The Effect of Sample Bias and Experimental Artefacts on the Statistical Phylogenetic Analysis of Picornaviruses
Abstract
1. Introduction
2. Materials and Methods
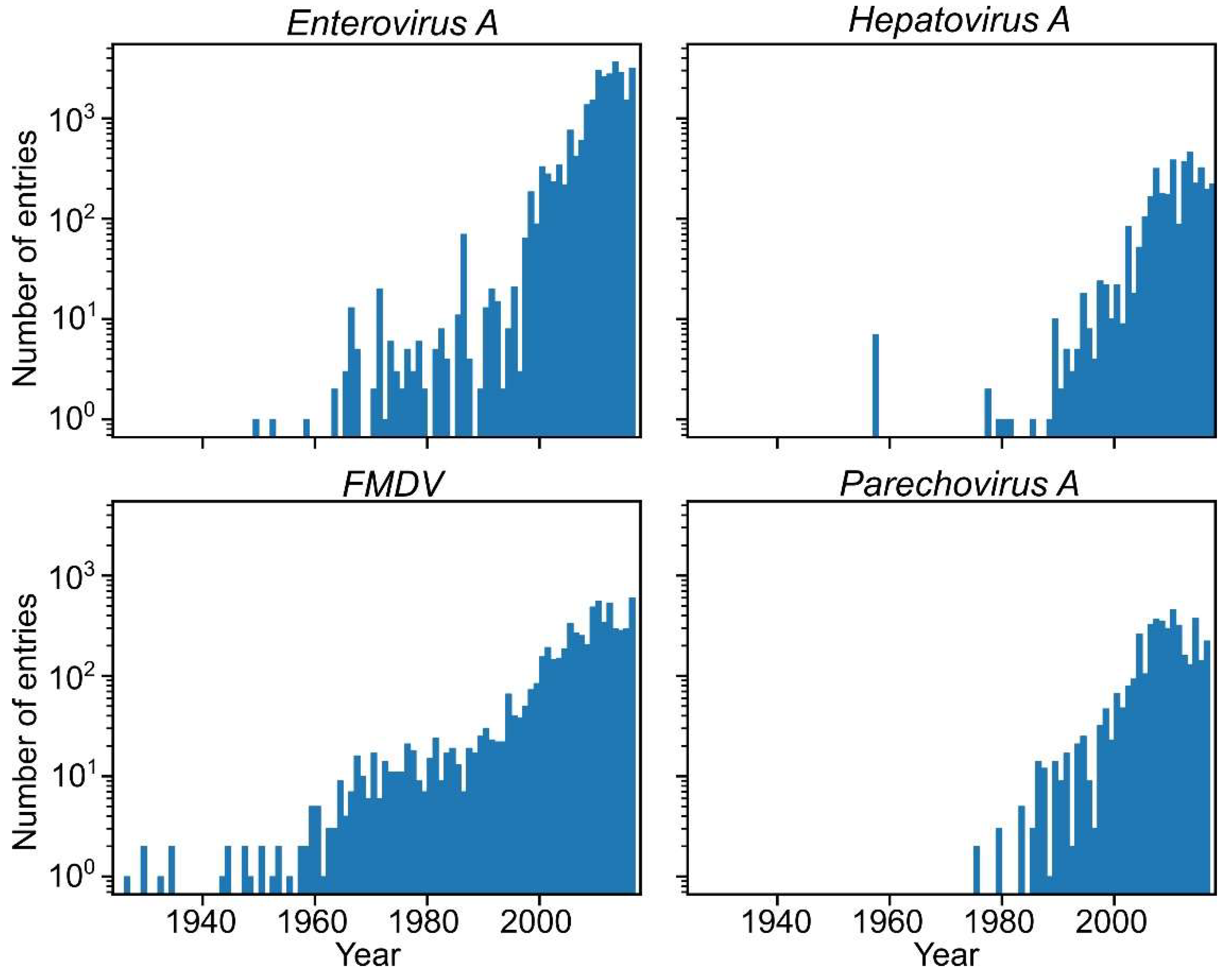
2.1. Distribution of Picornavirus Collection Dates
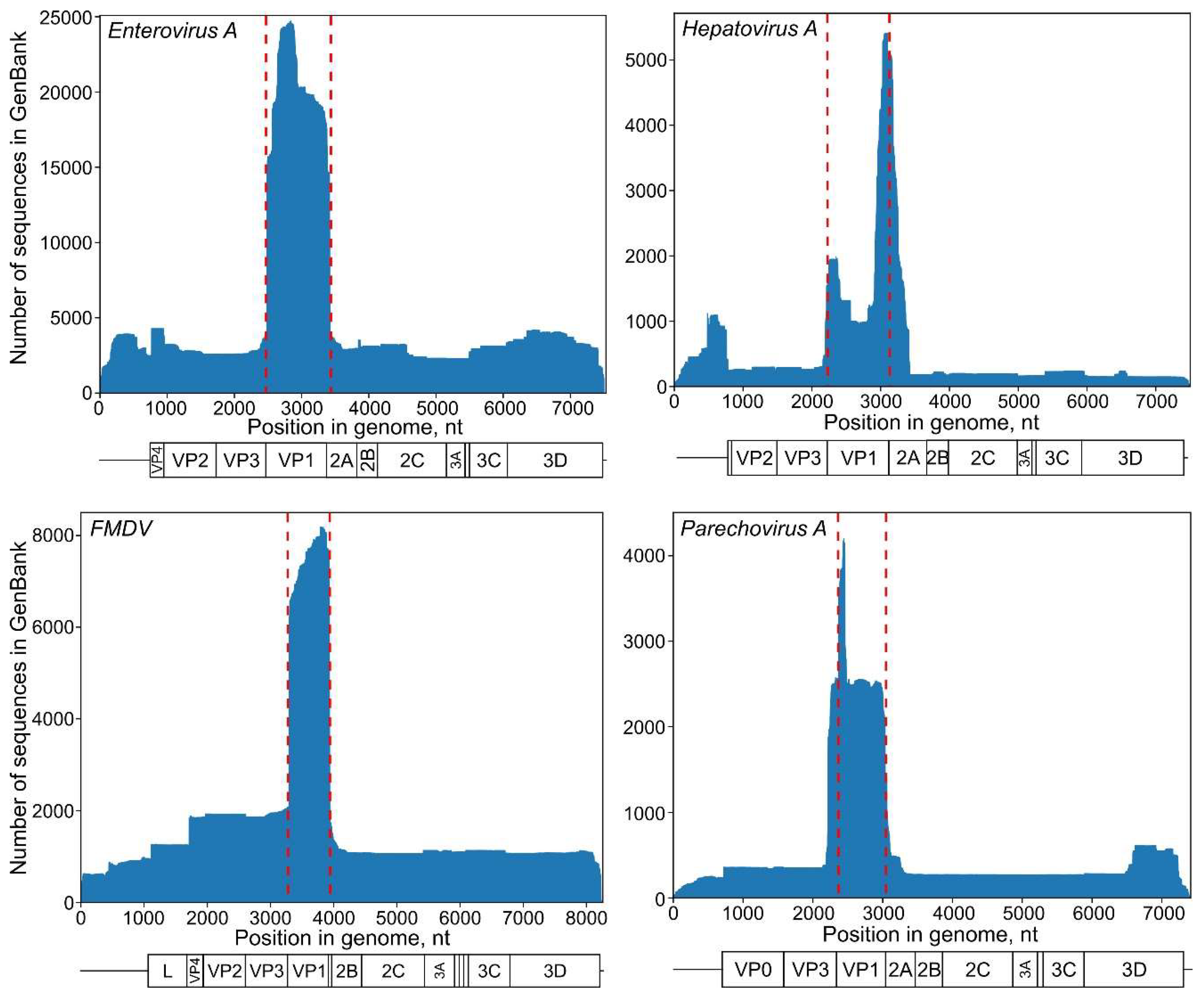
2.2. Distribution of Genome Fragments Deposited in GenBank along the Genome
2.3. Preparation of the EV-A71 Reference Alignment
2.4. Generating Random Sequence Sets from the Reference Alignment
- Random sampling of data subsets corresponding to distinct studies (“random groups”). All sequences in the reference alignment were partitioned into groups based on the first five characters of the accession number. A random group was then chosen, and all sequences from this group were added to the alignment. This step was repeated until the number of sequences reached the defined value. This algorithm reproduced the situation with enterovirus sampling 15 years ago, when only a few studies were done, or the currently available sample for less common types and species.
- Random sampling of sequences (“random single”). The selected number of sequences was randomly picked from the initial data set.
