; Liu, Q.; Bu, G.; Baskakov, I.V.; Moestrup, S.; McGuinness, L.; et al. Neuronal low-density lipoprotein receptor-related protein 1 binds and endocytoses prion fibrils via receptor cluster 4. J. Cell Sci. 2010, 123, 246–255. [Google Scholar] [CrossRef]
- Wolf, H.; Grassmann, A.; Bester, R.; Hossinger, A.; Mohl, C.; Paulsen, L.; Groschup, M.H.; Schatzl, H.; Vorberg, I. Modulation of glycosaminoglycans affects prpsc metabolism but does not block prpsc uptake. J. Virol. 2015, 89, 9853–9864. [Google Scholar] [CrossRef]
- Tixador, P.; Herzog, L.; Reine, F.; Jaumain, E.; Chapuis, J.; Le Dur, A.; Laude, H.; Beringue, V. The physical relationship between infectivity and prion protein aggregates is strain-dependent. PLoS Pathog. 2010, 6, e1000859. [Google Scholar] [CrossRef]
- Laferriere, F.; Tixador, P.; Moudjou, M.; Chapuis, J.; Sibille, P.; Herzog, L.; Reine, F.; Jaumain, E.; Laude, H.; Rezaei, H.; et al. Quaternary structure of pathological prion protein as a determining factor of strain-specific prion replication dynamics. PLoS Pathog. 2013, 9, e1003702. [Google Scholar] [CrossRef]
- Bett, C.; Lawrence, J.; Kurt, T.D.; Orru, C.; Aguilar-Calvo, P.; Kincaid, A.E.; Surewicz, W.K.; Caughey, B.; Wu, C.; Sigurdson, C.J. Enhanced neuroinvasion by smaller, soluble prions. Acta. Neuropathol. Commun. 2017, 5, 32. [Google Scholar] [CrossRef]
- Luhr, K.M.; Nordstrom, E.K.; Low, P.; Ljunggren, H.G.; Taraboulos, A.; Kristensson, K. Scrapie protein degradation by cysteine proteases in cd11c+ dendritic cells and gt1–1 neuronal cells. J. Virol. 2004, 78, 4776–4782. [Google Scholar] [CrossRef]
- Gilch, S.; Schmitz, F.; Aguib, Y.; Kehler, C.; Bulow, S.; Bauer, S.; Kremmer, E.; Schatzl, H.M. Cpg and lps can interfere negatively with prion clearance in macrophage and microglial cells. FEBS J. 2007, 274, 5834–5844. [Google Scholar] [CrossRef]
- Krejciova, Z.; Pells, S.; Cancellotti, E.; Freile, P.; Bishop, M.; Samuel, K.; Barclay, G.R.; Ironside, J.W.; Manson, J.C.; Turner, M.L.; et al. Human embryonic stem cells rapidly take up and then clear exogenous human and animal prions in vitro. J. Pathol. 2011, 223, 635–645. [Google Scholar] [CrossRef]
- Luhr, K.M.; Nordstrom, E.K.; Low, P.; Kristensson, K. Cathepsin b and l are involved in degradation of prions in gt1–1 neuronal cells. Neuroreport 2004, 15, 1663–1667. [Google Scholar] [CrossRef]
- Goold, R.; McKinnon, C.; Rabbanian, S.; Collinge, J.; Schiavo, G.; Tabrizi, S.J. Alternative fates of newly formed prpsc upon prion conversion on the plasma membrane. J. Cell Sci. 2013, 126, 3552–3562. [Google Scholar] [CrossRef]
- Vorberg, I.; Raines, A.; Priola, S.A. Acute formation of protease-resistant prion protein does not always lead to persistent scrapie infection in vitro. J. Biol. Chem. 2004, 279, 29218–29225. [Google Scholar] [CrossRef]
- Yamasaki, T.; Baron, G.S.; Suzuki, A.; Hasebe, R.; Horiuchi, M. Characterization of intracellular dynamics of inoculated prp-res and newly generated prp(sc) during early stage prion infection in neuro2a cells. Virology 2014, 450–451, 324–335. [Google Scholar] [CrossRef]
- Naslavsky, N.; Stein, R.; Yanai, A.; Friedlander, G.; Taraboulos, A. Characterization of detergent-insoluble complexes containing the cellular prion protein and its scrapie isoform. J. Biol. Chem. 1997, 272, 6324–6331. [Google Scholar] [CrossRef]
