Detection and Characterization of Human Enteroviruses, Human Cosaviruses, and a New Human Parechovirus Type in Healthy Individuals in Osun State, Nigeria, 2016/2017
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dunn, J.J. Enteroviruses and Parechoviruses. Microbiol. Spectr. 2016, 4. [Google Scholar] [CrossRef]
- Pons-Salort, M.; Parker, E.P.; Grassly, N.C. The epidemiology of non-polio enteroviruses: Recent advances and outstanding questions. Curr. Opin. Infect. Dis. 2015, 28, 479–487. [Google Scholar] [CrossRef]
- Harvala, H.; Simmonds, P. Human parechoviruses: Biology, epidemiology and clinical significance. J. Clin. Virol. 2009, 45, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kapoor, A.; Victoria, J.; Simmonds, P.; Slikas, E.; Chieochansin, T.; Naeem, A.; Shaukat, S.; Sharif, S.; Alam, M.M.; Angez, M.; et al. A highly prevalent and genetically diversified Picornaviridae genus in South Asian children. Proc. Natl. Acad. Sci. USA 2008, 105, 20482–20487. [Google Scholar] [CrossRef]
- Victoria, J.G.; Kapoor, A.; Li, L.; Blinkova, O.; Slikas, B.; Wang, C.; Naeem, A.; Zaidi, S.; Delwart, E. Metagenomic analyses of viruses in stool samples from children with acute flaccid paralysis. J. Virol. 2009, 83, 4642–4651. [Google Scholar] [CrossRef]
- Da Costa, A.C.; Luchs, A.; Milagres, F.A.P.; Komninakis, S.V.; Gill, D.E.; Lobato, M.; Brustulin, R.; das Chagas, R.T.; Abrao, M.; Soares, C.; et al. Near full length genome of a recombinant (E/D) cosavirus strain from a rural area in the central region of Brazil. Sci. Rep. 2018, 8, 12304. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Latorre, L.; Romero, B.; Bonifaz, E.; Timoneda, N.; Rusinol, M.; Girones, R.; Rios-Touma, B. Quito’s virome: Metagenomic analysis of viral diversity in urban streams of Ecuador’s capital city. Sci. Total Environ. 2018, 645, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Di Cristanziano, V.; Bottcher, S.; Diedrich, S.; Timmen-Wego, M.; Knops, E.; Lubke, N.; Kaiser, R.; Pfister, H.; Kabore, Y.; D’Alfonso, R. Erratum to “Detection and characterization of enteroviruses and parechoviruses in healthy people living in the South of Cote d’Ivoire”. J. Clin. Virol. 2015, 72, 153. [Google Scholar] [CrossRef] [PubMed]
