Emergence of Rare Bovine–Human Reassortant DS-1-Like Rotavirus A Strains with G8P[8] Genotype in Human Patients in the Czech Republic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Extraction and RT-PCR Detection
2.3. Phylogenetic Analysis
2.4. Whole-Genome Sequencing
3. Results and Discussion
3.1. Study Population
3.2. Rotavirus Genotypes
3.3. Specimens with G8 Genotype
3.4. Phylogenetic Analysis of VP7 Segment
3.5. Whole-Genome Sequencing
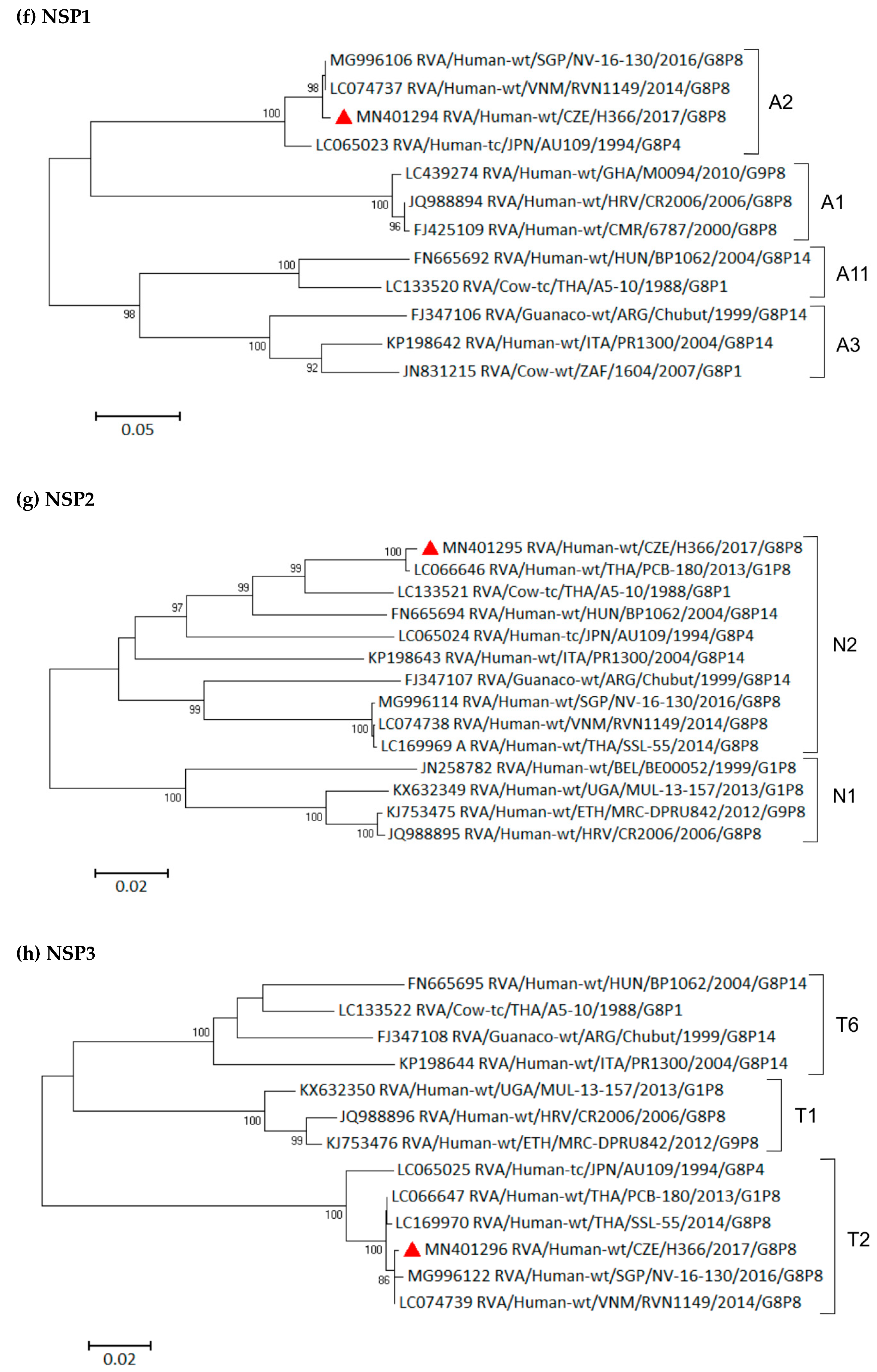
3.6. Phylogenetic Whole-Genome Analysis
3.7. Spread of G8P[8] Rotavirus Strains
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Soriano-Gabarró, M.; Mrukowicz, J.; Vesikari, T.; Verstraeten, T. Burden of rotavirus disease in European Union countries. Pediatr. Infect. Dis. J. 2006, 25, S7–S11. [Google Scholar] [CrossRef] [PubMed]
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, M.M.; Steele, D.; Gentsch, J.R.; Wecker, J.; Glass, R.I.; Parashar, U.D. Real-world impact of rotavirus vaccination. Pediatr. Infect. Dis. J. 2011, 30, S1–S5. [Google Scholar] [CrossRef] [PubMed]
- Clark, A.; Black, R.; Tate, J.; Roose, A.; Kotloff, K.; Lam, D.; Blackwelder, W.; Parashar, U.; Lanata, C.; Kang, G.; et al. Estimating global, regional and national rotavirus deaths in children aged <5 years: Current approaches, new analyses and proposed improvements. PLoS ONE 2017, 12, e0183392. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Parashar, U.D. World Health Organization-Coordinated Global Rotavirus Surveillance Network. Global, Regional, and National Estimates of Rotavirus Mortality in Children <5 Years of Age, 2000–2013. Clin. Infect. Dis. 2016, 62, S96–S105. [Google Scholar] [PubMed]
