Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses
Abstract
1. Introduction
2. Diversity of NLSs Identified in Viral Proteins
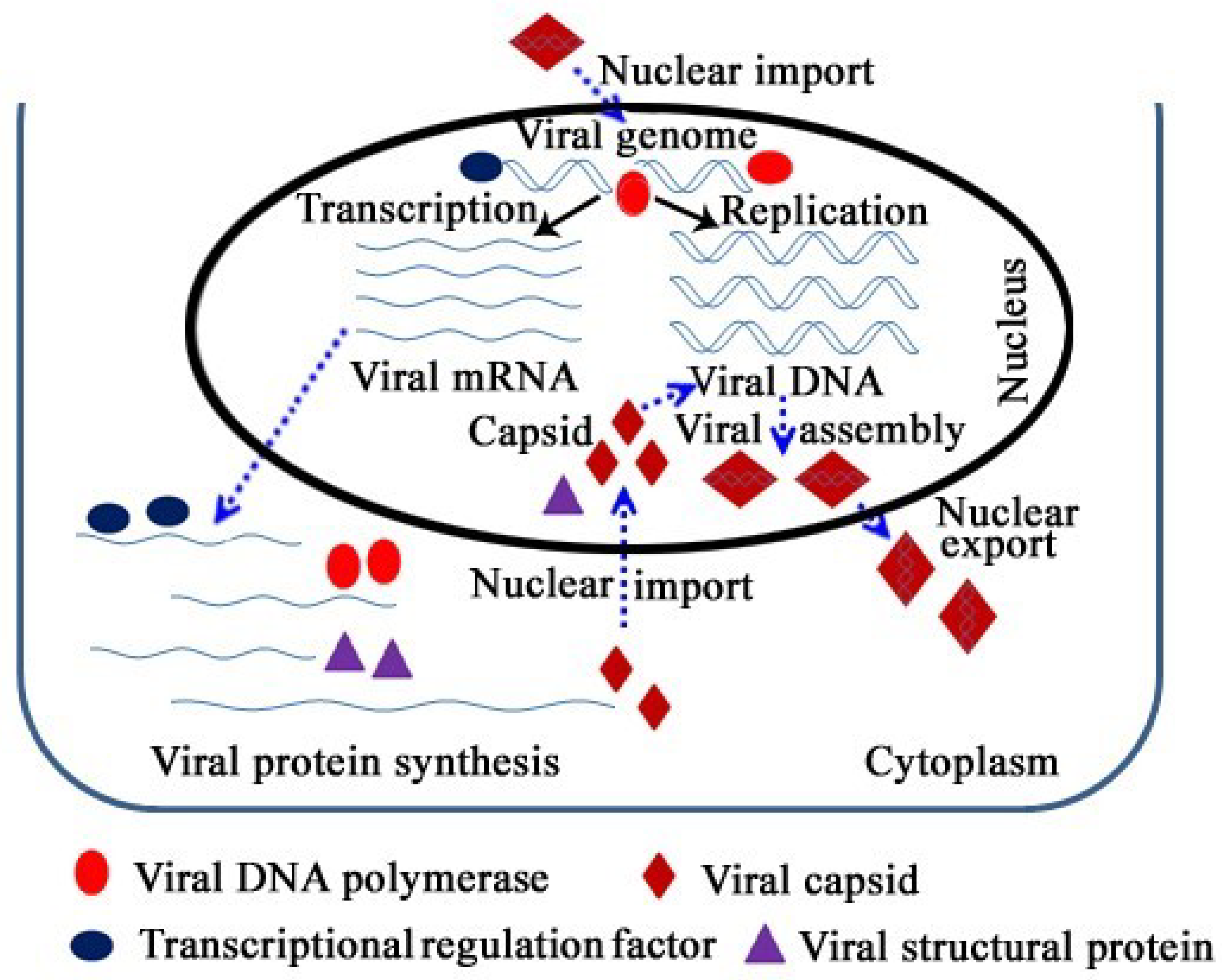
3. Entry of the Viral Genome and Capsid
3.1. Nuclear Import of the Viral Genome
3.2. Requirements for Nuclear Transport of Viral Capsids
4. Requirements for the Localization of Viral Self-Encoded DNA Polymerase
4.1. Replication of Double-Stranded DNA Viruses in the Cytoplasm
4.2. Replication of dsDNA Viruses in Host Cell Nuclei
4.3. Replication of ssDNA Viruses in Host Cell Nuclei
5. Mechanism of Nuclear Import of Viral Proteins Lacking NLSs
6. Conclusions and Future Studies
Funding
Conflicts of Interest
References
- Di Ventura, B.; Kuhlman, B. Go in! Go out! Inducible control of nuclear localization. Curr. Opin. Chem. Biol. 2016, 34, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.Y.H.; Aebi, U.; Fahrenkrog, B. Towards reconciling structure and function in the nuclear pore complex. Histochem. Cell Biol. 2008, 129, 105–116. [Google Scholar] [CrossRef] [PubMed]
- Dou, D.; Revol, R.; Östbye, H.; Wang, H.; Daniels, R. Influenza A Virus Cell Entry, Replication, Virion Assembly and Movement. Front. Immunol. 2018, 9, 1581. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Zhou, S.; Gao, S.; Deng, H. Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell 2019, 10, 315–326. [Google Scholar] [CrossRef]
- Schmid, M.; Speiseder, T.; Dobner, T.; Gonzalez, R.A. DNA virus replication compartments. J. Virol. 2014, 88, 1404–1420. [Google Scholar] [CrossRef]
- Au, H.H.; Jan, E. Novel viral translation strategies. Wiley Interdiscip. Rev. RNA 2014, 5, 779–801. [Google Scholar] [CrossRef]
- Roberts, L.; Wieden, H.-J. Viruses, IRESs, and a universal translation initiation mechanism. Biotechnol. Genet. Eng. Rev. 2018, 34, 60–75. [Google Scholar] [CrossRef]
- Tang, Q.; Wu, P.; Chen, H.; Li, G. Pleiotropic roles of the ubiquitin-proteasome system during viral propagation. Life Sci. 2018, 207, 350–354. [Google Scholar] [CrossRef]
- Lange, A.; Mills, R.E.; Lange, C.J.; Stewart, M.; Devine, S.E.; Corbett, A.H. Classical nuclear localization signals: Definition, function, and interaction with importin alpha. J. Biol. Chem. 2007, 282, 5101–5105. [Google Scholar] [CrossRef]
- Pouton, C.W.; Wagstaff, K.M.; Roth, D.M.; Moseley, G.W.; Jans, D.A. Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 2007, 59, 698–717. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, J.; Yang, B.; Zhou, L.; Hu, Y. The nuclear localization signal of the NS1 protein is essential for Periplaneta fuliginosa densovirus infection. Virus Res. 2009, 145, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Wulan, W.N.; Heydet, D.; Walker, E.J.; Gahan, M.E.; Ghildyal, R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. 2015, 6, 553. [Google Scholar] [CrossRef] [PubMed]
- Terry, L.J.; Shows, E.B.; Wente, S.R. Crossing the Nuclear Envelope: Hierarchical Regulation of Nucleocytoplasmic Transport. Science 2007, 318, 1412–1416. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Han, M.-E.; Oh, S.-O. The molecular mechanism for nuclear transport and its application. Anat. Cell Biol. 2017, 50, 77–85. [Google Scholar] [CrossRef]
- Macara, I.G. Transport into and out of the Nucleus. Microbiol. Mol. Biol. Rev. 2001, 65, 570–594. [Google Scholar] [CrossRef]