- Graul, S.; Bottcher, S.; Eibach, D.; Krumkamp, R.; Kasmaier, J.; Adu-Sarkodie, Y.; May, J.; Tannich, E.; Panning, M. High diversity of human parechovirus including novel types in stool samples from Ghanaian children. J. Clin. Virol. 2017, 96, 116–119. [Google Scholar] [CrossRef]
- Faleye, T.O.; Adewumi, M.O.; Coker, B.A.; Nudamajo, F.Y.; Adeniji, J.A. Direct Detection and Identification of Enteroviruses from Faeces of Healthy Nigerian Children Using a Cell-Culture Independent RT-Seminested PCR Assay. Adv. Virol. 2016, 2016, 1412838. [Google Scholar] [CrossRef]
- Oyero, O.G.; Adu, F.D. Non-polio enteroviruses serotypes circulating in Nigeria. Afr. J. Med. Med. Sci. 2010, 39, 201–208. [Google Scholar]
- Oyero, O.G.; Adu, F.D.; Ayukekbong, J.A. Molecular characterization of diverse species enterovirus-B types from children with acute flaccid paralysis and asymptomatic children in Nigeria. Virus Res. 2014, 189, 189–193. [Google Scholar] [CrossRef]
- Adeniji, J.A.; Faleye, T.O. Enterovirus C strains circulating in Nigeria and their contribution to the emergence of recombinant circulating vaccine-derived polioviruses. Arch. Virol. 2015, 160, 675–683. [Google Scholar] [CrossRef]
- Adeniji, J.A.; Oragwa, A.O.; George, U.E.; Ibok, U.I.; Faleye, T.O.C.; Adewumi, M.O. Preponderance of enterovirus C in RD-L20B-cell-culture-negative stool samples from children diagnosed with acute flaccid paralysis in Nigeria. Arch. Virol. 2017, 162, 3089–3101. [Google Scholar] [CrossRef] [PubMed]
- Kapusinszky, B.; Phan, T.G.; Kapoor, A.; Delwart, E. Genetic diversity of the genus Cosavirus in the family Picornaviridae: A new species, recombination, and 26 new genotypes. PLoS ONE 2012, 7, e36685. [Google Scholar] [CrossRef] [PubMed]
