Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast
Abstract
1. Introduction
2. Prion Variants in Yeast
3. Requirement for Hsp104
4. Hsp70s
4.1. Ssa1-4 (Free Cytosolic Hsp70s)
4.2. Ssb1/2 (Ribosome-Associated Hsp70s)
5. Hsp40s (J-Domain Proteins or J-Proteins)
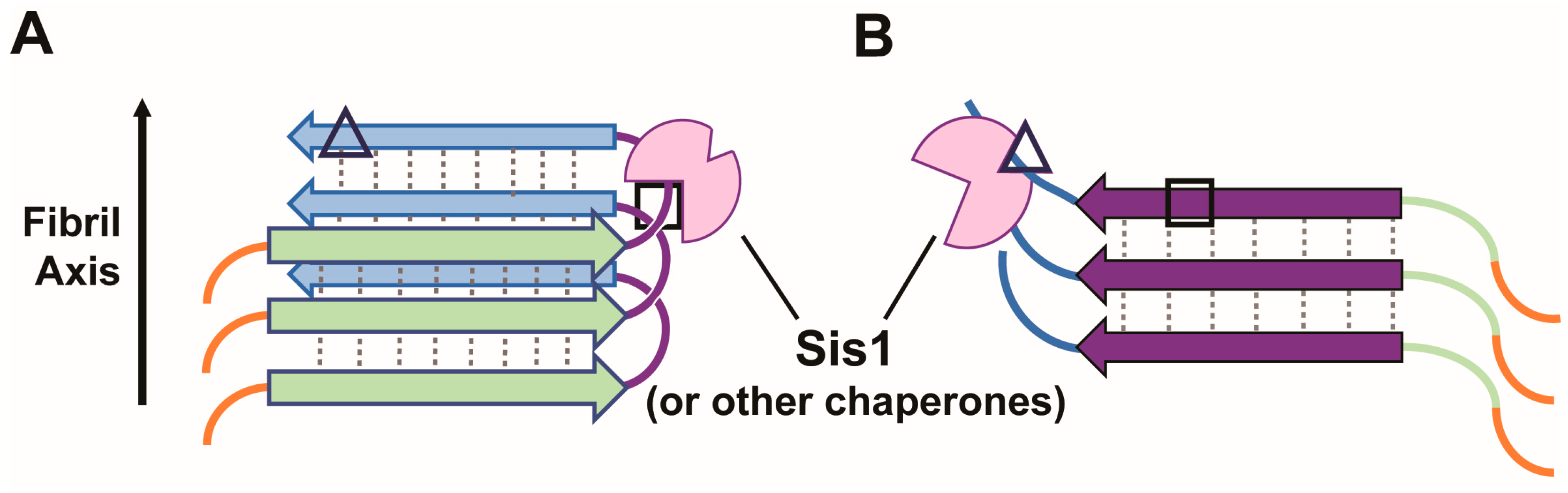
5.1. Sis1
5.1.1. Sis1 Deletions, Truncations, and Repression
5.1.2. Sis1 Orthologs in Other Species
5.1.3. Examples of Yeast Genetic Background Effects, Variant Switching, and Changes in Variant Dominance
5.2. Other Hsp40s: Ydj1, Apj1, and Swa2
6. Sse1 (Hsp110, Nucleotide Exchange Factor)
7. Hsp90 and its Cochaperones
8. Hsp104-Mediated Prion Elimination
9. Chaperone-Sorting Factors
10. Amyloid Variation is Bounded by Chaperone Activities and Other Systems
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wickner, R.B.; Edskes, H.K.; Shewmaker, F.; Nakayashiki, T. Prions of Fungi: Inherited Structures and Biological Roles. Nat. Rev. Microbiol. 2007, 5, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Liebman, S.W.; Chernoff, Y.O. Prions in Yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Satpute-Krishnan, P.; Langseth, S.X.; Serio, T.R. Hsp104-dependent remodeling of prion complexes mediates protein-only inheritance. PLoS Biol. 2007, 5, e24. [Google Scholar] [CrossRef] [PubMed]
- Higurashi, T.; Hines, J.K.; Sahi, C.; Aron, R.; Craig, E.A. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA 2008, 105, 16596–16601. [Google Scholar] [CrossRef] [PubMed]
- Tipton, K.A.; Verges, K.J.; Weissman, J.S. In Vivo Monitoring of the Prion Replication Cycle Reveals a Critical Role for Sis1 in Delivering Substrates to Hsp104. Mol. Cell 2008. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012, 198, 387–404. [Google Scholar] [CrossRef]
- Prusiner, S.B. Biology and Genetics of Prions Causing Neurodegeneration. Annu. Rev. Genet. 2013, 47, 601–623. [Google Scholar] [CrossRef]
- Safar, J.G. Molecular pathogenesis of sporadic prion diseases in man. Prion 2012, 6, 108–115. [Google Scholar] [CrossRef]
- Eisenberg, D.; Jucker, M. The Amyloid State of Proteins in Human Diseases. Cell 2012, 148, 1188–1203. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Chernoff, Y.O.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [PubMed]
- Wickner, R.B.; Edskes, H.K.; Kryndushkin, D.; McGlinchey, R.; Bateman, D.; Kelly, A. Prion diseases of yeast: Amyloid structure and biology. Semin. Cell Dev. Biol. 2011, 22, 469–475. [Google Scholar] [CrossRef]
- Hines, J.K.; Higurashi, T.; Srinivasan, M.; Craig, E.A. Influence of prion variant and yeast strain variation on prion-molecular chaperone requirements. Prion 2011, 5, 238–244. [Google Scholar] [CrossRef][Green Version]
- Stein, K.C.; True, H.L. Prion strains and amyloid polymorphism influence phenotypic variation. PLoS Pathog. 2014, 10, e1004328. [Google Scholar] [CrossRef]
- Toyama, B.H.; Kelly, M.J.S.; Gross, J.D.; Weissman, J.S. The structural basis of yeast prion strain variants. Nature 2007. [Google Scholar] [CrossRef]