- Desselberger, U. Rotaviruses. Virus Res. 2014, 190, 75–96. [Google Scholar] [CrossRef] [Green Version]
- Rotavirus Classification Working Group. Newly Assigned Genotypes. List of Accepted Genotypes. Available online: https://rega.kuleuven.be/cev/viralmetagenomics/virus-classification (accessed on 11 July 2019).
- Ianiro, G.; Micolano, R.; Di Bartolo, I.; Scavia, G.; Monini, M. RotaNet-Italy Study Group. Group A rotavirus surveillance before vaccine introduction in Italy, September 2014 to August 2017. Eurosurveillance 2019, 24, 1800418. [Google Scholar] [CrossRef]
- Iturriza-Gómara, M.; Dallman, T.; Bányai, K.; Bӧttiger, B.; Buesa, J.; Diedrich, S.; Fiore, L.; Johansen, K.; Koopmans, M.; Korsun, N.; et al. Rotavirus genotypes co-circulating in Europe between 2006 and 2009 as determined by EuroRotaNet, a pan-European collaborative strain surveillance network. Epidemiol. Infect. 2011, 139, 895–909. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Rahman, M.; Attoui, H.; Bányai, K.; Estes, M.K.; Gentsch, J.R.; Iturriza-Gómara, M.; Kirkwood, C.; Martella, V.; et al. Recommendations for the classification of group A rotaviruses using all 11 genomic RNA segments. Arch. Virol. 2008, 153, 1621–1629. [Google Scholar] [CrossRef] [Green Version]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.T.; et al. Full Genome-Based Classification of Rotaviruses Reveals a Common Origin between Human Wa-Like and Porcine rotavirus Strains and Human DS-1-Like and Bovine Rotavirus Strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef]