- World Health Organisation Global Action Plan to minimize poliovirus facility-associated risk after type-specific eradication of wild polioviruses and sequential cessation of oral polio vaccine use. 2014. Available online: http://polioeradication.org/wp-content/uploads/2016/12/GAPIII_2014.pdf (accessed on 31 July 2019).
- Kuryk, L.; Wieczorek, M.; Diedrich, S.; Bottcher, S.; Witek, A.; Litwinska, B. Genetic analysis of poliovirus strains isolated from sewage in Poland. J. Med. Virol. 2014, 86, 1243–1248. [Google Scholar] [CrossRef]
- Benschop, K.; Minnaar, R.; Koen, G.; van Eijk, H.; Dijkman, K.; Westerhuis, B.; Molenkamp, R.; Wolthers, K. Detection of human enterovirus and human parechovirus (HPeV) genotypes from clinical stool samples: Polymerase chain reaction and direct molecular typing, culture characteristics, and serotyping. Diagn. Microbiol. Infect. Dis 2010, 68, 166–173. [Google Scholar] [CrossRef]
- Nix, W.A.; Oberste, M.S.; Pallansch, M.A. Sensitive, seminested PCR amplification of VP1 sequences for direct identification of all enterovirus serotypes from original clinical specimens. J. Clin. Microbiol. 2006, 44, 2698–2704. [Google Scholar] [CrossRef]
- Bottcher, S.; Obermeier, P.E.; Neubauer, K.; Diedrich, S. Recombinant Enterovirus A71 Subgenogroup C1 Strains, Germany, 2015. Emerg. Infect. Dis. 2016, 22, 1843–1846. [Google Scholar] [CrossRef] [PubMed]
- Leitch, E.C.; Harvala, H.; Robertson, I.; Ubillos, I.; Templeton, K.; Simmonds, P. Direct identification of human enterovirus serotypes in cerebrospinal fluid by amplification and sequencing of the VP1 region. J. Clin. Virol. 2009, 44, 119–124. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, D.R.; Iber, J.C.; Chen, Q.; Ching, K.; Yang, S.J.; De, L.; Mandelbaum, M.D.; Emery, B.; Campagnoli, R.; Burns, C.C.; et al. Poliovirus serotype-specific VP1 sequencing primers. J. Virol. Methods 2011, 174, 128–130. [Google Scholar] [CrossRef]