- Frederick, K.K.; Debelouchina, G.T.; Kayatekin, C.; Dorminy, T.; Jacavone, A.C.; Griffin, R.G.; Lindquist, S. Distinct prion strains are defined by amyloid core structure and chaperone binding site dynamics. Chem. Biol. 2014, 21, 295–305. [Google Scholar] [CrossRef]
- Tanaka, M.; Collins, S.R.; Toyama, B.H.; Weissman, J.S. The physical basis of how prion conformations determine strain phenotypes. Nature 2006, 442, 585–589. [Google Scholar] [CrossRef]
- Silveira, J.R.; Raymond, G.J.; Hughson, A.G.; Race, R.E.; Sim, V.L.; Hayes, S.F.; Caughey, B. The most infectious prion protein particles. Nature 2005, 437, 257–261. [Google Scholar] [CrossRef]
- Legname, G.; Nguyen, H.-O.B.; Peretz, D.; Cohen, F.E.; DeArmond, S.J.; Prusiner, S.B. Continuum of prion protein structures enciphers a multitude of prion isolate-specified phenotypes. Proc. Natl. Acad. Sci. USA 2006, 103, 19105–19110. [Google Scholar] [CrossRef]
- Colby, D.W.; Giles, K.; Legname, G.; Wille, H.; Baskakov, I.V.; DeArmond, S.J.; Prusiner, S.B. Design and construction of diverse mammalian prion strains. Proc. Natl. Acad. Sci 2009, 106, 20417–20422. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Zhou, P.; Chernoff, Y.O.; Liebman, S.W. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 1997, 147, 507–519. [Google Scholar]
- Derkatch, I.L.; Bradley, M.E.; Hong, J.Y.; Liebman, S.W. Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Bradley, M.E.; Edskes, H.K.; Hong, J.Y.; Wickner, R.B.; Liebman, S.W. Interactions among prions and prion “‘strains’” in yeast. Proc. Natl. Acad. Sci. USA 2002, 99, 16392–16399. [Google Scholar] [CrossRef]
- Bradley, M.E.; Liebman, S.W. Destabilizing Interactions Among [PSI] and [PIN] Yeast Prion Variants. Genetics 2003, 165, 1675–1685. [Google Scholar]
- Stein, K.C.; True, H.L. Extensive diversity of prion strains is defined by differential chaperone interactions and distinct amyloidogenic regions. PLoS Genet. 2014, 10, e1004337. [Google Scholar] [CrossRef]
- Huang, V.J.; Stein, K.C.; True, H.L. Spontaneous variants of the [RNQ+] prion in yeast demonstrate the extensive conformational diversity possible with prion proteins. PLoS ONE 2013, 8, e79582. [Google Scholar] [CrossRef][Green Version]
- Schlumpberger, M.; Prusiner, S.B.; Herskowitz, I. Induction of Distinct [URE3] Yeast Prion Strains. Mol. Cell. Biol. 2001, 21, 7035–7046. [Google Scholar] [CrossRef]
- Brachmann, A.; Baxa, U.; Wickner, R.B. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005, 24, 3082–3092. [Google Scholar] [CrossRef]
- Edskes, H.K.; McCann, L.M.; Hebert, A.M.; Wickner, R.B. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics 2009, 181, 1159–1167. [Google Scholar] [CrossRef]
- McGlinchey, R.P.; Kryndushkin, D.; Wickner, R.B. Suicidal [PSI + ] is a lethal yeast prion. Proc. Natl. Acad. Sci. USA 2011, 108, 5337–5341. [Google Scholar] [CrossRef]
- Gates, S.N.; Yokom, A.L.; Lin, J.; Jackrel, M.E.; Rizo, A.N.; Kendsersky, N.M.; Buell, C.E.; Sweeny, E.A.; Mack, K.L.; Chuang, E.; et al. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science 2017, 357, 273–279. [Google Scholar] [CrossRef]