- Nakagomi, T.; Doan, Y.H.; Dove, W.; Ngwira, B.; Iturriza-Gómara, M.; Nakagomi, O.; Cunliffe, N.A. G8 rotaviruses with conserved genotype constellations detected in Malawi over 10 years (1997–2007) display frequent gene reassortment among strains co-circulating in humans. J. Gen. Virol. 2013, 94, 1273–1295. [Google Scholar] [CrossRef] [PubMed]
- Santos, N.; Lima, R.C.; Pereira, C.F.; Gouvea, V. Detection of rotavirus types G8 and G10 among Brazilian children with diarrhea. J. Clin. Microbiol. 1998, 36, 2727–2729. [Google Scholar] [PubMed]
- Gómez, M.M.; Volotão, E.M.; Lima de Mendonça, M.C.; Tort, L.F.L.; da Silva, M.F.M.; Leite, J.P.G. Detection of uncommon rotavirus A strains P [8] G8 and P [4] G8 in the city of Rio de Janeiro, 2002. J. Med. Virol. 2010, 82, 1272–1276. [Google Scholar] [CrossRef] [PubMed]
- Lucero, Y.; O’Ryan, M.; Liparoti, G.; Huerta, N.; Mamani, N.; Ramani, S.; Lagomarcino, A.J.; Del Canto, F.; Quense, J. Predominance of Rotavirus G8P [8] in a city of Chile, a country without rotavirus vaccination. J. Pediatr. 2019, 204, 298–300. [Google Scholar] [CrossRef]
- Tacharoenmuang, R.; Komoto, S.; Guntapong, R.; Ide, T.; Sinchai, P.; Upachai, S.; Yoshikawa, T.; Tharmaphornpilas, P.; Sangkitporn, S.; Taniguchi, K. Full Genome Characterization of Novel DS-1-Like G8P[8] Rotavirus Strains that Have Emerged in Thailand: Reassortment of Bovine and Human Rotavirus Gene Segments in Emerging DS-1-Like Intergenogroup Reassortant Strains. PLoS ONE 2016, 11, e0165826. [Google Scholar] [CrossRef]