- Hawkins, J.; Davis, L.; Bodén, M. Predicting nuclear localization. J. Proteome Res. 2007, 6, 1402–1409. [Google Scholar] [CrossRef]
- Kalderon, D.; Roberts, B.L.; Richardson, W.D.; Smith, A.E. A short amino acid sequence able to specify nuclear location. Cell 1984, 39, 499–509. [Google Scholar] [CrossRef]
- Robbins, J.; Dilwortht, S.M.; Laskey, R.A.; Dingwall, C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell 1991, 64, 615–623. [Google Scholar] [CrossRef]
- Lombardo, E.; Ramírez, J.C.; Agbandje-McKenna, M.; Almendral, J.M. A Beta-Stranded Motif Drives Capsid Protein Oligomers of the Parvovirus Minute Virus of Mice into the Nucleus for Viral Assembly. J. Virol. 2000, 74, 3804–3814. [Google Scholar] [CrossRef]
- Pillet, S.; Annan, Z.; Fichelson, S.; Morinet, F. Identification of a nonconventional motif necessary for the nuclear import of the human parvovirus B19 major capsid protein (VP2). Virology 2003, 306, 25–32. [Google Scholar] [CrossRef]
- Shin, H.Y.; Reich, N.C. Dynamic trafficking of STAT5 depends on an unconventional nuclear localization signal. J. Cell Sci. 2013, 126, 3333–3343. [Google Scholar] [CrossRef] [PubMed]
- Jarvis, D.L.; Bohlmeyer, D.A.; García, A. Requirements for nuclear localization and supramolecular assembly of a baculovirus polyhedrin protein. Virology 1991, 185, 795–810. [Google Scholar] [CrossRef]
- Wu, W.; Liang, H.; Kan, J.; Liu, C.; Yuan, M.; Liang, C.; Yang, K.; Pang, Y. Autographa californica Multiple Nucleopolyhedrovirus 38K Is a Novel Nucleocapsid Protein That Interacts with VP1054, VP39, VP80, and Itself. J. Virol. 2008, 82, 12356–12364. [Google Scholar] [CrossRef] [PubMed]
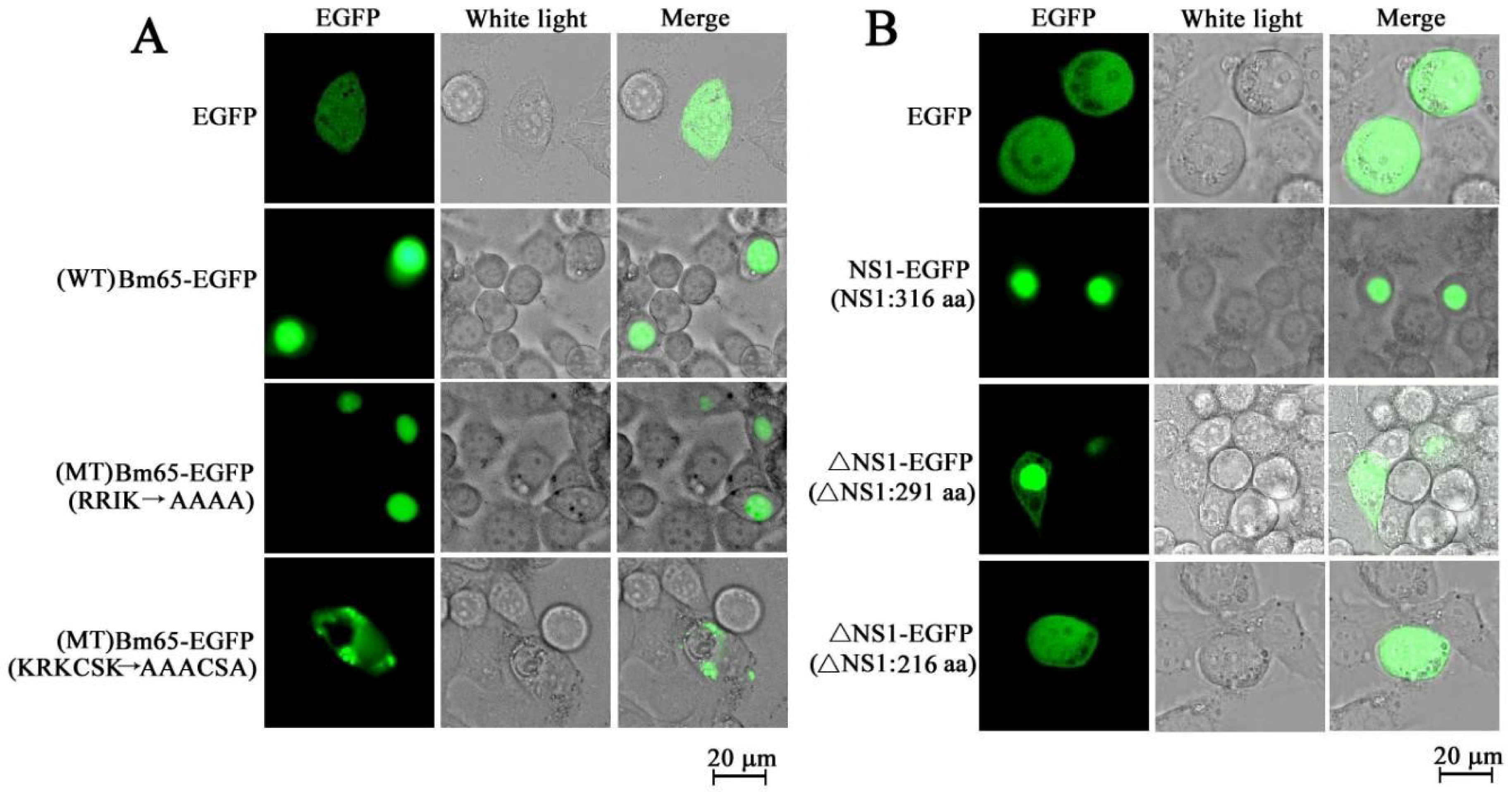
- Tang, Q.; Hu, Z.; Yang, Y.; Wu, H.; Qiu, L.; Chen, K.; Li, G. Overexpression of Bm65 correlates with reduced susceptibility to inactivation by UV light. J. Invertebr. Pathol. 2015, 127, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Dong, Z.; Hu, N.; Hu, Z.; Dong, F.; Jiang, Y.; Li, J.; Chen, P.; Lu, C.; Pan, M. Baculovirus LEF-11 nuclear localization signal is important for viral DNA replication. Virus Res. 2017, 238, 133–140. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Wu, P.; Hu, Z.; Yang, Y.; Qiu, L.; Liu, H.; Zhu, S.; Guo, Z.; Xia, H.; Chen, K.; et al. Evidence for the role of BmNPV Bm65 protein in the repair of ultraviolet-induced DNA damage. J. Invertebr. Pathol. 2017, 149, 82–86. [Google Scholar] [CrossRef]
- Olson, V.A.; Wetter, J.A.; Friesen, P.D. Baculovirus Transregulator IE1 Requires a Dimeric Nuclear Localization Element for Nuclear Import and Promoter Activation. J. Virol. 2002, 76, 9505–9515. [Google Scholar] [CrossRef]
- Tang, Q.; Li, G.; Yao, Q.; Chen, L.; Feng, F.; Yuan, Y.; Chen, K. Bm65 is essential for the propagation of Bombyx mori nucleopolyhedrovirus. Curr. Microbiol. 2013, 66, 22–29. [Google Scholar] [CrossRef]
- Li, G.; Li, M.; Wang, P.; Hu, Z.; Yao, Q.; Tang, Q.; Chen, K. Characterization of recombinant expression of Bombyx mori bidensovirus ns1 using a modified vector. Acta Biochim. Pol. 2014, 61, 787–794. [Google Scholar] [CrossRef]
- Li, G.; Li, M.; Xu, W.; Zhou, Q.; Hu, Z.; Tang, Q.; Chen, K.; Yao, Q. Regulation of BmBDV NS1 by phosphorylation: Impact of mutagenesis at consensus phosphorylation sites on ATPase activity and cytopathic effects. J. Invertebr. Pathol. 2016, 133, 66–72. [Google Scholar] [CrossRef]