- Grimminger-Marquardt, V.; Lashuel, H.A. Structure and function of the molecular chaperone Hsp104 from yeast. Biopolymers 2010, 93, 252–276. [Google Scholar] [CrossRef]
- Parsell, D.A.; Kowal, A.S.; Singer, M.A.; Lindquist, S. Protein disaggregation mediated by heat-shock protein Hspl04. Nature 1994, 372, 475–478. [Google Scholar] [CrossRef]
- Sielaff, B.; Tsai, F.T.F. The M-domain controls Hsp104 protein remodeling activity in an Hsp70/Hsp40-dependent manner. J. Mol. Biol. 2010, 402, 30–37. [Google Scholar] [CrossRef]
- Tuite, M.F.; Mundy, C.R.; Cox, B.S. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 1981, 98, 691–711. [Google Scholar]
- Chernoff, Y.O.; Lindquist, S.L.; Ono, B.; Inge-Vechtomov, S.G.; Liebman, S.W. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science 1995, 268, 880–884. [Google Scholar] [CrossRef]
- Eaglestone, S.S.; Ruddock, L.W.; Cox, B.S.; Tuite, M.F. Guanidine hydrochloride blocks a critical step in the propagation of the prion-like determinant [PSI(+)] of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 240–244. [Google Scholar] [CrossRef]
- Jung, G.; Jones, G.; Masison, D.C. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc. Natl. Acad. Sci. USA 2002, 99, 9936–9941. [Google Scholar] [CrossRef]
- Grimminger, V.; Richter, K.; Imhof, A.; Buchner, J.; Walter, S. The Prion Curing Agent Guanidinium Chloride Specifically Inhibits ATP Hydrolysis by Hsp104. J. Biol. Chem. 2004, 279, 7378–7383. [Google Scholar] [CrossRef]
- Hines, J.K.; Li, X.; Du, Z.; Higurashi, T.; Li, L.; Craig, E.A. [SWI+], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in hsp70 chaperone system activity. PLoS Genet. 2011. [Google Scholar] [CrossRef]
- Harris, J.M.; Nguyen, P.P.; Patel, M.J.; Sporn, Z.A.; Hines, J.K. Functional Diversification of Hsp40: Distinct J-Protein Functional Requirements for Two Prions Allow for Chaperone-Dependent Prion Selection. PLoS Genet. 2014, 10, 1004510. [Google Scholar] [CrossRef][Green Version]
- Borchsenius, A.S.; Muïler, S.; Newnam, G.P.; Sergey, A.E.; Inge-Vechtomov, G.; Chernoff, Y.O. Prion variant maintained only at high levels of the Hsp104 disaggregase. Curr. Genet. 2006, 49, 21–29. [Google Scholar] [CrossRef]
- Dulle, J.E.; True, H.L. Low activity of select Hsp104 mutants is sufficient to propagate unstable prion variants. Prion 2013, 7, 394–403. [Google Scholar] [CrossRef][Green Version]
- Dulle, J.E.; Stein, K.C.; True, H.L. Regulation of the Hsp104 middle domain activity is critical for yeast prion propagation. PLoS ONE 2014, 9, e87521. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Craig, E.A. The Hsp70 chaperone machinery: J-proteins as drivers of functional specificity NIH Public Access. Nat. Rev. Mol. Cell Biol. 2010, 11, 579–592. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A. Hsp70 at the membrane: Driving protein translocation. BMC Biol. 2018, 16. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Andreasson, C.; Barducci, A.; Cheetham, M.E.; Cyr, D.; Emanuelsson, C.; Genevaux, P.; Gestwicki, J.E.; Goloubinoff, P.; Huerta-Cepas, J.; et al. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 2019, 24, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Boorstein, W.R.; Ziegelhoffer, T.; Craig, E.A. Molecular evolution of the HSP70 multigene family. J. Mol. Evol. 1994, 38, 1–17. [Google Scholar] [CrossRef]