- Magadan, J.G.; Barbieri, M.A.; Mesa, R.; Stahl, P.D.; Mayorga, L.S. Rab22a regulates the sorting of transferrin to recycling endosomes. Mol. Cell Biol. 2006, 26, 2595–2614. [Google Scholar] [CrossRef]
- Lippincott-Schwartz, J.; Yuan, L.; Tipper, C.; Amherdt, M.; Orci, L.; Klausner, R.D. Brefeldin a’s effects on endosomes, lysosomes, and the tgn suggest a general mechanism for regulating organelle structure and membrane traffic. Cell 1991, 67, 601–616. [Google Scholar] [CrossRef]
- Puri, C.; Vicinanza, M.; Ashkenazi, A.; Gratian, M.J.; Zhang, Q.; Bento, C.F.; Renna, M.; Menzies, F.M.; Rubinsztein, D.C. The rab11a-positive compartment is a primary platform for autophagosome assembly mediated by wipi2 recognition of pi3p-rab11a. Dev. Cell 2018, 45, 114–131.e8. [Google Scholar] [CrossRef]
- Cui, Y.; Carosi, J.M.; Yang, Z.; Ariotti, N.; Kerr, M.C.; Parton, R.G.; Sargeant, T.J.; Teasdale, R.D. Retromer has a selective function in cargo sorting via endosome transport carriers. J. Cell Biol. 2019, 218, 615–631. [Google Scholar] [CrossRef]
- Press, B.; Feng, Y.; Hoflack, B.; Wandinger-Ness, A. Mutant rab7 causes the accumulation of cathepsin d and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 1998, 140, 1075–1089. [Google Scholar] [CrossRef]
- Vilette, D.; Laulagnier, K.; Huor, A.; Alais, S.; Simoes, S.; Maryse, R.; Provansal, M.; Lehmann, S.; Andreoletti, O.; Schaeffer, L.; et al. Efficient inhibition of infectious prions multiplication and release by targeting the exosomal pathway. Cell Mol. Life. Sci. 2015, 72, 4409–4427. [Google Scholar] [CrossRef]
- Vitelli, R.; Santillo, M.; Lattero, D.; Chiariello, M.; Bifulco, M.; Bruni, C.B.; Bucci, C. Role of the small gtpase rab7 in the late endocytic pathway. J. Biol. Chem. 1997, 272, 4391–4397. [Google Scholar] [CrossRef]
- Bucci, C.; Thomsen, P.; Nicoziani, P.; McCarthy, J.; van Deurs, B. Rab7: A key to lysosome biogenesis. Mol. Biol. Cell 2000, 11, 467–480. [Google Scholar] [CrossRef]
- Ikonen, E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol 2008, 9, 125–138. [Google Scholar] [CrossRef]
- Rink, J.; Ghigo, E.; Kalaidzidis, Y.; Zerial, M. Rab conversion as a mechanism of progression from early to late endosomes. Cell 2005, 122, 735–749. [Google Scholar] [CrossRef]
- Arighi, C.N.; Hartnell, L.M.; Aguilar, R.C.; Haft, C.R.; Bonifacino, J.S. Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J. Cell Biol. 2004, 165, 123–133. [Google Scholar] [CrossRef]
- Rojas, R.; Kametaka, S.; Haft, C.R.; Bonifacino, J.S. Interchangeable but essential functions of snx1 and snx2 in the association of retromer with endosomes and the trafficking of mannose 6-phosphate receptors. Mol. Cell Biol. 2007, 27, 1112–1124. [Google Scholar] [CrossRef] [PubMed]
- Girard, E.; Chmiest, D.; Fournier, N.; Johannes, L.; Paul, J.L.; Vedie, B.; Lamaze, C. Rab7 is functionally required for selective cargo sorting at the early endosome. Traffic 2014, 15, 309–326. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, E.; Urbe, S.; Clague, M.J. Usp8 controls the trafficking and sorting of lysosomal enzymes. Traffic 2014, 15, 879–888. [Google Scholar] [CrossRef]