- Summers, M. Electron microscopic observations on granulosis virus entry, uncoating and replication processes during infection of the midgut cells of Trichoplusia ni. J. Ultrastruct. Res. 1971, 35, 606–625. [Google Scholar] [CrossRef]
- Au, S.; Wu, W.; Zhou, L.; Theilmann, D.A.; Panté, N. A new mechanism for nuclear import by actin-based propulsion used by a baculovirus nucleocapsid. J. Cell Sci. 2016, 129, 2905–2911. [Google Scholar] [CrossRef] [PubMed]
- Sodeik, B. Microtubule-mediated Transport of Incoming Herpes Simplex Virus 1Capsids to the Nucleus. J. Cell Biol. 1997, 136, 1007–1021. [Google Scholar] [CrossRef] [PubMed]
- Ojala, P.M.; Sodeik, B.; Ebersold, M.W.; Kutay, U.; Helenius, A. Herpes Simplex Virus Type 1 Entry into Host Cells: Reconstitution of Capsid Binding and Uncoating at the Nuclear Pore Complex In Vitro. Mol. Cell. Biol. 2000, 20, 4922–4931. [Google Scholar] [CrossRef] [PubMed]
- Trotman, L.C.; Mosberger, N.; Fornerod, M.; Stidwill, R.P.; Greber, U.F. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001, 3, 1092–1100. [Google Scholar] [CrossRef]
- Modis, Y.; Trus, B.; Harrison, S. Atomic model of the papillomavirus capsid. At. Model Papillomavirus Capsid 2002, 21, 4754–4762. [Google Scholar] [CrossRef]
- Li, Z.; Yan, X.; Yu, H.; Wang, D.; Song, S.; Li, Y.; He, M.; Hong, Q.; Zheng, Q.; Zhao, Q.; et al. The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction. Structure 2016, 24, 874–885. [Google Scholar] [CrossRef]
- Aydin, I.; Villalonga-Planells, R.; Greune, L.; Bronnimann, M.P.; Calton, C.M.; Becker, M.; Lai, K.-Y.; Campos, S.K.; Schmidt, M.A.; Schelhaas, M. A central region in the minor capsid protein of papillomaviruses facilitates viral genome tethering and membrane penetration for mitotic nuclear entry. PLoS Pathog. 2017, 13, 1006308. [Google Scholar] [CrossRef]
- Oess, S.; Hildt, E. Novel cell permeable motif derived from the PreS2-domain of hepatitis-B virus surface antigens. Gene Ther. 2000, 7, 750–758. [Google Scholar] [CrossRef]
- Brandenburg, B.; Stockl, L.; Gutzeit, C.; Roos, M.; Lupberger, J.; Schwartlander, R.; Gelderblom, H.; Sauer, I.M.; Hofschneider, P.H.; Hildt, E. A novel system for efficient gene transfer into primary human hepatocytes via cell-permeable hepatitis B virus-like particle. Hepatology 2005, 42, 1300–1309. [Google Scholar] [CrossRef]
- Kann, M.; Bischof, A.; Gerlich, W.H. In vitro model for the nuclear transport of the hepadnavirus genome. J. Virol. 1997, 71, 1310–1316. [Google Scholar] [PubMed]
- Lupberger, J.; Schaedler, S.; Peiran, A.; Hildt, E. Identification and characterization of a novel bipartite nuclear localization signal in the hepatitis B virus polymerase. World J. Gastroenterol. 2013, 19, 8000–8010. [Google Scholar] [CrossRef] [PubMed]
- Cotmore, S.F.; Tattersall, P. Parvovirus Diversity and DNA Damage Responses. Cold Spring Harb. Perspect. Biol. 2013, 5, a012989. [Google Scholar] [CrossRef]
- Cotmore, S.F.; Tattersall, P. Parvoviruses: Small Does Not Mean Simple. Annu. Rev. Virol. 2014, 1, 517–537. [Google Scholar] [CrossRef] [PubMed]
- Tu, M.; Liu, F.; Chen, S.; Wang, M.; Cheng, A. Role of capsid proteins in parvoviruses infection. Virol. J. 2015, 12, 114. [Google Scholar] [CrossRef]