- Bottcher, S.; Prifert, C.; Weissbrich, B.; Adams, O.; Aldabbagh, S.; Eis-Hubinger, A.M.; Diedrich, S. Detection of enterovirus D68 in patients hospitalised in three tertiary university hospitals in Germany, 2013 to 2014. Euro. Surveill. 2016, 21. [Google Scholar] [CrossRef]
- Kroneman, A.; Vennema, H.; Deforche, K.; Avoort, H.v.d.; Penaranda, S.; Oberste, M.S.; Vinje, J.; Koopmans, M. An automated genotyping tool for enteroviruses and noroviruses. J. Clin. Virol. 2011, 51, 121–125. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Bottcher, S.; Neubauer, K.; Baillot, A.; Rieder, G.; Adam, M.; Diedrich, S. Stool screening of Syrian refugees and asylum seekers in Germany, 2013/2014: Identification of Sabin like polioviruses. Int. J. Med. Microbiol. 2015, 305, 601–606. [Google Scholar] [CrossRef]
- Oberste, M.S.; Maher, K.; Kilpatrick, D.R.; Pallansch, M.A. Molecular evolution of the human enteroviruses: Correlation of serotype with VP1 sequence and application to picornavirus classification. J. Virol. 1999, 73, 1941–1948. [Google Scholar]
- Nix, W.A.; Maher, K.; Pallansch, M.A.; Oberste, M.S. Parechovirus typing in clinical specimens by nested or semi-nested PCR coupled with sequencing. J. Clin. Virol. 2010, 48, 202–207. [Google Scholar] [CrossRef]
- Enfissi, A.; Joffret, M.L.; Delaune, D.; Delpeyroux, F.; Rousset, D.; Bessaud, M. Coxsackievirus A24 Variant Associated with Acute Haemorrhagic Conjunctivitis Cases, French Guiana, 2017. Intervirology 2017, 60, 271–275. [Google Scholar] [CrossRef]
- Sadeuh-Mba, S.A.; Bessaud, M.; Massenet, D.; Joffret, M.-L.; Endegue, M.-C.; Njouom, R.; Reynes, J.-M.; Rousset, D.; Delpeyroux, F. High Frequency and Diversity of Species C Enteroviruses in Cameroon and Neighboring Countries. J. Clin. Microbiol. 2013, 51, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A.N.; Drexler, J.F.; Kotova, V.O.; Amjaga, E.N.; Reznik, V.I.; Gmyl, A.P.; Grard, G.; Taty Taty, R.; Trotsenko, O.E.; Leroy, E.M.; et al. Novel serotypes 105 and 116 are members of distinct subgroups of Human enterovirus C. J. Gen. Virol. 2012, 93, 2357–2362. [Google Scholar] [CrossRef] [PubMed]