- Identity filtration of sequences. Sequences that differed from any other entry in the dataset by less than the selected percentage of the nucleotide sequence were omitted. The comparison of sequences by the script started from the first sequence in the dataset; therefore, the initial alignment was shuffled prior to each repetition of sampling.
- “Smart picking”. All sequences in the initial alignment were partitioned into groups based on the first five characters of the accession number. Sequences from the subsets with a size that did not exceed the user-defined threshold were all included in the final dataset; for bigger subsets, one sequence or a defined fraction of randomly chosen sequences was added to the reduced dataset. This sampling algorithm allowed the inclusion of unique sequences from small studies and reduced the number of sequences from massive epidemiological investigations. For each Genbank number range, at least two sequences were included, or 1% from the larger studies. These conditions were necessary to sufficiently reduce the large EV-A71 dataset; less stringent parameters may be recommended for taxa with fewer studies presented in GenBank.
2.5. Bayesian Phylogenetic Analysis
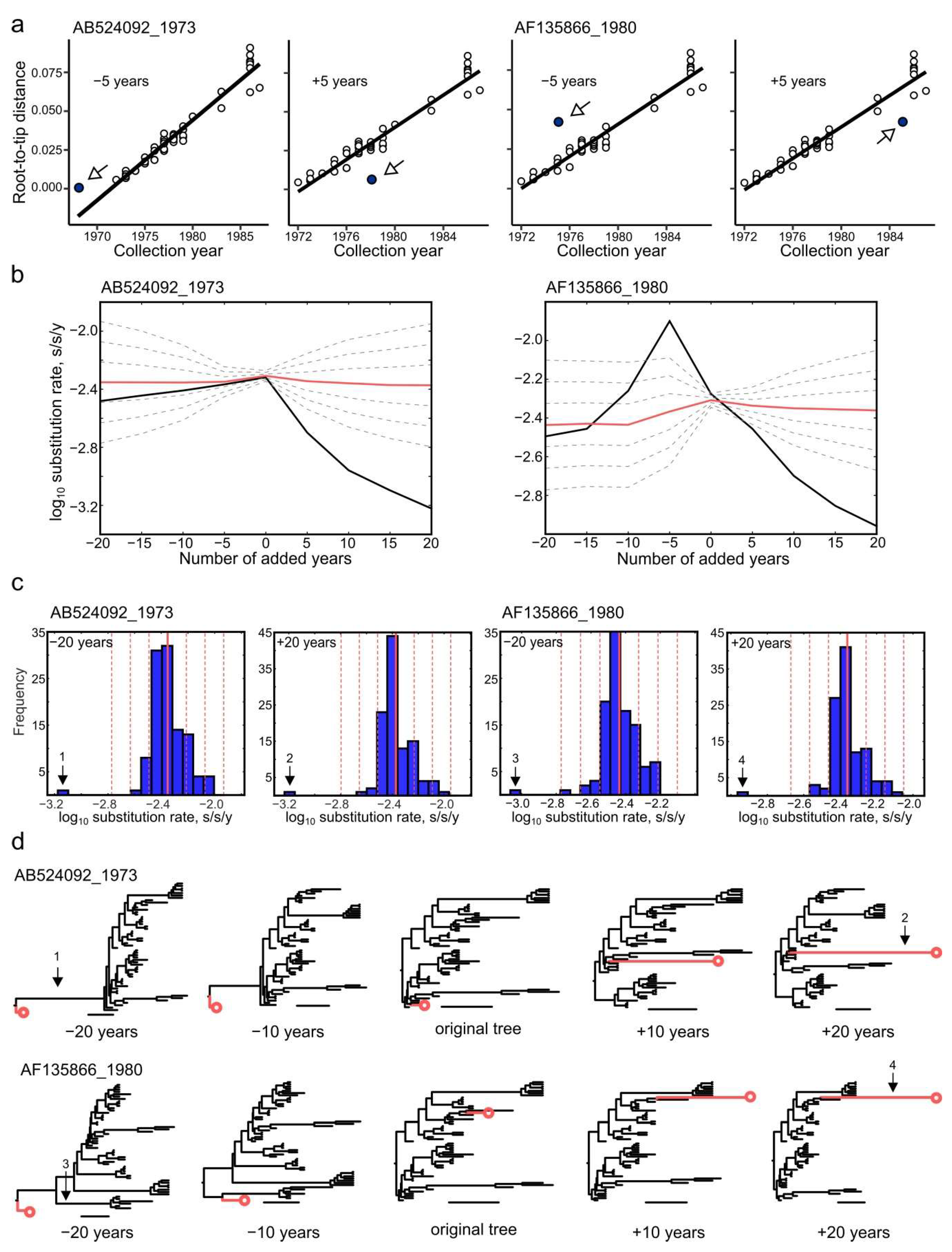
2.6. Effect of Sequencing Errors and Errors in Annotation on Evolutionary Estimates
3. Results and Discussion
3.1. Selection of a Genome Region and Recombination Analysis
3.2. Selection of a Taxon
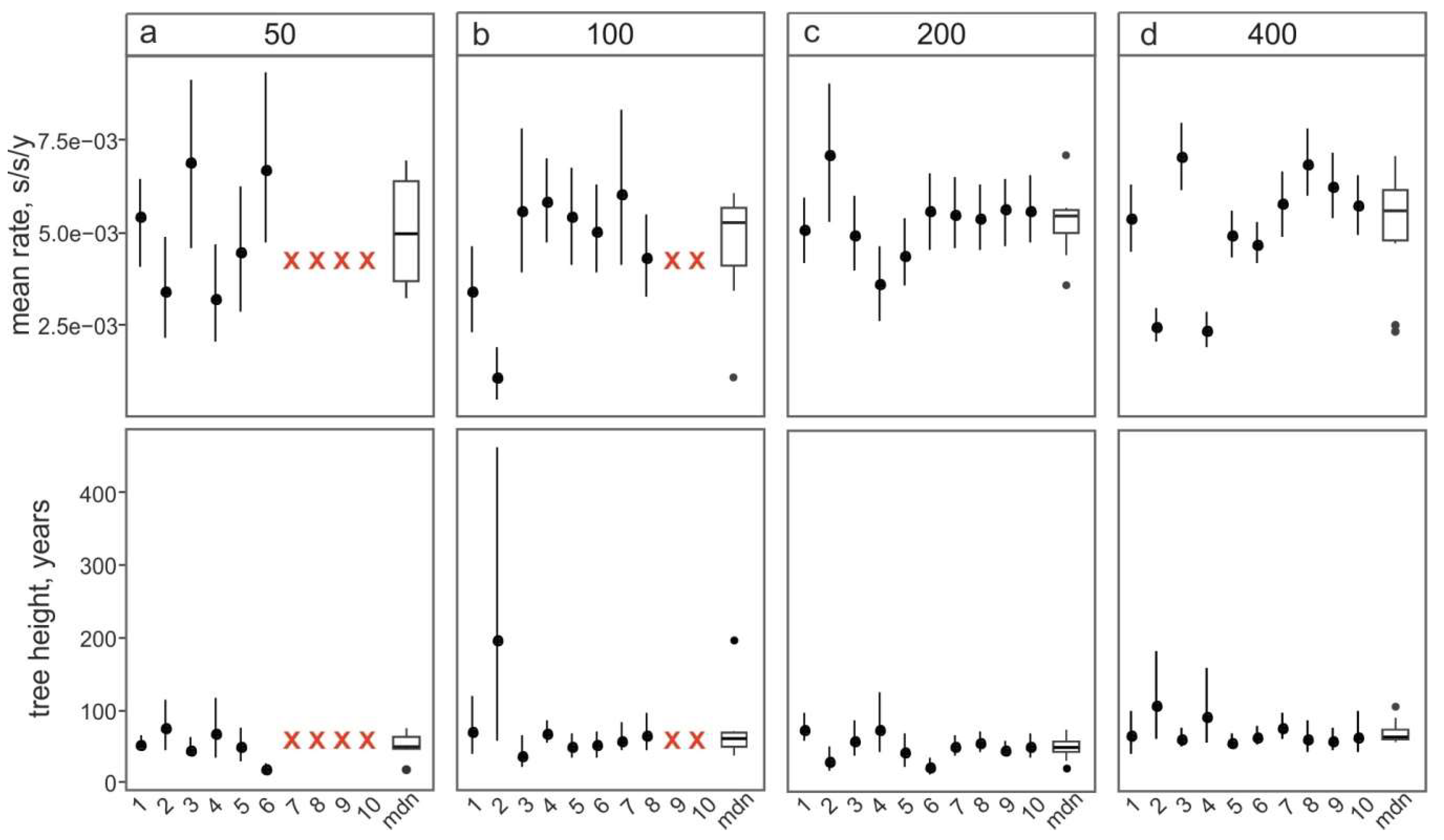
3.3. Effect of Dataset Size and Sample Bias on Evolutionary Estimates
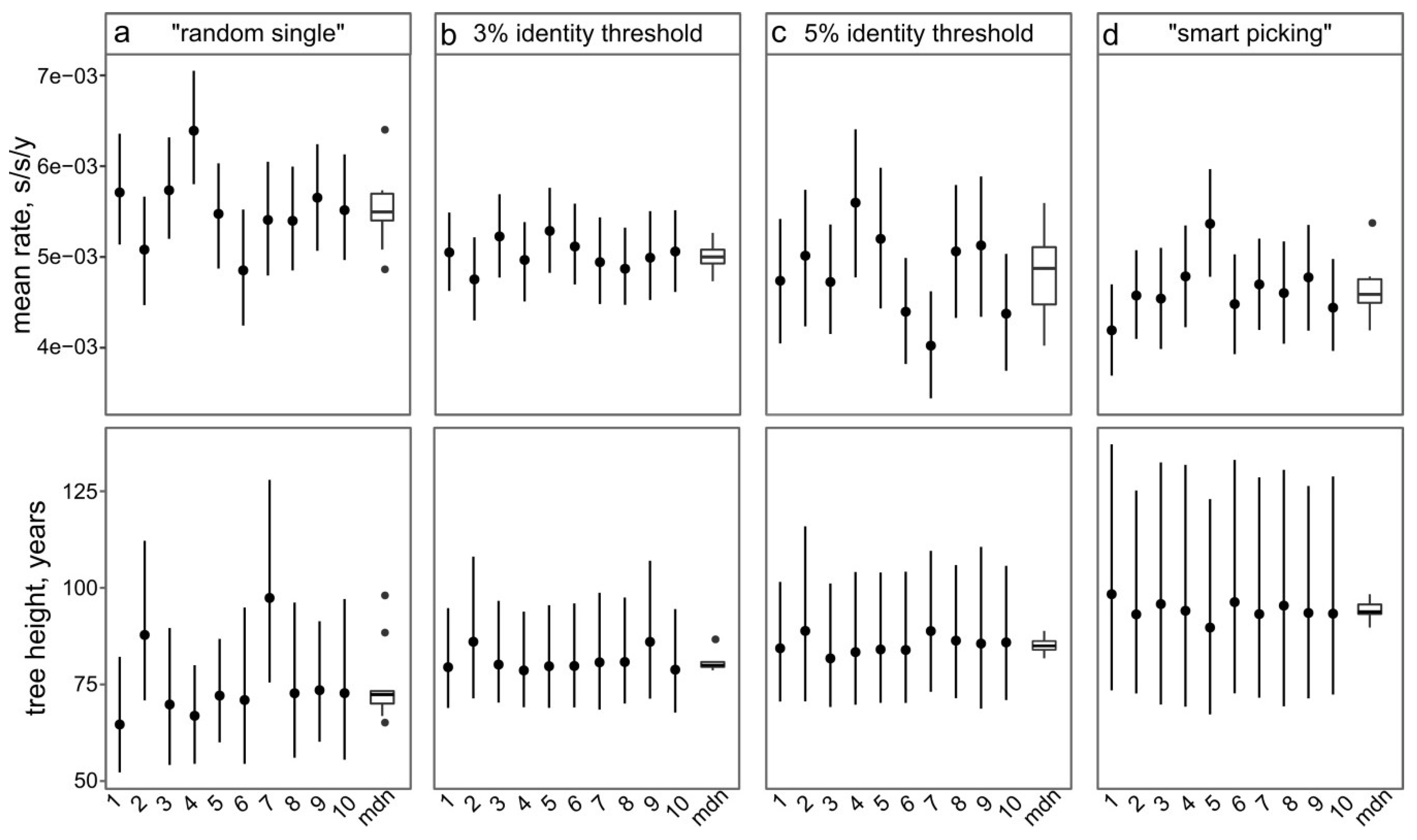
3.4. Approaches to Reducing the Dataset
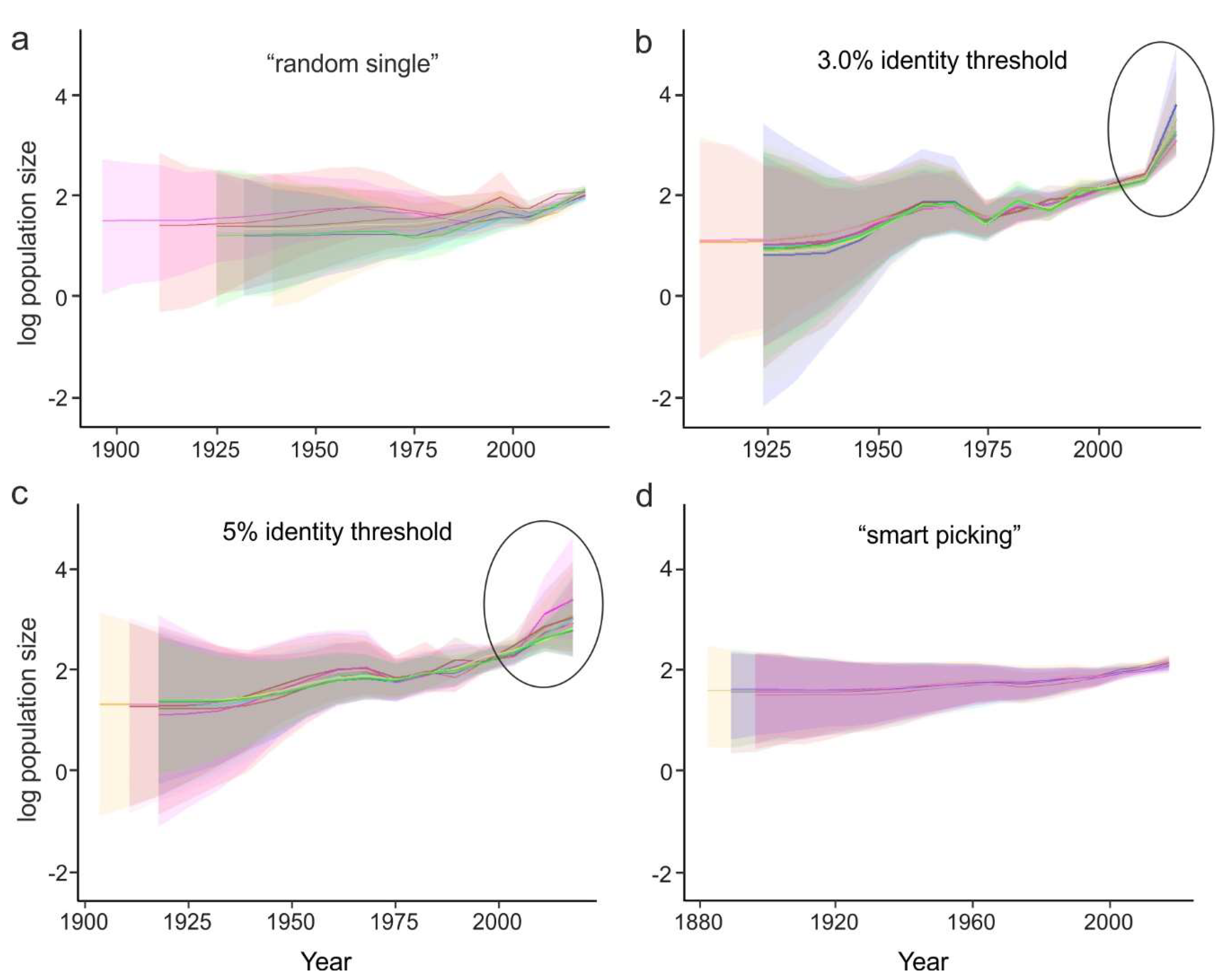
- Random sampling of sequences. In this case, the sequences from large studies (sometimes originating from narrow samples, such as outbreaks) would be over-represented, while rare (but very informative) sequences may be lost. This results in a significant variation of the key evolutionary estimates (Figure 4a) and the Bayesian estimation of past population dynamics using Bayesian Skygrid analysis (Figure 5a).
- Identity filtration—discarding sequences that are almost identical to each other. This can be done using Jalview [51], CD-HIT [52], UCLUST [53], the skipredundant tool in EMBOSS software [54], or in-house scripts. This approach ensures that the reduced dataset is as informative in terms of genetic diversity as the original one because all