- Vihinen-Ranta, M.; Kakkola, L.; Kalela, A.; Vilja, P.; Vuento, M.; Vihinen-Ranta, M. Characterization of a Nuclear Localization Signal of Canine Parvovirus Capsid Proteins. JBIC J. Biol. Inorg. Chem. 1997, 250, 389–394. [Google Scholar] [CrossRef] [PubMed]
- Boisvert, M.; Bouchard-Lévesque, V.; Fernandes, S.; Tijssen, P. Classic Nuclear Localization Signals and a Novel Nuclear Localization Motif Are Required for Nuclear Transport of Porcine Parvovirus Capsid Proteins. J. Virol. 2014, 88, 11748–11759. [Google Scholar] [CrossRef]
- Kozlov, E.N.; Mukha, D.V. Mammalian cell culture as a model for studying the intracellular traffic of densovirus proteins. Genetika 2015, 51, 218–222. [Google Scholar] [CrossRef]
- Kozlov, E.N.; Martynova, E.U.; Popenko, V.I.; Schal, C.; Mukha, D.V. Intracellular Localization of Blattella germanica Densovirus (BgDV1) Capsid Proteins. Viruses 2018, 10, 370. [Google Scholar] [CrossRef]
- Adams, M.J.; Carstens, E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2012). Arch. Virol. 2012, 157, 1411–1422. [Google Scholar] [CrossRef]
- Hu, Z.; Li, G.; Li, G.; Yao, Q.; Chen, K. Bombyx mori bidensovirus: The type species of the new genus Bidensovirus in the new family Bidnaviridae. Chin. Sci. Bull. 2013, 58, 4528–4532. [Google Scholar] [CrossRef]
- Hayakawa, T.; Kojima, K.; Nonaka, K.; Nakagaki, M.; Sahara, K.; Asano, S.I.; Iizuka, T.; Bando, H. Analysis of proteins encoded in the bipartite genome of a new type of parvo-like virus isolated from silkworm—Structural protein with DNA polymerase motif. Virus Res. 2000, 66, 101–108. [Google Scholar] [CrossRef]
- Wang, Y.J.; Yao, Q.; Chen, K.P.; Wang, Y.; Lu, J.; Han, X. Characterization of the genome structure of Bombyx mori densovirus (China isolate). Virus Genes 2007, 35, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Gupta, T.; Ito, K.; Kadono-Okuda, K.; Murthy, G.N.; Gowri, E.V.; Ponnuvel, K.M. Characterization and genome comparison of an Indian isolate of bidensovirus infecting the silkworm Bombyx mori. Arch. Virol. 2018, 163, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Xing, Y.; Hu, Z.; Yang, Y.; Pan, Y.; Chen, K.; Zhu, F.; Zhou, Y.; Chen, K.; Yao, Q. A characterization of structural proteins expressed by Bombyx mori bidensovirus. J. Invertebr. Pathol. 2017, 144, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Colson, P.; De Lamballerie, X.; Yutin, N.; Asgari, S.; Bigot, Y.; Bideshi, D.K.; Cheng, X.W.; Federici, B.A.; Van Etten, J.L.; Koonin, E.V.; et al. “Megavirales”, a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch. Virol. 2013, 158, 2517–2521. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhang, J.; Liu, H.; Gao, Y.; Li, X.; Zheng, L.; Cui, R.; Yao, Q.; Rong, L.; Huang, Z.; et al. Unique 5′-P recognition and basis for dG:dGTP misincorporation of ASFV DNA polymerase X. PLoS Biol. 2017, 15, 1002599. [Google Scholar] [CrossRef] [PubMed]
- Czarnecki, M.W.; Traktman, P. The vaccinia virus DNA polymerase and its processivity factor. Virus Res. 2017, 234, 193–206. [Google Scholar] [CrossRef]
- Oliveros, M.; Yáñez, R.J.; Salas, M.L.; Salas, J.; Viñuela, E.; Blanco, L. Characterization of an African Swine Fever Virus 20-kDa DNA Polymerase Involved in DNA Repair. J. Biol. Chem. 1997, 272, 30899–30910. [Google Scholar] [CrossRef]