- Piper, R.C.; Katzmann, D.J. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 2007, 23, 519–547. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, E.M.; Schultz, S.W.; Schink, K.O.; Pedersen, N.M.; Nahse, V.; Carlson, A.; Brech, A.; Stenmark, H.; Raiborg, C. Concerted escrt and clathrin recruitment waves define the timing and morphology of intraluminal vesicle formation. Nat. Commun. 2018, 9, 2932. [Google Scholar] [CrossRef]
- Beranger, F.; Mange, A.; Goud, B.; Lehmann, S. Stimulation of prp(c) retrograde transport toward the endoplasmic reticulum increases accumulation of prp(sc) in prion-infected cells. J. Biol. Chem. 2002, 277, 38972–38977. [Google Scholar] [CrossRef]
- Vilette, D.; Courte, J.; Peyrin, J.M.; Coudert, L.; Schaeffer, L.; Andreoletti, O.; Leblanc, P. Cellular mechanisms responsible for cell-to-cell spreading of prions. Cell Mol. Life. Sci. 2018, 75, 2557–2574. [Google Scholar] [CrossRef]
| Cell Line | Origin | Mouse-Adapted TSE Strain | References | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scrapie | Other TSE | |||||||||||
| Ch./RML | 79A | 139A | 22L | ME7 | 87V | Fu-1 b | M1000 c | 301C d | ||||
| N2a | Mouse neuroblastoma | [33,34,57,58] | ||||||||||
| SMB | Mouse brain cells | [39] | ||||||||||
| GT-1 | Mouse hypothalamic neurons | [40,44,59] | ||||||||||
| CAD5 | Mouse catecholaminergic neurons | [41,60] | ||||||||||
| SN56 | Mouse septal neurons | [61,62] | ||||||||||
| L929 | Mouse fibroblasts | [41,43,63] | ||||||||||
| RK13 | Rabbit kidney epithelial (moPrP) a | [42,64] | ||||||||||
| Cell Line | Clone | Strain | PM | Flotillin-1 | Clathrin | Caveolin-1 | Giantin | TGN38 | CI-M6PR | Rab4 | Rab5 | EEA-1 | Tfn/Tfr | Rab11 | Rab7 | Rab9 | Lamp-1 | Limp-2 | LYAAT-1 | LBPA | Cathepsin D | LC3 | Detection | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N2a | H6 | 22L | + c | + c | + c | IF | [103] b | |||||||||||||||||
| H6 | 22L | + d | + c | + | EM | [103] | ||||||||||||||||||
| 22L | + b | - | + | +++ | + | IF | [85] e | |||||||||||||||||
| 3 | 22L | + | +++ | (+) | + | + | + | + | + | ++ | IF | [104] | ||||||||||||
| 3 | 22L | ++ | ++ | +++ | IF | [106] | ||||||||||||||||||
| C24 a | 22L | - | + | + | + | +++ | ++ | ++ | IF | [102] c | ||||||||||||||
| 22L | + | + | ++ | IF | [95] c | |||||||||||||||||||
| 3 | 22L, 72 h | (+) | +++ | +++ | (+) | IF | [106] | |||||||||||||||||
| a | Ch./RML | - | - | - | +++ | - | IF | [84] c | ||||||||||||||||
| Ch./RML | ++ | +++ | + | IF | [100] f | |||||||||||||||||||
| C24 a | Ch./RML | + | + | ++ | +++ | ++ | ++ | IF | [102] c | |||||||||||||||
| Ch./RML | ++ | IF | [101] c | |||||||||||||||||||||
| GT-1 | 22L | ++ | IF | [101] c | ||||||||||||||||||||
| 7 | 22L | (+) | (+) | + | + | ++ | ++ | ++ | + | + | IF | [105] c | ||||||||||||
| GT1 | 7 | Ch./RML | ++ | - | +++ | - | IF | [84] c | ||||||||||||||||
| Ch./RML | + | - | + | ++ | + | IF | [85] e | |||||||||||||||||
| Ch./RML | + | + | IF | [91] e | ||||||||||||||||||||
| Ch./RML | ++ | IF | [101] c | |||||||||||||||||||||
| SMB | 139A | - | + | + | ++ | IF | [95] c |
| Cell Line | Strain | Manipulation | PrPSc Colocalization | Total PrPSc | Observed Effect on Endosomes, Control Cargo | Ref. |
|---|---|---|---|---|---|---|
| N2a | Ch./RML | Rab4 S22N | n.d. * | Increase | n.d. | [140] |
| Rab6 Q72L | n.d. * | Increase | n.d. | [140] | ||