- Kondo, K.; Tsugawa, T.; Ono, M.; Ohara, T.; Fujibayashi, S.; Tahara, Y. Clinical and molecular characteristics of human Rotavirus G8P [8] outbreak strain, Japan, 2014. Emerg. Infect. Dis. 2017, 23, 968–972. [Google Scholar] [CrossRef]
- Hoa-Tran, T.N.; Nakagomi, T.; Vu, H.M.; Do, L.P.; Gauchan, P.; Agbemabiese, C.A.; Nguyen, T.T.T.; Nakagomi, O.; Thanh, N.T.H. Abrupt emergence and predominance in Vietnam of rotavirus A strains possessing a bovine-like G8 on a DS-1-like background. Arch. Virol. 2016, 161, 479–482. [Google Scholar] [CrossRef]
- Lee, S.K.; Choi, S.; Shin, S.H.; Lee, E.J.; Hyun, J.; Kim, J.S.; Kim, H.S. Emergence of G8P [6] rotavirus strains in Korean neonates. Gut Pathog. 2018, 10, 27. [Google Scholar] [CrossRef]
- Delogu, R.; Lo Presti, A.; Ruggeri, F.M.; Cella, E.; Giovanetti, M.; Ciccozzi, M.; Ljubin-Sternak, S.; Bukovski-Simonoski, S.; Lukic-Grlic, A.; Ianiro, G.; et al. Full-genome characterization of a G8P[8] rotavirus that emerged among children with diarrhea in Croatia in 2006. J. Clin. Microbiol. 2013, 51, 1583–1588. [Google Scholar] [CrossRef]
- Steele, A.D.; Parker, S.P.; Peenze, I.; Pager, C.T.; Taylor, M.B.; Cubitt, W.D. Comparative studies of human rotavirus serotype G8 strains recovered in South Africa and the United Kingdom. J. Gen. Virol. 1999, 80, 3029–3034. [Google Scholar] [CrossRef]
- Steyer, A.; Poljsak-Prijatelj, M.; Bufon, T.L.; Marcun-Varda, N.; Marin, J. Rotavirus genotypes in Slovenia: Unexpected detection of G8P [8] and G12P [8] genotypes. J. Med. Virol. 2007, 79, 626–632. [Google Scholar] [CrossRef] [PubMed]