- García-Escudero, R.; García-Díaz, M.; Salas, M.L.; Blanco, L.; Salas, J. DNA Polymerase X of African Swine Fever Virus: Insertion Fidelity on Gapped DNA substrates and AP lyase Activity Support a Role in Base Excision Repair of Viral DNA. J. Mol. Biol. 2003, 326, 1403–1412. [Google Scholar] [CrossRef]
- Sobhy, H. A comparative review of viral entry and attachment during large and giant dsDNA virus infections. Arch. Virol. 2017, 162, 3567–3585. [Google Scholar] [CrossRef] [PubMed]
- Radziwill, G.; Tucker, W.; Schaller, H. Mutational analysis of the hepatitis B virus P gene product: Domain structure and RNase H activity. J. Virol. 1990, 64, 613–620. [Google Scholar] [PubMed]
- Chen, G.; Yan, Q.; Fang, Y.; Wu, L.; Krell, P.J.; Feng, G. The N Terminus of Autographa californica Multiple Nucleopolyhedrovirus DNA Polymerase Is Required for Efficient Viral DNA Replication and Virus and Occlusion Body Production. J. Virol. 2018, 92, e00398-18. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Fang, Y.; Wu, L.; Yan, Q.; Krell, P.J.; Feng, G. A betabaculovirus DNA polymerase cannot substitute for the DNA polymerase of the alphabaculovirus Autographa californica nucleopolyhedrovirus. Arch. Virol. 2017, 162, 3487–3492. [Google Scholar] [CrossRef]
- Zarrouk, K.; Piret, J.; Boivin, G. Herpesvirus DNA polymerases: Structures, functions and inhibitors. Virus Res. 2017, 234, 177–192. [Google Scholar] [CrossRef]
- Lawler, J.L.; Mukherjee, P.; Coen, D.M. Herpes Simplex Virus 1 DNA Polymerase RNase H Activity Acts in a 3′-to-5′ Direction and Is Dependent on the 3′-to-5′ Exonuclease Active Site. J. Virol. 2018, 92, e01813-17. [Google Scholar] [CrossRef]
- Menéndez-Arias, L.; Alvarez, M.; Pacheco, B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase: Mechanism of action and resistance. Curr. Opin. Virol. 2014, 8, 1–9. [Google Scholar] [CrossRef]
- Mak, L.-Y.; Seto, W.-K.; Lai, C.-L.; Yuen, M.-F. DNA polymerase inhibitors for treating hepatitis B: A safety evaluation. Expert Opin. Drug Saf. 2016, 15, 383–392. [Google Scholar] [CrossRef]
- Ramachandra, M.; Padmanabhan, R. Expression, Nuclear Transport, and Phosphorylation of Adenovirus DNA Replication Proteins. Curr. Top. Microbiol. Immunol. 1995, 199, 49–88. [Google Scholar]
- Loregian, A.; Piaia, E.; Cancellotti, E.; Papini, E.; Marsden, H.S.; Palù, G. The Catalytic Subunit of Herpes Simplex Virus Type 1 DNA Polymerase Contains a Nuclear Localization Signal in the UL42-Binding Region. Virology 2000, 273, 139–148. [Google Scholar] [CrossRef]
- Alvisi, G.; Ripalti, A.; Ngankeu, A.; Giannandrea, M.; Caraffi, S.G.; Dias, M.M.; Jans, D.A. Human Cytomegalovirus DNA Polymerase Catalytic Subunit pUL54 Possesses Independently Acting Nuclear Localization and ppUL44 Binding Motifs. Traffic 2006, 7, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Alvisi, G.; Musiani, D.; Jans, D.A.; Ripalti, A. An importin alpha/beta-recognized bipartite nuclear localization signal mediates targeting of the human herpes simplex virus type 1 DNA polymerase catalytic subunit pUL30 to the nucleus. Biochemistry 2007, 46, 9155–9163. [Google Scholar] [CrossRef]