- Brown, B.A.; Maher, K.; Flemister, M.R.; Naraghi-Arani, P.; Uddin, M.; Oberste, M.S.; Pallansch, M.A. Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J. Gen. Virol. 2009, 90, 1713–1723. [Google Scholar] [CrossRef] [PubMed]
- Krumbholz, A.; Egerer, R.; Braun, H.; Schmidtke, M.; Rimek, D.; Kroh, C.; Hennig, B.; Groth, M.; Sauerbrei, A.; Zell, R. Analysis of an echovirus 18 outbreak in Thuringia, Germany: Insights into the molecular epidemiology and evolution of several enterovirus species B members. Med. Microbiol. Immunol. 2016, 205, 471–483. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Guo, J.; Xu, W.; Sun, S.; Xie, Z. An outbreak of echovirus 18 encephalitis/meningitis in children in Hebei Province, China, 2015. Emerg. Microbes. Infect. 2017, 6, e54. [Google Scholar] [CrossRef]
- Sadeuh-Mba, S.A.; Bessaud, M.; Joffret, M.-L.; Endegue Zanga, M.-C.; Balanant, J.; Mpoudi Ngole, E.; Njouom, R.; Reynes, J.-M.; Delpeyroux, F.; Rousset, D. Characterization of Enteroviruses from Non-Human Primates in Cameroon Revealed Virus Types Widespread in Humans along with Candidate New Types and Species. PLOS Negl. Trop. Dis. 2014, 8, e3052. [Google Scholar] [CrossRef]
- Bessaud, M.; Joffret, M.L.; Blondel, B.; Delpeyroux, F. Exchanges of genomic domains between poliovirus and other cocirculating species C enteroviruses reveal a high degree of plasticity. Sci. Rep. 2016, 6, 38831. [Google Scholar] [CrossRef]
- Burns, C.C.; Shaw, J.; Jorba, J.; Bukbuk, D.; Adu, F.; Gumede, N.; Pate, M.A.; Abanida, E.A.; Gasasira, A.; Iber, J.; et al. Multiple Independent Emergences of Type 2 Vaccine-Derived Polioviruses during a Large Outbreak in Northern Nigeria. J. Virol. 2013, 87, 4907–4922. [Google Scholar] [CrossRef]