- Jung, G.; Jones, G.; Wegrzyn, R.D.; Masison, D.C. A role for cytosolic hsp70 in yeast [PSI(+)] prion propagation and [PSI(+)] as a cellular stress. Genetics 2000, 156, 559–570. [Google Scholar]
- Masison, D.C.; Kirkland, P.A.; Sharma, D. Influence of Hsp70 and Its Regulators on Yeast Prion Propagation. Prion. 2009, 3, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Werner-Washburne, M.; Stone, D.E.; Craig, E.A. Complex Interactions among Members of an Essential Subfamily of hsp7O Genes in Saccharomyces Cerevisiae. Mol. Cell Biol. 1987, 7, 2568–2577. [Google Scholar] [CrossRef] [PubMed]
- Newnam, G.P.; Wegrzyn, R.D.; Lindquist, S.L.; Chernoff, Y.O. Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell Biol. 1999, 19, 1325–1333. [Google Scholar] [CrossRef]
- Allen, K.D.; Wegrzyn, R.D.; Chernova, T.A.; Müller, S.; Newnam, G.P.; Winslett, P.A.; Wittich, K.B.; Wilkinson, K.D.; Chernoff, Y.O. Hsp70 Chaperones as Modulators of Prion Life Cycle: Novel Effects of Ssa and Ssb on the Saccharomyces cerevisiae Prion [PSI]. Genetics 2005, 169, 1227–1242. [Google Scholar] [CrossRef]
- Mathur, V.; Hong, J.Y.; Liebman, S.W. Ssa1 overexpression and [PIN+] variants cure [PSI+] by dilution of aggregates. J. Mol. Biol. 2009, 390, 155–167. [Google Scholar] [CrossRef][Green Version]
- Reidy, M.; Masison, D.C. Sti1 Regulation of Hsp70 and Hsp90 Is Critical for Curing of Saccharomyces cerevisiae [PSI+] Prions by Hsp104. Mol. Cell. Biol. 2010, 30, 3542–3552. [Google Scholar] [CrossRef]
- Newnam, G.P.; Birchmore, J.L.; Chernoff, Y.O. Destabilization and recovery of a yeast prion after mild heat shock. J. Mol. Biol. 2011, 408, 432–448. [Google Scholar] [CrossRef]
- Yu, C.-I.; King, C.-Y. Forms and abundance of chaperone proteins influence yeast prion variant competition. Mol. Microbiol. 2019, 111, 798–810. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Gorkovskiy, A.; Bezsonov, E.E.; Stroobant, E.E. Yeast and Fungal Prions: Amyloid-Handling Systems, Amyloid Structure, and Prion Biology. Adv. Genet. 2016, 93, 191–236. [Google Scholar]
- Nelson, R.J.; Ziegelhoffer, T.; Nicolet, C.; Werner-Washburne, M.; Craig, E.A. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 1992, 71, 97–105. [Google Scholar] [CrossRef]
- Kushnirov, V.V.; Kryndushkin, D.S.; Boguta, M.; Smirnov, V.N.; Ter-Avanesyan, M.D. Chaperones that cure yeast artificial [PSI+] and their prion-specific effects. Curr. Biol. 2000, 10, 1443–1446. [Google Scholar] [CrossRef]
- Chacinska, A.; Szczesniak, B.; Kochneva-Pervukhova, N.V.; Kushnirov, V.V.; Ter-Avanesyan, M.D.; Boguta, M. Ssb1 chaperone is a [PSI+] prion-curing factor. Curr. Genet. 2001, 39, 62–67. [Google Scholar] [CrossRef]