- Feng, G.; Krell, P.J. Autographa californica Multiple Nucleopolyhedrovirus DNA Polymerase C Terminus Is Required for Nuclear Localization and Viral DNA Replication. J. Virol. 2014, 88, 10918–10933. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, G.; Fang, Y.; Hu, Z.; Krell, P.J.; Feng, G. Rescue of dnapol-null Autographa californica multiple nucleopolyhedrovirus with DNA polymerase (DNApol) of Spodoptera litura nucleopolyhedrovirus (SpltNPV) and identification of a nuclear localization signal in SpltNPV DNApol. J. Gen. Virol. 2016, 97, 1968–1980. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.-P.; Du, W.-J.; Huang, L.-P.; Wei, Y.-W.; Wu, H.-L.; Feng, L.; Liu, C.-M. The Pseudorabies Virus DNA Polymerase Accessory Subunit UL42 Directs Nuclear Transport of the Holoenzyme. Front. Microbiol. 2016, 7, 547. [Google Scholar] [CrossRef]
- Krupovič, M.; Koonin, E.V. Evolution of eukaryotic single-stranded DNA viruses of the Bidnaviridae family from genes of four other groups of widely different viruses. Sci. Rep. 2014, 4, 5347. [Google Scholar] [CrossRef]
- Tijssen, P.; Bergoin, M. Densonucleosis viruses constitute an increasingly diversified subfamily among the parvoviruses. Semin. Virol. 1995, 6, 347–355. [Google Scholar] [CrossRef]
- Krupovič, M. Networks of evolutionary interactions underlying the polyphyletic origin of ssDNA viruses. Curr. Opin. Virol. 2013, 3, 578–586. [Google Scholar] [CrossRef]
- Schildgen, O.; Qiu, J.; Söderlund-Venermo, M. Genomic features of the human bocaviruses. Futur. Virol. 2012, 7, 31–39. [Google Scholar] [CrossRef]
- Guido, M.; Tumolo, M.R.; Verri, T.; Romano, A.; Serio, F.; De Giorgi, M.; De Donno, A.; Bagordo, F.; Zizza, A. Human bocavirus: Current knowledge and future challenges. World J. Gastroenterol. 2016, 22, 8684–8697. [Google Scholar] [CrossRef]
- Qiu, J.; Söderlund-Venermo, M.; Young, N.S. Human Parvoviruses. Clin. Microbiol. Rev. 2017, 30, 43–113. [Google Scholar] [CrossRef] [PubMed]
- Wiersbitzky, S.; Bruns, R.; Ballke, E.-H.; Wiersbitzky, H.; Schwarz, T.F.; Roggendorf, M.; Deinhardt, F. Acute obstructive respiratory diseases in infants and children associated with parvovirus B19 infection. Infection 1991, 19, 252. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Zhu, Q.-R.; Wang, X.-H.; Yu, H.; Shen, J. Human bocavirus in children with respiratory tract infection in Shanghai: A retrospective study. World J. Pediatr. 2010, 6, 65–70. [Google Scholar] [CrossRef]
- Kerr, J.R. The role of parvovirus B19 in the pathogenesis of autoimmunity and autoimmune disease. J. Clin. Pathol. 2016, 69, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Hii, H.; Chiu, C.; Lin, D.; Shi, Y.; Hsu, T.; Tzang, B. Selective activation of inflammation factors by human parvovirus B19 and human bocavirus VP1 unique region on H9c2 cardiomyocyte. Mol. Med. Rep. 2018, 18, 4072–4078. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, D.; Kanda, T.; Murata, T.; Saito, S.; Sugimoto, A.; Narita, Y.; Tsurumi, T. Nuclear Transport of Epstein-Barr Virus DNA Polymerase Is Dependent on the BMRF1 Polymerase Processivity Factor and Molecular Chaperone Hsp90. J. Virol. 2013, 87, 6482–6491. [Google Scholar] [CrossRef] [PubMed]
- Olesen, A.S.; Lohse, L.; Dalgaard, M.D.; Woźniakowski, G.; Belsham, G.J.; Bøtner, A.; Rasmussen, T.B. Complete genome sequence of an African swine fever virus (ASFV POL/2015/Podlaskie) determined directly from pig erythrocyte-associated nucleic acid. J. Virol. Methods 2018, 261, 14–16. [Google Scholar] [CrossRef]