- Pietsch, C.; Petersen, L.; Patzer, L.; Liebert, U.G. Molecular characteristics of German G8P [4] rotavirus strain GER1H-09 suggest that a genotyping and subclassification update is required for G8. J. Clin. Microbiol. 2009, 47, 3569–3576. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, G.; Delogu, R.; Bonomo, P.; Castiglia, P.; Ruggeri, F.M.; Fiore, L. Molecular characterization of human G8P [4] rotavirus strains in Italy: Proposal of a more complete subclassification of the G8 genotype in three major lineages. Infect. Genet. Evol. 2014, 21, 129–133. [Google Scholar] [CrossRef] [PubMed]
- Moutelíková, R.; Dvořáková Heroldová, M.; Holá, V.; Sauer, P.; Prodělalová, J. Human rotavirus A detection: Comparison of enzymatic immunoassay and rapid chromatographic test with two quantitative RT-PCR assays. Epidemiol. Mikrobiol. Imunol. 2018, 67, 110–113. [Google Scholar]
- Fujii, Y.; Shimoike, T.; Takagi, H.; Murakami, K.; Todaka-Takai, R.; Park, Y.B.; Katayama, K. Amplification of all 11 RNA segments of group A rotaviruses based on reverse transcription polymerase chain reaction. Microbiol. Immunol. 2012, 56, 630–638. [Google Scholar] [CrossRef]
- Gentsch, J.R.; Glass, R.I.; Woods, P.; Gouvea, V.; Gorziglia, M.; Flores, J.; Das, B.K.; Bhan, M.K. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 1992, 30, 1365–1373. [Google Scholar] [Green Version]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar]
- Maes, P.; Matthijnssens, J.; Rahman, M.; Van Ranst, M. RotaC: A web-based tool for the complete genome classification of group A rotaviruses. BMC Microbiol. 2009, 9, 238. [Google Scholar] [CrossRef]