- Chernoff, Y.O.; Newnam, G.P.; Kumar, J.; Allen, K.; Zink, A.D. Evidence for a Protein Mutator in Yeast: Role of the Hsp70-Related Chaperone Ssb in Formation, Stability, and Toxicity of the [PSI] Prion. Mol. Cell. Biol. 1999, 19, 8103–8112. [Google Scholar] [CrossRef]
- Kiktev, D.A.; Melomed, M.M.; Lu, C.D.; Newnam, G.P.; Chernoff, Y.O. Feedback control of prion formation and propagation by the ribosome-associated chaperone complex. Mol. Microbiol. 2015, 96, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Amor, A.J.; Castanzo, D.T.; Delany, S.P.; Selechnik, D.M.; van Ooy, A.; Cameron, D.M. The ribosome-associated complex antagonizes prion formation in yeast. Prion 2015, 9, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Sondheimer, N.; Lopez, N.; Craig, E.A.; Lindquist, S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001, 20, 2435–2442. [Google Scholar] [CrossRef] [PubMed]
- Stein, K.C.; Bengoechea, R.; Harms, M.B.; Weihl, C.C.; True, H.L. Myopathy-causing Mutations in an HSP40 Chaperone Disrupt Processing of Specific Client Conformers. J. Biol. Chem. 2014, 289, 21120–21130. [Google Scholar] [CrossRef]
- Stein, K.C.; True, H.L. Structural variants of yeast prions show conformer-specific requirements for chaperone activity. Mol. Microbiol. 2014, 93, 1156–1171. [Google Scholar] [CrossRef]
- Astor, M.T.; Kamiya, E.; Sporn, Z.A.; Berger, S.E.; Hines, J.K. Variant-specific and reciprocal Hsp40 functions in Hsp104-mediated prion elimination. Mol. Microbiol. 2018, 109, 41–62. [Google Scholar] [CrossRef] [PubMed]
- Verma, A.K.; Diwan, D.; Raut, S.; Dobriyal, N.; Brown, R.E.; Gowda, V.; Hines, J.K.; Sahi, C. Evolutionary Conservation and Emerging Functional Diversity of the Cytosolic Hsp70:J Protein Chaperone Network of Arabidopsis thaliana. G3 2017, 7, 1941–1954. [Google Scholar] [CrossRef]
- Reidy, M.; Sharma, R.; Roberts, B.-L.; Masison, D.C. Human J-protein DnaJB6b Cures a Subset of Saccharomyces cerevisiae Prions and Selectively Blocks Assembly of Structurally Related Amyloids. J. Biol. Chem. 2016, 291, 4035–4047. [Google Scholar] [CrossRef] [PubMed]
- Sporn, Z.A.; Hines, J.K. Hsp40 function in yeast prion propagation: Amyloid diversity necessitates chaperone functional complexity. Prion 2015, 9, 80–89. [Google Scholar] [CrossRef]
- Lancaster, D.L.; Dobson, C.M.; Rachubinski, R.A. Chaperone proteins select and maintain [PIN+] prion conformations in Saccharomyces cerevisiae. J. Biol. Chem. 2013, 288, 1266–1276. [Google Scholar] [CrossRef]
- Sahi, C.; Craig, E.A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 2007, 24, 7163–7168. [Google Scholar] [CrossRef]
- Schilke, B.A.; Ciesielski, S.J.; Ziegelhoffer, T.; Kamiya, E.; Tonelli, M.; Lee, W.; Cornilescu, G.; Hines, J.K.; Markley, J.L.; Craig, E.A. Broadening the functionality of a J-protein/ Hsp70 molecular chaperone system. PLoS Genet. 2017, 13, e1007084. [Google Scholar] [CrossRef]
- Reidy, M.; Sharma, R.; Shastry, S.; Roberts, B.-L.; Albino-Flores, I.; Wickner, S.; Masison, D.C. Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines. PLoS Genet. 2014, 10, 1004720. [Google Scholar] [CrossRef]