- Zou, W.; Wang, Z.; Xiong, M.; Chen, A.Y.; Xu, P.; Ganaie, S.S.; Badawi, Y.; Kleiboeker, S.; Nishimune, H.; Ye, S.Q.; et al. Human Parvovirus B19 Utilizes Cellular DNA Replication Machinery for Viral DNA Replication. J. Virol. 2018, 92, e01881-17. [Google Scholar] [CrossRef]
- Zhang, J.; Dong, Z.-Q.; Zhang, C.-D.; He, Q.; Chen, X.-M.; Cao, M.-Y.; Li, H.-Q.; Xiao, W.-F.; Lu, C.; Pan, M.-H. Identification of a novel nuclear localization signal of baculovirus late expression factor 11. Virus Res. 2014, 184, 111–119. [Google Scholar] [CrossRef]
- Wang, D.; Álvarez-Cabrera, A.L.; Chen, X.S. Study of SV40 large T antigen nucleotide specificity for DNA unwinding. Virol. J. 2017, 14, 79. [Google Scholar] [CrossRef]
- Murakami, I.; Egawa, N.; Griffin, H.; Yin, W.; Kranjec, C.; Nakahara, T.; Kiyono, T.; Doorbar, J. Roles for E1-independent replication and E6-mediated p53 degradation during low-risk and high-risk human papillomavirus genome maintenance. PLoS Pathog. 2019, 15, e1007755. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; De Jong, J.; Nagy, É.; Theilmann, D.A.; Krell, P.J. Nuclear Translocation Sequence and Region in Autographa californica Multiple Nucleopolyhedrovirus ME53 That Are Important for Optimal Baculovirus Production. J. Virol. 2016, 90, 3953–3965. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-H.; Lin, B.Y.; Deng, W.; Broker, T.R.; Chow, L.T. Mitogen-Activated Protein Kinases Activate the Nuclear Localization Sequence of Human Papillomavirus Type 11 E1 DNA Helicase to Promote Efficient Nuclear Import. J. Virol. 2007, 81, 5066–5078. [Google Scholar] [CrossRef] [PubMed]
- Bergvall, M.; Melendy, T.; Archambault, J. The E1 proteins. Virology 2013, 445, 35–56. [Google Scholar] [CrossRef] [PubMed]
- Jehle, J.A.; Blissard, G.W.; Bonning, B.C.; Cory, J.S.; Herniou, E.A.; Rohrmann, G.F.; Theilmann, D.A.; Thiem, S.M.; Vlak, J.M.; Herniou, E. On the classification and nomenclature of baculoviruses: A proposal for revision. Arch. Virol. 2006, 151, 1257–1266. [Google Scholar] [CrossRef]
- Miele, S.A.B.; Garavaglia, M.J.; Belaich, M.N.; Ghiringhelli, P.D. Baculovirus: Molecular Insights on Their Diversity and Conservation. Int. J. Evol. Biol. 2011, 2011, 1–15. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Suryamohan, K.; Kuriakose, B.; Janakiraman, V.; Reichelt, M.; Chaudhuri, S.; Guillory, J.; Divakaran, N.; Rabins, P.E.; Goel, R.; et al. Comprehensive analysis of single molecule sequencing-derived complete genome and whole transcriptome of Hyposidra talaca nuclear polyhedrosis virus. Sci. Rep. 2018, 8, 8924. [Google Scholar] [CrossRef]
- Chen, Z.; Carstens, E.B. Identification of Domains in Autographa californica Multiple Nucleopolyhedrovirus Late Expression Factor 3 Required for Nuclear Transport of P143. J. Virol. 2005, 79, 10915–10922. [Google Scholar] [CrossRef]
- Au, V.; Yu, M.; Carstens, E.B. Characterization of a baculovirus nuclear localization signal domain in the late expression factor 3 protein. Virology 2009, 385, 209–217. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Liang, C.; Song, J.; Li, N.; Shi, H.; Chen, X. Autographa californica Multiple Nucleopolyhedrovirus Nucleocapsid Protein BV/ODV-C42 Mediates the Nuclear Entry of P78/83. J. Virol. 2008, 82, 4554–4561. [Google Scholar] [CrossRef]