| Rab9 WT | n.d. | Reduction | n.d. | [89] | ||
| 22L | Rab4 S22N | n.d. * | Increase | n.d. | [140] | |
| Rab4 N121I | n.d. | No effect | n.d. | [85] | ||
| Rab6 Q72L | n.d. * | Increase | n.d. | [140] | ||
| Rab22a WT | n.d. | No effect | No inhibition of transport Tfn from EE to ERC | [85] | ||
| siRNA Hrs | n.d. | Reduction | n.d. | [95,141] | ||
| siRNA Tsg101 | n.d. | Reduction | n.d. | [95] | ||
| siRNA Clint-1 | Redistribution PrPSc from Tfn-positive vesicles to Lamp-1-positive vesicles | No effect | n.d. | [105] | ||
| siRNA Ap1g1 | n.d. | Increase | n.d. | [105] | ||
| GT1 | Ch./RML | Rab4 N121I | n.d. | No effect | n.d. | [85] |
| Rab11 S25N | Higher PrPSc levels in cellular compartment positive for Tfn Rare PrPSc in LBPA-positive endosomes | Increase | No inhibition of transport Tfn from EE to ERC Impaired recycling Tfn from ERC to PM Strong colocalization Tfn with Rab11 S25N-GFP | [85] | ||
| Rab22a WT | PrP enriched in EEA-1-positive endosomes (no denaturation step) No PrPSc in GFP-Rab22a-positive cells after 6 days | Reduction | Enlarged EEA-1/Rab22a-double positive endosomes Normal internalization of Tfn Tfn retained in GFP-Rab22a-positive endosomes | [85] | ||
| siRNA Alix | Strong colocalization Tfn and PrPSc | Increase | Less LBPA-positive endosomes Less lysosensor-positive endosomes | [85] | ||
| SMB | 139A | Rab5 Q71L | Colocalization with EEA-1/Lamp-1 double-positive endosomes | Reduction | EEA-1/Lamp-1 double-positive endosomes | [95] |
| Rab7a T22N | Colocalization with enlarged Lamp-1-positive endosomes | Reduction | Enlarged Lamp-1/CI-M6PR double-positive endosomes | |||
| Rab7a WT | No change in PrPSc distribution | No effect | No enlarged Lamp-1-positive endosomes | |||
| Rab7a Q67L | No change in PrPSc distribution | No effect | No enlarged Lamp-1-positive endosomes | |||
| Rab22a Q64L | n.d. | Reduction | Enlarged EEA-1/Lamp-1-double-positive endosomes | |||
| Rab22a WT | n.d. | Reduction | EEA-1/Lamp-1 double-positive endosomes | |||
| siRNA Hrs | Initial colocalization with EEA-1/Lamp-1-positive endosomes | Reduction | EEA-1/Lamp-1 double-positive endosomes | |||
| siRNA Tsg101 | Initial colocalization with EEA-1/Lamp-1-positive endosomes | Reduction | EEA-1/Lamp-1 double-positive endosomes | |||
| siRNA AILIX | Unchanged colocalization with Lamp-1-positive endosomes | Increase | n.d. | |||
| siRNA SNX2 (retromer) | n.d. | Increase | n.d. | |||
| siRNA Vps26 (retromer) | Partial colocalization with enlarged Lamp-1/CI-M6PR endosomes | Increase | Enlarged Lamp-1/CI-M6PR double-positive endosomes |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vorberg, I.M. All the Same? The Secret Life of Prion Strains within Their Target Cells. Viruses 2019, 11, 334. https://doi.org/10.3390/v11040334
Vorberg IM. All the Same? The Secret Life of Prion Strains within Their Target Cells. Viruses. 2019; 11(4):334. https://doi.org/10.3390/v11040334
Chicago/Turabian StyleVorberg, Ina M. 2019. "All the Same? The Secret Life of Prion Strains within Their Target Cells" Viruses 11, no. 4: 334. https://doi.org/10.3390/v11040334
APA StyleVorberg, I. M. (2019). All the Same? The Secret Life of Prion Strains within Their Target Cells. Viruses, 11(4), 334. https://doi.org/10.3390/v11040334