rare sequences are preserved. Identity filtration can be considered in most phylogenetic studies because it is simple and results in a much more limited variation of evolutionary estimates than random picking (Figure 4b). However, an overly stringent reduction can lead to increased deviations of evolutionary estimates (Figure 4c). In the case of statistical phylogenetic studies, identity filtration can introduce bias by itself. Most prominently, it can result in artefacts in the Bayesian Skygrid analysis, because artificial removal of similar sequences universally discards the most recent tree nodes and thus simulates explosive population growth at a time that corresponds to the cut-off threshold (Figure 5b,c, circled).
- “Smart picking” first identifies the sequences that most likely belong to distinct studies based on the first five characters (two letters and three digits) of the GenBank accession number. All small studies are then included, while datasets from larger studies are reduced by random sampling proportionally to their size. In this way, rare sequences are less likely to be lost, and no bias should be introduced. Indeed, the variation of evolutionary estimates in this case was low (Figure 4d), and no artefacts were apparent in the Skygrid analysis (Figure 5d). This algorithm was implemented using in-house scripts (available at https://github.com/v-julia/sample_bias). This method requires more manual tuning than the identity filtration to obtain the desired dataset size, and overly complicated datasets, such as EV-A71, may be difficult to reduce. However, smart sampling is also less prone to introducing additional bias in population dynamics analysis (Figure 5d in comparison to Figure 5b,c, circled), and produces reproducible population size estimates (Figure 5d).
3.5. Managing Ambiguous Sequence Characters
- Discard sequences with too many ambiguous characters, which may be a sign of poor sequence quality rather than natural heterogeneity (0.1%–0.2%, or one position in the full VP1, should have minimal effect on the analysis, see below);
- In the remaining sequences, identify a region (e.g., 100 nt) with an ambiguous character and blast it against the dataset;
- Identify the frequency of different nucleotides at this position in the most closely related sequences;
- Replace the ambiguous character with the most common nucleotide among the most closely related sequences.