- Hines, J.K.; Craig, E.A. The sensitive [SWI+] prion New perspectives on yeast prion diversity. Prion 2011, 5, 164–168. [Google Scholar] [CrossRef]
- Troisi, E.M.; Rockman, M.E.; Nguyen, P.P.; Oliver, E.E.; Hines, J.K. Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant [URE3-1]. Mol. Microbiol. 2015, 97, 926–941. [Google Scholar] [CrossRef]
- Oliver, E.E.; Troisi, E.M.; Hines, J.K. Prion-specific Hsp40 function: The role of the auxilin homolog Swa2. Prion 2017, 11, 174–185. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Smirnov, V.N.; Ter-Avanesyan, M.D.; Kushnirov, V. Increased Expression of Hsp40 Chaperones, Transcriptional Factors, and Ribosomal Protein Rpp0 Can Cure Yeast Prions. J. Biol. Chem. 2002, 277, 23702–23708. [Google Scholar] [CrossRef]
- Xiao, J.; Kim, L.S.; Graham, T.R. Dissection of Swa2p/auxilin domain requirements for cochaperoning Hsp70 clathrin-uncoating activity in vivo. Mol. Biol. Cell 2006, 17, 3281–3290. [Google Scholar] [CrossRef]
- Fan, Q.; Park, K.-W.; Du, Z.; Morano, K.A.; Li, L. The Role of Sse1 in the de Novo Formation and Variant Determination of the [PSI+] Prion. Genetics 2007, 177, 1583–1593. [Google Scholar] [CrossRef]
- Kryndushkin, D.; Wickner, R.B. Nucleotide Exchange Factors for Hsp70s Are Required for [URE3] Prion Propagation in Saccharomyces cerevisiae. Mol. Biol. Cell 2007, 18, 2149–2154. [Google Scholar] [CrossRef]
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef]
- Moosavi, B.; Wongwigkarn, J.; Tuite, M.F. Hsp70/Hsp90 co-chaperones are required for efficient Hsp104-mediated elimination of the yeast [PSI+] prion but not for prion propagation. Yeast 2010, 27, 167–179. [Google Scholar] [CrossRef]
- Kumar, N.; Gaur, D.; Gupta, A.; Puri, A.; Sharma, D. Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in Saccharomyces cerevisiae. PLOS Genet. 2015, 11, e1005567. [Google Scholar] [CrossRef]
- Cox, B.; Tuite, M. The life of [PSI]. Curr. Genet. 2018, 64, 1–8. [Google Scholar] [CrossRef]
- Matveenko, A.G.; Barbitoff, Y.A.; Jay-Garcia, L.M.; Chernoff, Y.O.; Zhouravleva, G.A. Differential effects of chaperones on yeast prions: CURrent view. Curr. Genet. 2018, 64, 317–325. [Google Scholar] [CrossRef]
- Greene, L.E.; Zhao, X.; Eisenberg, E. Curing of [PSI+] by Hsp104 Overexpression: Clues to solving the puzzle. Prion 2018, 12, 9–15. [Google Scholar] [CrossRef]
- Zenthon, J.F.; Ness, F.; Cox, B.; Tuite, M.F. The [PSI+] prion of Saccharomyces cerevisiae can be propagated by an Hsp104 orthologue from Candida albicans. Eukaryot. Cell 2006, 5, 217–225. [Google Scholar] [CrossRef]
- Zhao, X.; Rodriguez, R.; Silberman, R.E.; Ahearn, J.M.; Saidha, S.; Cummins, K.C.; Eisenberg, E.; Greene, L.E. Heat shock protein 104 (Hsp104)-mediated curing of [PSI+] yeast prions depends on both [PSI+] conformation and the properties of the Hsp104 homologs. J. Biol. Chem. 2017, 292, 8630–8641. [Google Scholar] [CrossRef]
- Tanaka, M.; Chien, P.; Naber, N.; Cooke, R.; Weissman, J.S. Conformational variations in an infectious protein determine prion strain differences. Nature 2004, 428, 323–328. [Google Scholar] [CrossRef]