- Hepp, S.E.; Borgo, G.M.; Ticau, S.; Ohkawa, T.; Welch, M.D. Baculovirus AC102 Is a Nucleocapsid Protein That Is Crucial for Nuclear Actin Polymerization and Nucleocapsid Morphogenesis. J. Virol. 2018, 92, e00111-18. [Google Scholar] [CrossRef] [PubMed]
- Marek, M.; Merten, O.-W.; Galibert, L.; Vlak, J.M.; Van Oers, M.M. Baculovirus VP80 Protein and the F-Actin Cytoskeleton Interact and Connect the Viral Replication Factory with the Nuclear Periphery. J. Virol. 2011, 85, 5350–5362. [Google Scholar] [CrossRef] [PubMed]
- Ginzberg, D.; Wong, R.J.; Gish, R. Global HBV burden: Guesstimates and facts. Hepatol. Int. 2018, 12, 315–329. [Google Scholar] [CrossRef] [PubMed]

| Num | Virus Name | Viral Genome (size) | dsDNA or ssDNA | Viral Polymerase | NLS Motif | References |
|---|---|---|---|---|---|---|
| 1 | VACV | 192 kb | dsDNA | E9 | No NLS | [58] |
| 2 | ASFV | 189 kb | dsDNA | PolX | No NLS | [25,87] |
| 3 | HAd2V | 36 kb | dsDNA | Ad2V Pol | RARR11, RRRVR29, RARRRR46 | [69] |
| 4 | HCMV | 236 kb | dsDNA | UL54 | NLSA (PAKKRAR1159), NLSB (PRRLHL1227) | [71] |
| 5 | HSV-1 | 153 kb | dsDNA | UL30 | RRMLHR1229, PRRSRLW130, PAKRPRETPSPADPPGGASKPRK1136 | [70,72] |
| 6 | HBV | 3.2 kb | dsDNA | P protein | a bipartite nuclear localization signal (residues K90-K91, K104-R106) | [34] |
| 7 | AcMNPV | 134 kb | dsDNA | DNApol | DNPGKKRKSTDDNEGPSPKRRVIT827, CSVKRKRDDD948 | [73] |
| 8 | SpltNPV | 139 kb | dsDNA | DNApol | QE PPA KRARMPT838 | [74] |
| 9 | PRV | 143 kb | dsDNA | UL42 | KRPAAPRMYTPIAKRPR370 | [75] |
| 10 | BmBDV | VD1 (6543 nts); VD2 (6022 nts). | ssDNA | BmBDV pPolB | Unclear | [51] |
| 11 | B19 virus | 5.6 nts | ssDNA | No | No | [81,88] |
| 12 | HBoV1 | 5.3 nts | ssDNA | No | No | [79,80] |
| Num | Target Proteins | Predicted MW | AcMNPV ORF | Homlogs in BmNPV | Potential NLS |
|---|---|---|---|---|---|
| 1 | VP39 | 39 kDa | 89 | BmNPV Orf76 | 52HLIKRFKMS |
| 2 | 38K | 38 kDa | 98 | BmNPV Orf86 | 13RLNDAIIKRHVLVLSEYADLKYLGFEKYKFFEY |
| 3 | BV/ODV-EC27 | 34 kDa | 144 | BmNPV Orf128 | 2KRIKCNKVRTVTEIVNSDEKIQKTYEL |
| 4 | VP80 | 80 kDa | 104 | BmNPV Orf92 | 424KRSAEDDLLPTRSSKR; 464YEKESKRRKLEDEDF |
| 5 | VLF-1 | 44 kDa | 77 | BmNPV Orf67 | 225LIKRGKLHSDTINLKRKRSRNN |
| 6 | BV/ODV-C42 | 42 kDa | 101 | BmNPV Orf 89 | 357KRKK |
| 7 | P78/83 | 61 kDa | 9 | BmNPV Orf2 | No NLS |
| 8 | 49K | 55 kDa | 142 | BmNPV Orf126 | No NLS |
| 9 | Ac109 | 45 kDa | 109 | BmNPV Orf96 | No NLS |
| 10 | VP1054 | 42 kDa | 54 | BmNPV Orf46 | No NLS |
| 11 | Ac102 | 13 kDa | 102 | BmNPV Orf90 | No NLS |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, G.; Qi, X.; Hu, Z.; Tang, Q. Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses. Viruses 2019, 11, 1035. https://doi.org/10.3390/v11111035
Li G, Qi X, Hu Z, Tang Q. Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses. Viruses. 2019; 11(11):1035. https://doi.org/10.3390/v11111035
Chicago/Turabian StyleLi, Guohui, Xinyu Qi, Zhaoyang Hu, and Qi Tang. 2019. "Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses" Viruses 11, no. 11: 1035. https://doi.org/10.3390/v11111035
APA StyleLi, G., Qi, X., Hu, Z., & Tang, Q. (2019). Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses. Viruses, 11(11), 1035. https://doi.org/10.3390/v11111035