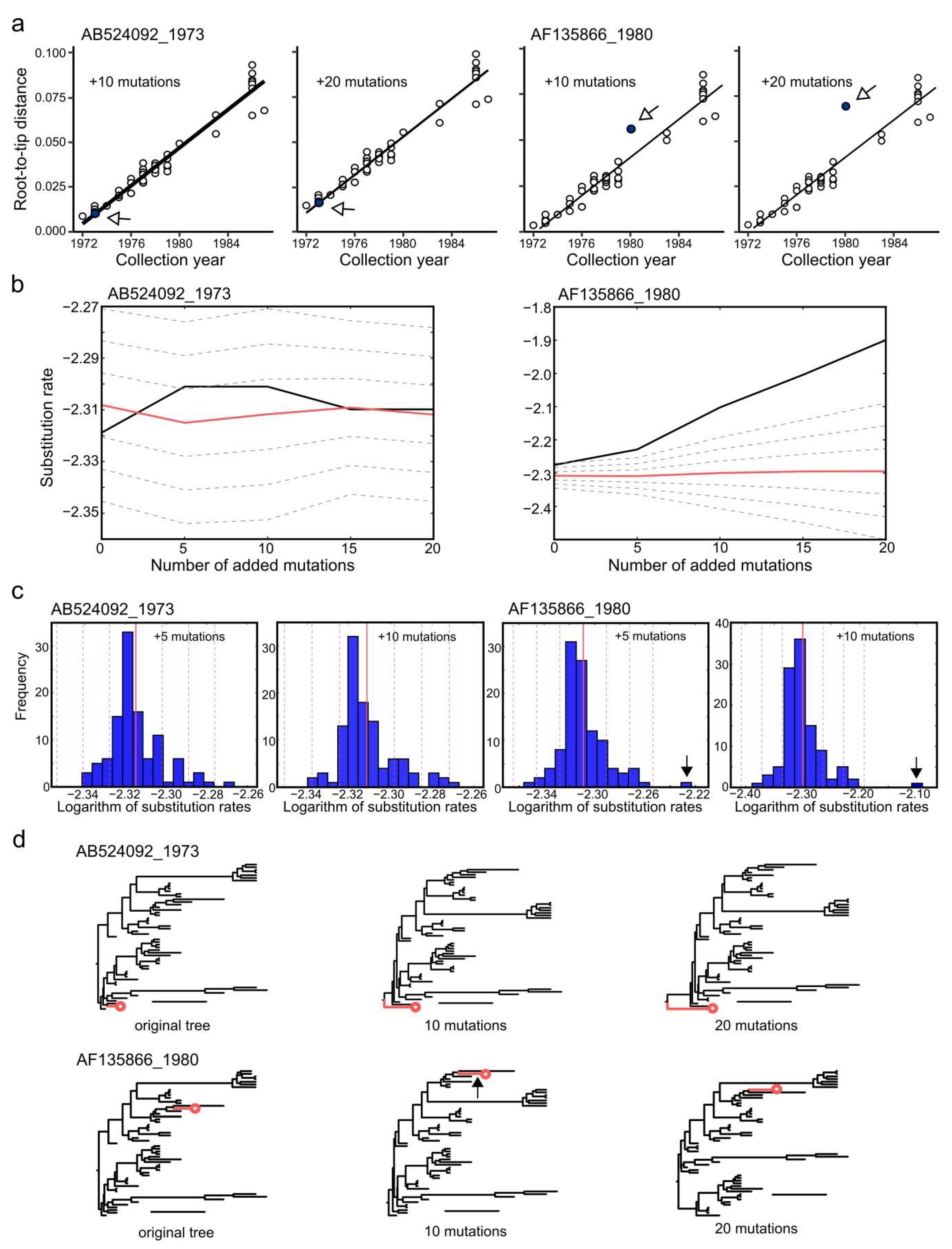
3.6. Effect of Sequencing Errors on Evolutionary Estimates
3.7. Effect of Annotation Errors on Evolutionary Estimates
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Moratorio, G.; Costa-Mattioli, M.; Piovani, R.; Romero, H.; Musto, H.; Cristina, J. Bayesian coalescent inference of hepatitis A virus populations: Evolutionary rates and patterns. J. Gen. Virol. 2007, 88, 3039–3042. [Google Scholar] [CrossRef]
- Cella, E.; Riva, E.; Angeletti, S.; Fogolari, M.; Blasi, A.; Scolamacchia, V.; Spoto, S.; Bazzardi, R.; Lai, A.; Sagnelli, C.; et al. Genotype I hepatitis A virus introduction in Italy: Bayesian phylogenetic analysis to date different epidemics. J. Med. Virol. 2018, 90, 1493–1502. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.-Y.; Zheng, H.-H.; Cao, J.-Y.; Zhou, W.-T.; Bi, S.-L. Evolution and genetic characterization of hepatitis A virus isolates in China. Int. J. Infect. Dis. 2015, 33, 156–158. [Google Scholar] [CrossRef]
- Ma, X.; Sheng, Z.; Huang, B.; Qi, L.; Li, Y.; Yu, K.; Liu, C.; Qin, Z.; Wang, D.; Song, M.; et al. Molecular Evolution and Genetic Analysis of the Major Capsid Protein VP1 of Duck Hepatitis A Viruses: Implications for Antigenic Stability. PLoS ONE 2015, 10, e0132982. [Google Scholar] [CrossRef]
- Brito, B.P.; Mohapatra, J.K.; Subramaniam, S.; Pattnaik, B.; Rodriguez, L.L.; Moore, B.R.; Perez, A.M. Dynamics of widespread foot-and-mouth disease virus serotypes A, O and Asia-1 in southern Asia: A Bayesian phylogenetic perspective. Transbound. Emerg. Dis. 2018, 65, 696–710. [Google Scholar] [CrossRef]
- Subramaniam, S.; Mohapatra, J.K.; Sharma, G.K.; Das, B.; Dash, B.B.; Sanyal, A.; Pattnaik, B. Phylogeny and genetic diversity of foot and mouth disease virus serotype Asia1 in India during 1964–2012. Vet. Microbiol. 2013, 167, 280–288. [Google Scholar] [CrossRef]
- Omondi, G.; Alkhamis, M.A.; Obanda, V.; Gakuya, F.; Sangula, A.; Pauszek, S.; Perez, A.; Ngulu, S.; van Aardt, R.; Arzt, J.; et al. Phylogeographical and cross-species transmission dynamics of SAT1 and SAT2 foot-and-mouth disease virus in Eastern Africa. Mol. Ecol. 2019, 28, 2903–2916. [Google Scholar] [CrossRef]
- Faria, N.R.; De Vries, M.; Van Hemert, F.J.; Benschop, K.; van der Hoek, L. Rooting human parechovirus evolution in time. BMC Evol. Biol. 2009, 9, 164. [Google Scholar] [CrossRef]
- Lukashev, A.; Vakulenko, Y. Molecular evolution of types in non-polio enteroviruses. J. Gen. Virol. 2017, 98, 2968–2981. [Google Scholar] [CrossRef] [PubMed]
- Hicks, A.L.; Duffy, S. Genus-Specific Substitution Rate Variability among Picornaviruses. J. Virol. 2011, 85, 7942–7947. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bessaud, M.; Razafindratsimandresy, R.; Nougairède, A.; Joffret, M.L.; Deshpande, J.M.; Dubot-Pérès, A.; Héraud, J.M.; De Lamballerie, X.; Delpeyroux, F.; Bailly, J.L. Molecular comparison and evolutionary analyses of VP1 nucleotide sequences of new African human enterovirus 71 isolates reveal a wide genetic diversity. PLoS ONE 2014, 9, e90624. [Google Scholar] [CrossRef] [PubMed]