- Jiang, P.; Faase, J.A.J.; Toyoda, H.; Paul, A.; Wimmer, E.; Gorbalenya, A.E. Evidence for emergence of diverse polioviruses from C-cluster coxsackie A viruses and implications for global poliovirus eradication. Proc. Natl. Acad. Sci. USA 2007, 104, 9457–9462. [Google Scholar] [CrossRef]
- Adeniji, A.J.; Onoja, A.B.; Adewumi, M.O. Polio virus neutralizing antibody dynamics among children in a north-central and South-Western Nigeria state. J. Immunoass. Immunochem. 2015, 36, 45–53. [Google Scholar] [CrossRef]
- Adeniji, J.A.; Osundare, F.A.; Adewumi, O.M.; Onoja, A.B.; Fagbami, A.H. Immunity to poliovirus serotypes in children population of selected communities in south-west, Nigeria. Afr. J. Infect. Dis. 2015, 9, 1–5. [Google Scholar] [CrossRef][Green Version]
- Kadambari, S.; Harvala, H.; Simmonds, P.; Pollard, A.J.; Sadarangani, M. Strategies to improve detection and management of human parechovirus infection in young infants. Lancet Infect. Dis. 2019, 19, e51–e58. [Google Scholar] [CrossRef]
- Rezig, D.; Ben Farhat, E.; Touzi, H.; Meddeb, Z.; Ben Salah, A.; Triki, H. Prevalence of human cosaviruses in Tunisia, North Africa. J. Med. Virol. 2015, 87, 940–943. [Google Scholar] [CrossRef]
- Ayouni, S.; Estienney, M.; Hammami, S.; Neji Guediche, M.; Pothier, P.; Aouni, M.; Belliot, G.; de Rougemont, A. Cosavirus, Salivirus and Bufavirus in Diarrheal Tunisian Infants. PLoS ONE 2016, 11, e0162255. [Google Scholar] [CrossRef] [PubMed]
- Cinek, O.; Kramna, L.; Mazankova, K.; Kunteová, K.; Chudá, K.; C. J. Claas, E.; Stene, L.C.; Tapia, G. Virus genotyping by massive parallel amplicon sequencing: Adenovirus and enterovirus in the Norwegian MIDIA study. J. Med. Virol. 2019, 91, 606–614. [Google Scholar] [CrossRef] [PubMed]
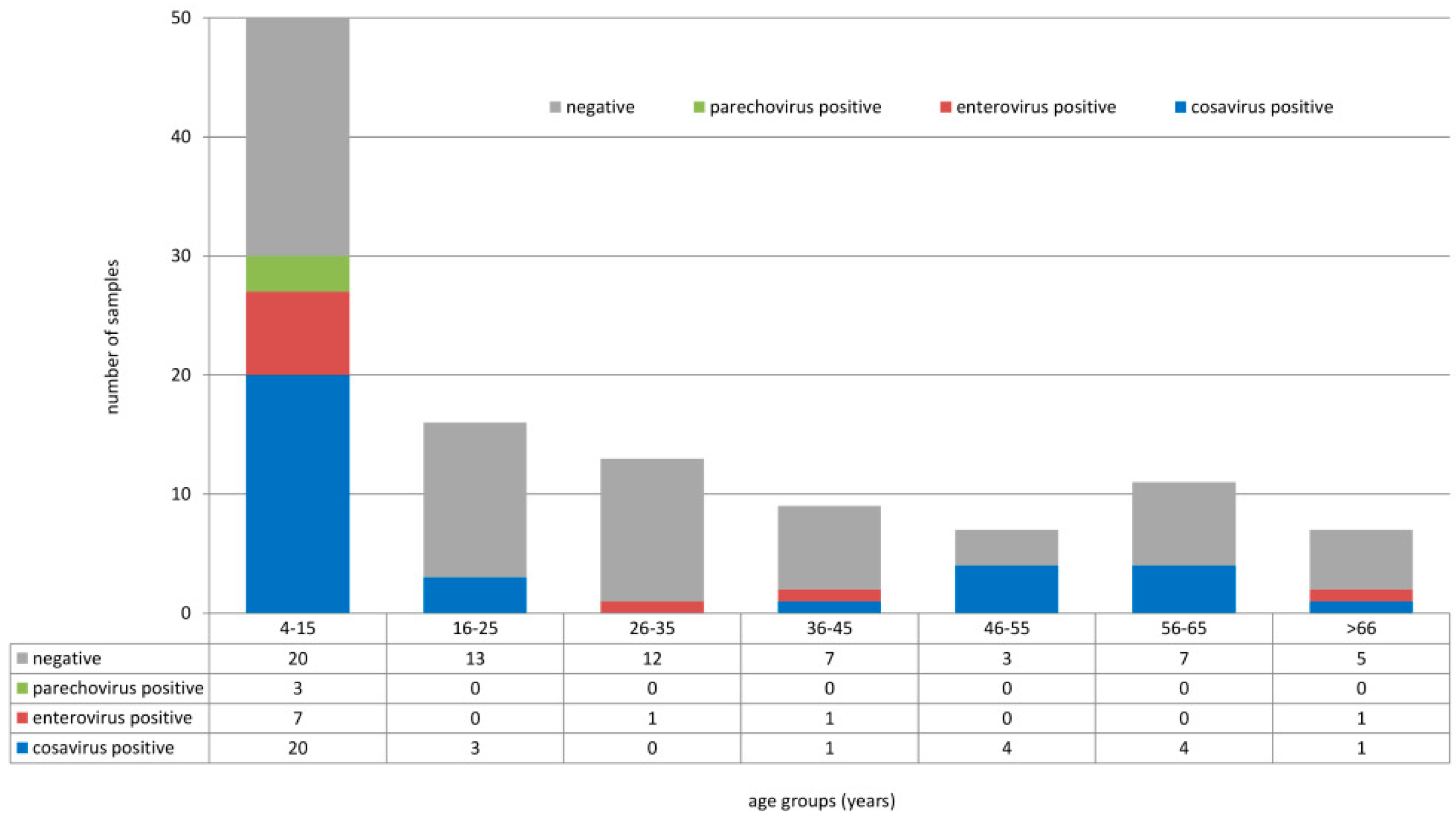
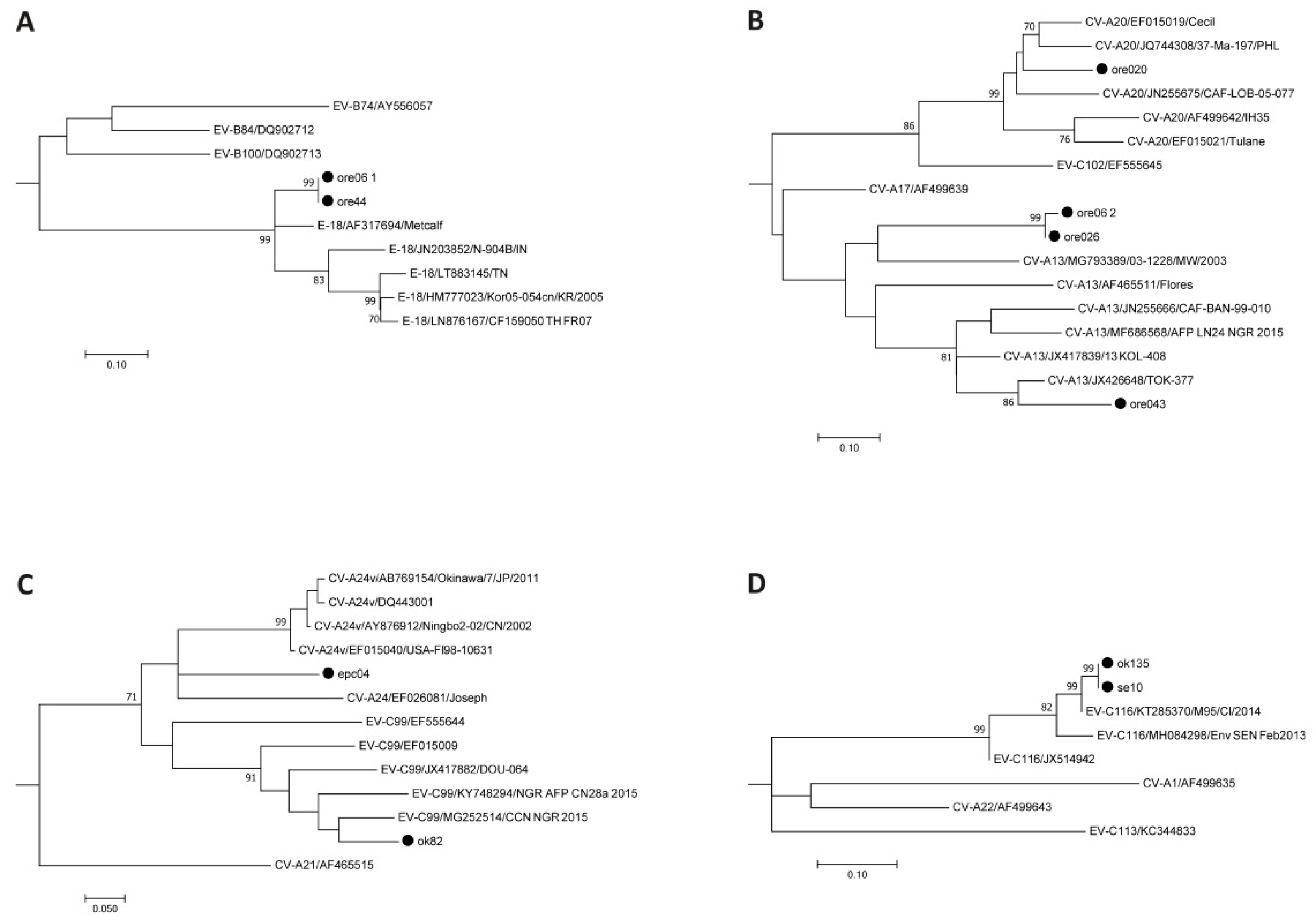
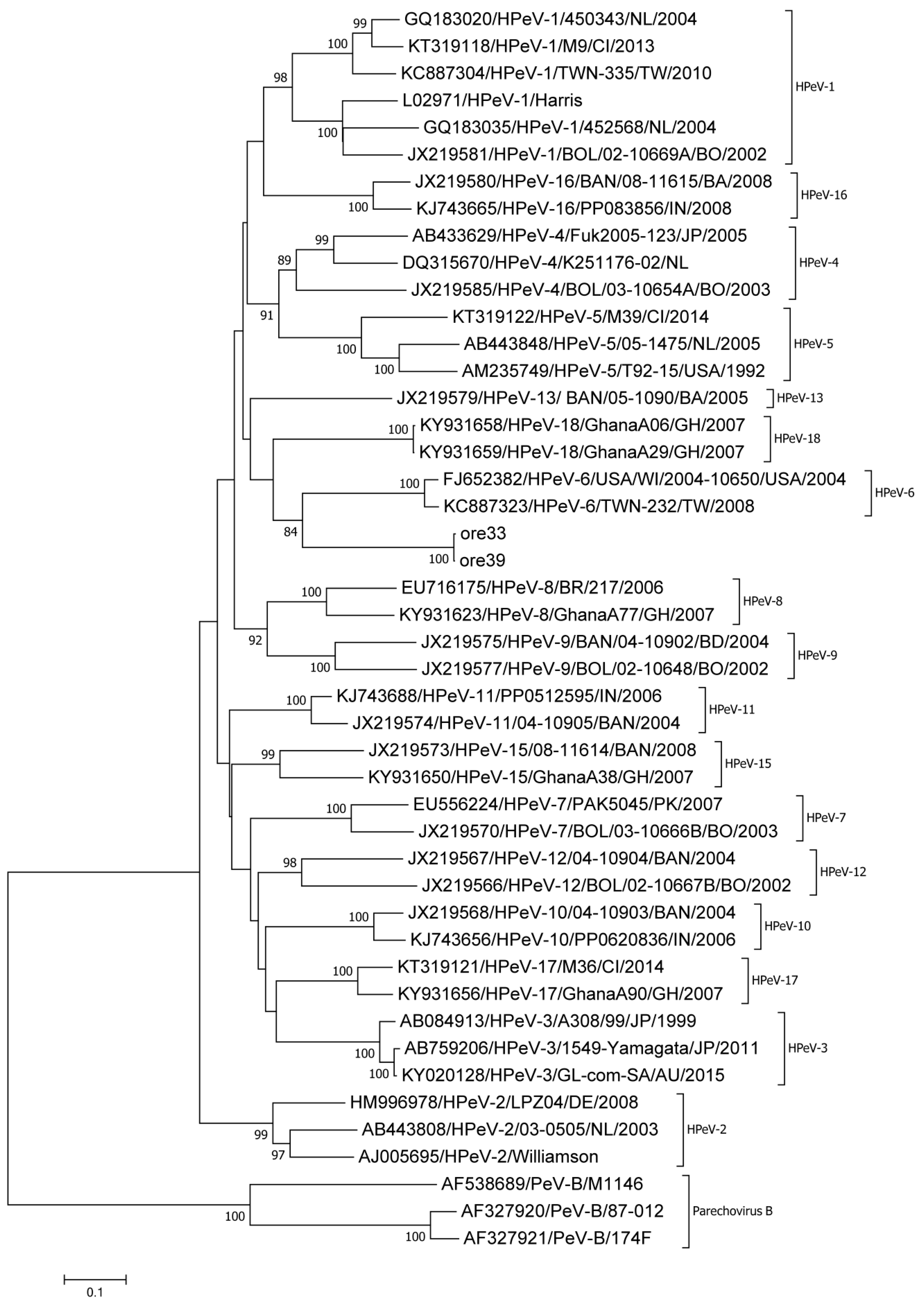
| Sample ID | Name of Location | Type of Location | Age (Y) | Sex | Collection Date | 5’-NCR seq | VP1 seq |
|---|---|---|---|---|---|---|---|
| epc 04 | Ede | Urban | 28 | Male | September 2016 | Enterovirus C | CV-A24 |