- Theuns, S.; Conceiçāo-Neto, N.; Zeller, M.; Heylen, E.; Roukaerts, I.D.M.; Desmarets, L.M.B.; Van Ranst, M.; Nauwynck, H.J.; Matthijnssens, J. Characterization of a genetically heterogeneous porcine rotavirus C, and other viruses present in the fecal virome of a non-diarrheic Belgian piglet. Infect. Genet. Evol. 2016, 43, 135–145. [Google Scholar] [CrossRef]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef]
- Dóró, R.; László, B.; Martella, V.; Leshem, E.; Gentsch, J.; Parashar, U.; Bányai, K. Review of global rotavirus strain prevalence data from six years post vaccine licensure surveillance: Is there evidence of strain selection from vaccine pressure? Infect. Genet. Evol. 2014, 28, 446–461. [Google Scholar] [CrossRef] [PubMed]
- Chia, G.; Ho, H.J.; Ng, C.G.; Neo, F.J.X.; Win, M.K.; Cui, L.; Leo, Y.S.; Chow, A. An unusual outbreak of rotavirus G8P [8] gastroenteritis in adults in an urban community, Singapore, 2016. J. Clin. Virol. 2018, 105, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Agbemabiese, C.A.; Nakagomi, T.; Doan, Y.H.; Nakagomi, O. Whole genomic constellation of the first human G8 rotavirus strain detected in Japan. Infect. Genet. Evol. 2015, 35, 184–193. [Google Scholar] [CrossRef] [PubMed]
- Banyai, K.; Papp, H.; Dandar, E.; Molnar, P.; Mihaly, I.; Van Ranst, M.; Martella, V.; Matthijnssens, J. Whole genome sequencing and phylogenetic analysis of a zoonotic human G8P [14] rotavirus strain. Infect. Genet. Evol. 2010, 10, 1140–1144. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Rahman, M.; Yang, X.; Delbeke, T.; Arijs, I.; Kabue, J.P.; Muyembe, J.J.; Van Ranst, M. G8 rotavirus strains isolated in the Democratic Republic of Congo belong to the DS-1-like genogroup. J. Clin. Microbiol. 2006, 44, 1801–1809. [Google Scholar] [CrossRef]