- Tee, K.K.; Lam, T.T.-Y.; Chan, Y.F.; Bible, J.M.; Kamarulzaman, A.; Tong, C.Y.W.; Takebe, Y.; Pybus, O.G. Evolutionary genetics of human enterovirus 71: Origin, population dynamics, natural selection, and seasonal periodicity of the VP1 gene. J. Virol. 2010, 84, 3339–3350. [Google Scholar] [CrossRef] [PubMed]
- Jorba, J.; Campagnoli, R.; De, L.; Kew, O. Calibration of multiple poliovirus molecular clocks covering an extended evolutionary range. J. Virol. 2008, 82, 4429–4440. [Google Scholar] [CrossRef]
- Cano-Gómez, C.; Palero, F.; Buitrago, M.D.; García-Casado, M.A.; Fernández-Pinero, J.; Fernández-Pacheco, P.; Agüero, M.; Gómez-Tejedor, C.; Jiménez-Clavero, M.Á. Analyzing the genetic diversity of teschoviruses in Spanish pig populations using complete VP1 sequences. Infect. Genet. Evol. 2011, 11, 2144–2150. [Google Scholar] [CrossRef]
- Möller, S.; du Plessis, L.; Stadler, T. Impact of the tree prior on estimating clock rates during epidemic outbreaks. Proc. Natl. Acad. Sci. USA 2018, 115, 4200–4205. [Google Scholar] [CrossRef]
- Boskova, V.; Stadler, T.; Magnus, C. The influence of phylodynamic model specifications on parameter estimates of the Zika virus epidemic. Virus Evol. 2018, 4, 1–14. [Google Scholar] [CrossRef]
- Baele, G.; Lemey, P.; Bedford, T.; Rambaut, A.; Suchard, M.A.; Alekseyenko, A.V. Improving the Accuracy of Demographic and Molecular Clock Model Comparison While Accommodating Phylogenetic Uncertainty. Mol. Biol. Evol. 2012, 29, 2157–2167. [Google Scholar] [CrossRef]
- Russel, P.M.; Brewer, B.J.; Klaere, S.; Bouckaert, R.R. Model Selection and Parameter Inference in Phylogenetics Using Nested Sampling. Syst. Biol. 2019, 68, 219–233. [Google Scholar] [CrossRef]
- Nascimento, F.F.; dos Reis, M.; Yang, Z. A biologist’s guide to Bayesian phylogenetic analysis. Nat. Ecol. Evol. 2017, 1, 1446–1454. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A.; Vakulenko, Y.; Turbabina, N.; Deviatkin, A.; Drexler, J. Molecular epidemiology and phylogenetics of human enteroviruses: Is there a forest behind the trees? Rev. Med. Virol. 2018, 28, e2002. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kazunori, Y.D.; Tomii, K.; Katoh, K. Application of the MAFFT sequence alignment program to large data—Reexamination of the usefulness of chained guide trees. Bioinformatics 2016, 32, 3246–3251. [Google Scholar]
- Suchard, M.; Lemey, P.; Baele, G.; Ayres, D.; Drummond, A.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef]
- Shapiro, B.; Rambaut, A.; Drummond, A.J. Choosing appropriatesubstitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006, 23, 7–9. [Google Scholar] [CrossRef]
- Gill, M.S.; Lemey, P.; Faria, N.R.; Rambaut, A.; Shapiro, B.; Suchard, A.M. Improving Bayesian Population Dynamics Inference: A Coalescent-Based Model for Multiple Loci. Mol. Biol. Evol. 2013, 30, 713–714. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.; Xie, D.; Baele, G.; Suchard, M. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- FigTree 1.4.4. Available online: https://github.com/rambaut/figtree/releases (accessed on 1 June 2019).