| se 10 | Ede | Urban | 45 | Male | September 2016 | Enterovirus C | EV-C116 |
| ok 82 | Oke-Osun | Semi-Urban | 4 | Male | September 2016 | Enterovirus C | EV-C99 |
| ok 135 | Oke-Osun | Semi-Urban | 72 | Female | September 2016 | Enterovirus C | EV-C116 |
| ore 06 | Ore | Rural | 5 | Female | May 2017 | Enterovirus B | E-18,CV-A13 |
| ore 026 | Ore | Rural | 14 | Male | May 2017 | Enterovirus C | CV-A13 |
| ore 043 | Ore | Rural | 4 | Male | May 2017 | Enterovirus C | CV-A13 |
| ore 047 | Ore | Rural | 8 | Female | May 2017 | Enterovirus C | Negative |
| ore 20 | Ore | Rural | 11 | Female | May 2017 | Enterovirus C | CV-A20 |
| ore 44 | Ore | Rural | 5 | Female | May 2017 | Enterovirus B | E-18 |
| Sample ID | Sequencing Result | Cp Value Stool | CPE on RD-A | Cp Value RD-A Supernatant | CPE on CaCo-2 | Cp Value CaCo-2, Supernatant | CPE on Hep-2C | CP Value Hep-2C, Supernatant | CPE on L20B | Cp Value L20B Supernatant |
|---|---|---|---|---|---|---|---|---|---|---|
| epc 04 | CV-A24 | 29.15 | negative | 33.60 | negative | 38.46 | positive | 23.65 | negative * | 35.84 |
| se 10 | EV-C116 | 29.34 | negative | 36.77 | negative | 37.15 | negative | 36.01 | negative * | 32.22 |
| ok 82 | EV-C99 | 33.56 | negative | 40.0 | negative | 37.27 | negative | n.d. | negative * | 35.95 |
| ok 135 | EV-C116 | 40 | negative | n.d. | negative | n.d. | negative | n.d. | negative | n.d. |
| ore 06 | E-18, CV-A13 | n.e. | negative | 35.84 | negative | 37.67 | positive | 23.01 | negative | n.d. |
| ore 026 | CV-A13 | 33.11 | negative | 22.21 | negative | n.d. | positive | 24.95 | negative | n.d. |
| ore 043 | CV-A13 | 40 | negative | n.d. | negative | n.d. | negative | n.d. | negative | n.d. |
| ore 047 | Enterovirus C | 38.78 | negative | n.d. | negative | n.d. | negative | n.d. | negative * | n.d. |
| ore 20 | CV-A20 | 28.81 | negative | 26.97 | negative | 26.76 | positive | 23.04 | negative | n.d. |