- Gorkovskiy, A.; Reidy, M.; Masison, D.C.; Wickner, R.B. Hsp104 disaggregase at normal levels cures many [PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. Proc. Natl. Acad. Sci. USA 2017, 114, E4193–E4202. [Google Scholar] [CrossRef]
- Kirkland, P.A.; Reidy, M.; Masison, D.C. Functions of yeast Hsp40 chaperone Sis1p dispensable for prion propagation but important for prion curing and protection from prion toxicity. Genetics 2011, 188, 565–577. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Engel, A.; Edskes, H.; Wickner, R.B. Molecular Chaperone Hsp104 Can Promote Yeast Prion Generation. Genetics 2011. [Google Scholar] [CrossRef][Green Version]
- Sahi, C.; Kominek, J.; Ziegelhoffer, T.; Yu, H.Y.; Baranowski, M.; Marszalek, J.; Craig, E.A. Sequential duplications of an ancient member of the DnaJ-family expanded the functional chaperone network in the eukaryotic cytosol. Mol. Biol. Evol. 2013, 30, 985–998. [Google Scholar] [CrossRef]
- Kryndushkin, D.S.; Shewmaker, F.; Wickner, R.B. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008, 27, 2725–2735. [Google Scholar] [CrossRef]
- Wickner, R.B.; Bezsonov, E.; Bateman, D.A. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc. Natl. Acad. Sci. USA 2014, 111, E2711-20. [Google Scholar] [CrossRef]
- Malinovska, L.; Kroschwald, S.; Munder, M.C.; Richter, D.; Alberti, S. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell 2012, 23, 3041–3056. [Google Scholar] [CrossRef]
- Barbitoff, Y.A.; Matveenko, A.G.; Moskalenko, S.E.; Zemlyanko, O.M.; Newnam, G.P.; Patel, A.; Chernova, T.A.; Chernoff, Y.O.; Zhouravleva, G.A. To CURe or not to CURe? Differential effects of the chaperone sorting factor Cur1 on yeast prions are mediated by the chaperone Sis1. Mol. Microbiol. 2017, 105, 242–257. [Google Scholar] [CrossRef]
- Wickner, R.B.; Shewmaker, F.P.; Bateman, D.A.; Edskes, H.K.; Gorkovskiy, A.; Dayani, Y.; Bezsonov, E.E. Yeast Prions: Structure, Biology, and Prion-Handling Systems. Microbiol. Mol. Biol. Rev. 2015, 79, 1–17. [Google Scholar] [CrossRef]
- Killian, A.N.; Hines, J.K. Chaperone functional specificity promotes yeast prion diversity. PLoS Pathog. 2018, 14, 1–7. [Google Scholar] [CrossRef]
- Wickner, R. Anti-prion systems in yeast. J. Biol. Chem. 2019, 294, 1729–1738. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Killian, A.N.; Miller, S.C.; Hines, J.K. Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast. Viruses 2019, 11, 349. https://doi.org/10.3390/v11040349
Killian AN, Miller SC, Hines JK. Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast. Viruses. 2019; 11(4):349. https://doi.org/10.3390/v11040349
Chicago/Turabian StyleKillian, Andrea N., Sarah C. Miller, and Justin K. Hines. 2019. "Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast" Viruses 11, no. 4: 349. https://doi.org/10.3390/v11040349
APA StyleKillian, A. N., Miller, S. C., & Hines, J. K. (2019). Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast. Viruses, 11(4), 349. https://doi.org/10.3390/v11040349