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rambaut, A.; Lam, T.; Carvalho, L.; Pybus, O. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen). Virus Evol. 2016, 2, 2. [Google Scholar] [CrossRef]
- Simmonds, P. Recombination and selection in the evolution of picornaviruses and other Mammalian positive-stranded RNA viruses. J. Virol. 2006, 80, 11124–11140. [Google Scholar] [CrossRef] [PubMed]
- Lukashev, A. Recombination among picornaviruses. Rev. Med. Virol. 2010, 20, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bouslama, L.; Nasri, D.; Chollet, L.; Belguith, K.; Bourlet, T.; Aouni, M.; Pozzetto, B.; Pillet, S. Natural Recombination Event within the Capsid Genomic Region Leading to a Chimeric Strain of Human enterovirus B. J. Virol. 2007, 81, 8944–8952. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Drexler, J.F.; Belalov, I.S.; Eschbach-Bludau, M.; Baumgarte, S.; Drosten, C. Genetic variation and recombination in Aichi virus. J. Gen. Virol. 2012, 93, 1226–1235. [Google Scholar] [CrossRef]
- Belalov, I.S.; Isaeva, O.V.; Lukashev, A.N. Recombination in hepatitis A virus: Evidence for reproductive isolation of genotypes. J. Gen. Virol. 2011, 92, 860–872. [Google Scholar] [CrossRef]
- Xia, X. DAMBE7: New and Improved Tools for Data Analysis in Molecular Biology and Evolution. Mol. Biol. Evol. 2018, 35, 1550–1552. [Google Scholar] [CrossRef]
- Duchêne, S.; Ho, S.; Holmes, E.C. Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models. BMC Evol. Biol. 2015, 15, 36. [Google Scholar] [CrossRef]
- Duchêne, S.; Duchêne, D.; Holmes, E.C.; Ho, S.Y.W. The Performance of the Date-Randomization Test in Phylogenetic Analyses of Time-Structured Virus Data. Mol. Biol. Evol. 2015, 32, 1895–1906. [Google Scholar] [CrossRef]
- Murray, G.G.R.; Wang, F.; Harrison, E.M.; Paterson, G.K.; Mather, A.E.; Harris, S.R.; Holmes, M.A.; Rambaut, A.; Welch, J.J. The effect of genetic structure on molecular dating and tests for temporal signal. Methods Ecol. Evol. 2016, 7, 80–89. [Google Scholar] [CrossRef]
- Rieux, A.; Khatchikian, C.E. TipDatingBeast: An R package to assist the implementation of phylogenetic tip-dating tests using BEAST. Mol. Ecol. Resour. 2017, 17, 608–613. [Google Scholar] [CrossRef] [PubMed]
- Ballinger, M.J.; Bruenn, J.A.; Kotov, A.A.; Taylor, D.J. Selectively maintained paleoviruses in Holarctic water fleas reveal an ancient origin for phleboviruses. Virology 2013, 446, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Aiewsakun, P.; Katzourakis, A. Endogenous viruses: Connecting recent and ancient viral evolution. Virology 2015, 479–480, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Membrebe, J.V.; Suchard, M.A.; Rambaut, A.; Baele, G.; Lemey, P. Bayesian Inference of Evolutionary Histories under Time-Dependent Substitution Rates. Mol. Biol. Evol. 2019, 36, 1793–1803. [Google Scholar] [CrossRef] [PubMed]
- Smura, T.; Savolainen-Kopra, C.; Roivainen, M. Evolution of newly described enteroviruses. Future Virol. 2011, 6, 109–131. [Google Scholar] [CrossRef]
- Solomon, T.; Lewthwaite, P.; Perera, D.; Cardosa, M.J.; McMinn, P.; Ooi, M.H. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 2010, 10, 778–790. [Google Scholar] [CrossRef]
- Saxena, V.K.; Sane, S.; Nadkarni, S.S.; Sharma, D.K.; Deshpande, J.M. Genetic Diversity of Enterovirus A71, India. Emerg. Infect. Dis. 2015, 21, 123–126. [Google Scholar] [CrossRef]
- McMinn, P.C. Recent advances in the molecular epidemiology and control of human enterovirus 71 infection. Curr. Opin. Virol. 2012, 2, 199–205. [Google Scholar] [CrossRef]
- Yi, E.-J.; Shin, Y.-J.; Kim, J.-H.; Kim, T.-G.; Chang, S.-Y. Enterovirus 71 infection and vaccines. Clin. Exp. Vaccine Res. 2017, 6, 4. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 6, 276–277. [Google Scholar] [CrossRef]
- Vakulenko, Y.; Deviatkin, A.; Lukashev, A. Using Statistical Phylogenetics for Investigation of Enterovirus 71 Genotype A Reintroduction into Circulation. Viruses 2019, 10, 895. [Google Scholar] [CrossRef]
- Famulare, M.; Chang, S.; Iber, J.; Zhao, K.; Adeniji, J.A.; Bukbuk, D.; Baba, M. Sabin Vaccine Reversion in the Field: A Comprehensive Analysis of Sabin-Like Poliovirus Isolates in Nigeria. J. Virol. 2016, 90, 317–331. [Google Scholar] [CrossRef]
- Colbère-Garapin, F.; Jacques, S.; Drillet, A.; Pavio, N.; Couderc, T.; Blondel, B.; Pelletier, I. Poliovirus persistence in human cells in vitro. Dev. Biol. 2001, 105, 99–104. [Google Scholar]
| Virus | Rate, ×10−3 s/s/y | Reference |
|---|---|---|
| Hepatitis A virus | 1.0 | [3] |
| 1.21–2.0 | [4] | |
| 0.6 | [5] | |
| Duck Hepatitis A Virus | 0.6–1.9 | [6] |
| FMDV | serotype O–6.0 serotype A–11.9 serotype Asia-1–3.1 | [7] |
| serotype Asia1–5.9 | [8] | |
| serotype SAT1–3.00 serotype SAT2–4.0 | [9] | |
| Parechovirus | 2.8 | [10] |
| Non-polio enteroviruses | 6.0–11.0 | [11] |
| 3.40–11.9 | [12] | |
| Enterovirus A71 | 3.6–5.3 | [13] |
| 4.2–4.6 | [14] | |
| Poliovirus | 10.0 | [15] |
| Teschovirus A | 1.62 | [12] |
| 2.46 | [16] | |
| Cardiovirus A | 1.61 | [12] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vakulenko, Y.; Deviatkin, A.; Lukashev, A. The Effect of Sample Bias and Experimental Artefacts on the Statistical Phylogenetic Analysis of Picornaviruses. Viruses 2019, 11, 1032. https://doi.org/10.3390/v11111032
Vakulenko Y, Deviatkin A, Lukashev A. The Effect of Sample Bias and Experimental Artefacts on the Statistical Phylogenetic Analysis of Picornaviruses. Viruses. 2019; 11(11):1032. https://doi.org/10.3390/v11111032
Chicago/Turabian StyleVakulenko, Yulia, Andrei Deviatkin, and Alexander Lukashev. 2019. "The Effect of Sample Bias and Experimental Artefacts on the Statistical Phylogenetic Analysis of Picornaviruses" Viruses 11, no. 11: 1032. https://doi.org/10.3390/v11111032
APA StyleVakulenko, Y., Deviatkin, A., & Lukashev, A. (2019). The Effect of Sample Bias and Experimental Artefacts on the Statistical Phylogenetic Analysis of Picornaviruses. Viruses, 11(11), 1032. https://doi.org/10.3390/v11111032