| ore 44 | E-18 | 25.06 | negative | 31.50 | negative | 26.50 | negative | 36.48 | negative | n.d. |
| Sample ID | Name of Location | Location | Age (Years) | Sex | Collection Date | 5’-NCR Seq | VP1 Seq |
|---|---|---|---|---|---|---|---|
| ore 44 | Ore | Rural | 5 | female | May 2017 | Parechovirus A | negative |
| ore 39 | Ore | Rural | 8 | male | May 2017 | Parechovirus A | unassigned |
| ore 33 | Ore | Rural | 10 | male | May 2017 | Parechovirus A | unassigned |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Osundare, F.A.; Opaleye, O.O.; Akindele, A.A.; Adedokun, S.A.; Akanbi, O.A.; Bock, C.-T.; Diedrich, S.; Böttcher, S. Detection and Characterization of Human Enteroviruses, Human Cosaviruses, and a New Human Parechovirus Type in Healthy Individuals in Osun State, Nigeria, 2016/2017. Viruses 2019, 11, 1037. https://doi.org/10.3390/v11111037
Osundare FA, Opaleye OO, Akindele AA, Adedokun SA, Akanbi OA, Bock C-T, Diedrich S, Böttcher S. Detection and Characterization of Human Enteroviruses, Human Cosaviruses, and a New Human Parechovirus Type in Healthy Individuals in Osun State, Nigeria, 2016/2017. Viruses. 2019; 11(11):1037. https://doi.org/10.3390/v11111037
Chicago/Turabian StyleOsundare, Folakemi Abiodun, Oladele Oluyinka Opaleye, Akeem Abiodun Akindele, Samuel Adeyinka Adedokun, Olusola Anuoluwapo Akanbi, Claus-Thomas Bock, Sabine Diedrich, and Sindy Böttcher. 2019. "Detection and Characterization of Human Enteroviruses, Human Cosaviruses, and a New Human Parechovirus Type in Healthy Individuals in Osun State, Nigeria, 2016/2017" Viruses 11, no. 11: 1037. https://doi.org/10.3390/v11111037
APA StyleOsundare, F. A., Opaleye, O. O., Akindele, A. A., Adedokun, S. A., Akanbi, O. A., Bock, C.-T., Diedrich, S., & Böttcher, S. (2019). Detection and Characterization of Human Enteroviruses, Human Cosaviruses, and a New Human Parechovirus Type in Healthy Individuals in Osun State, Nigeria, 2016/2017. Viruses, 11(11), 1037. https://doi.org/10.3390/v11111037