- Esona, M.D.; Geyer, A.; Page, N.; Trabelsi, A.; Fodha, I.; Aminu, M.; Agbaya, V.A.; Tsion, B.; Kerin, T.K.; Armah, G.E.; et al. Genomic characterization of human rotavirus G8 strains from the African rotavirus network: Relationship to animal rotaviruses. J. Med. Virol. 2009, 81, 937–951. [Google Scholar] [CrossRef]
- Chitambar, S.D.; Arora, R.; Kolpe, A.B.; Yadav, M.M.; Raut, C.G. Molecular characterization of unusual bovine group A rotavirus G8P [14] strains identified in western India: Emergence of P [14] genotype. Vet. Microbiol. 2011, 148, 384–388. [Google Scholar] [CrossRef]
- Komoto, S.; Pongsuwanna, Y.; Tacharoenmuang, R.; Guntapong, R.; Ide, T.; Higo-Moriguchi, K.; Tsuji, T.; Yoshikawa, T.; Taniguchi, K. Whole genomic analysis of bovine group A rotavirus strains A5-10 and A5-13 provides evidence for close evolutionary relationship with human rotaviruses. Vet. Microbiol. 2016, 195, 37–57. [Google Scholar] [CrossRef]
- Komoto, S.; Tacharoenmuang, R.; Guntapong, R.; Ide, T.; Haga, K.; Katayama, K.; Kato, T.; Ouchi, Y.; Kurahashi, H.; Tsuji, T.; et al. Emergence and characterization of unusual DS-1-like G1P[8] Rotavirus strains in children with diarrhea in Thailand. PLoS ONE 2015, 10, e141739. [Google Scholar] [CrossRef]
- Wikipedia. Overseas Vietnamese. Available online: https://en.wikipedia.org/wiki/Overseas_Vietnamese (accessed on 20 September 2019).
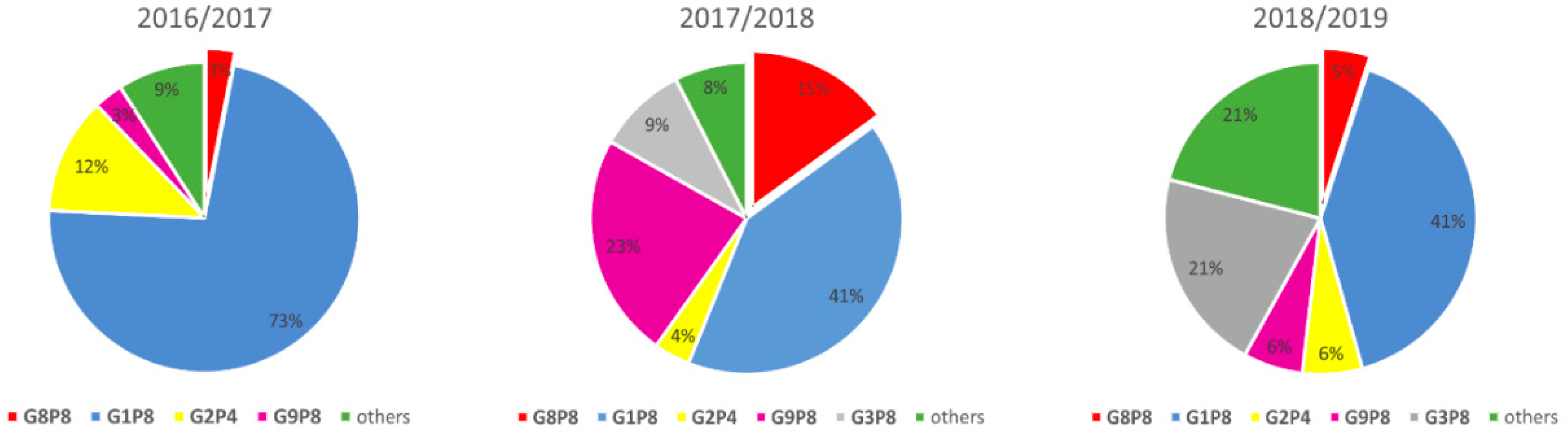
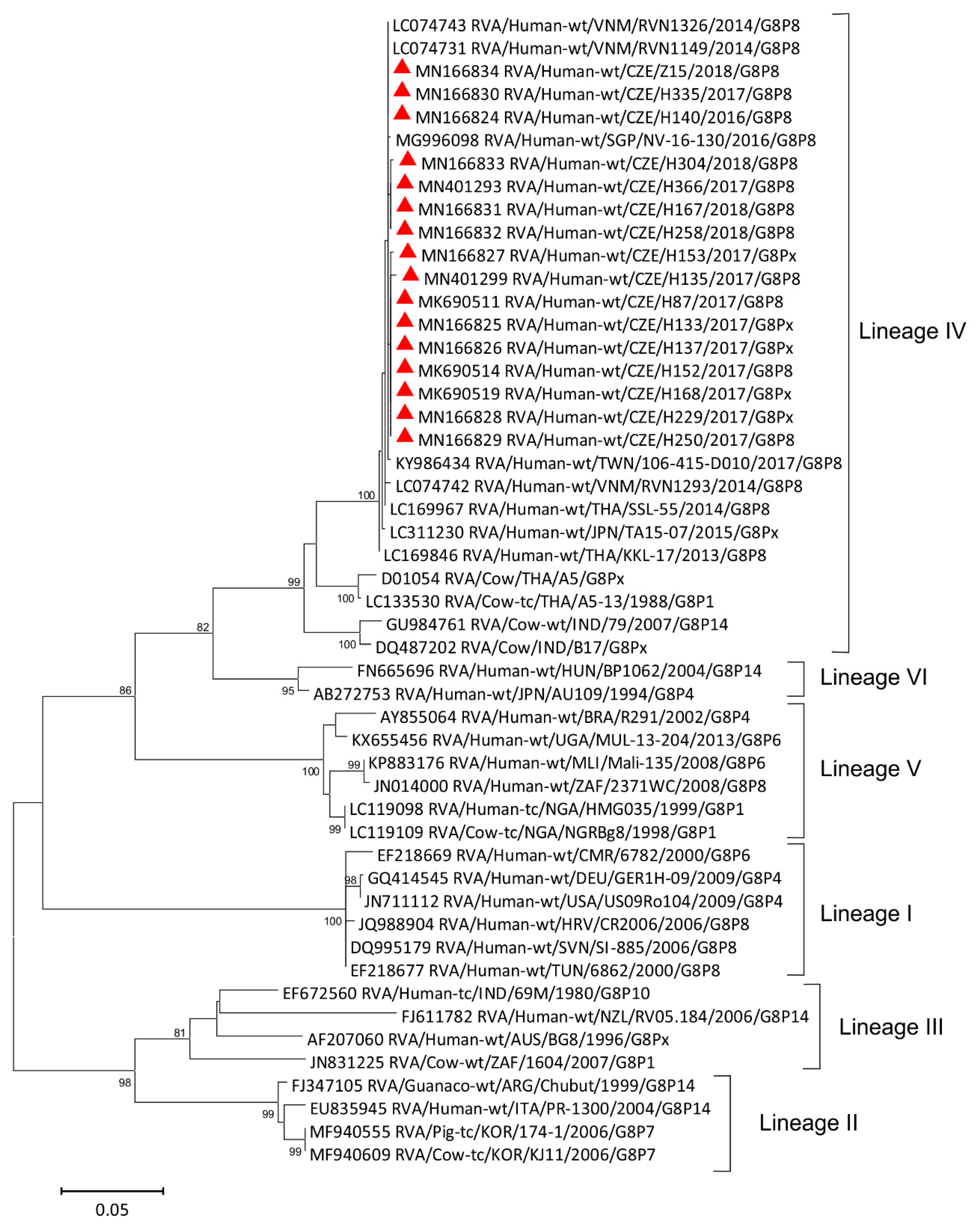
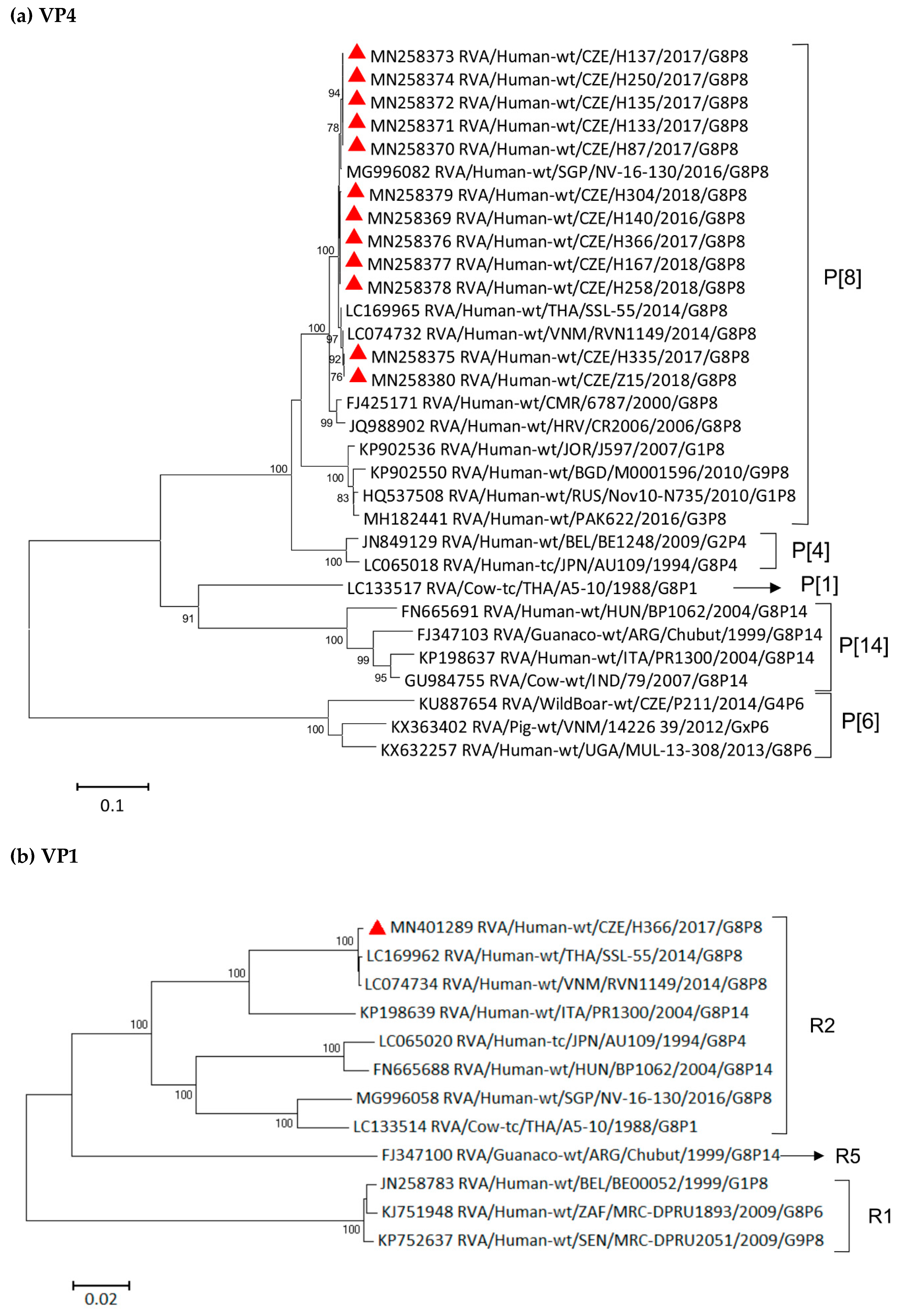
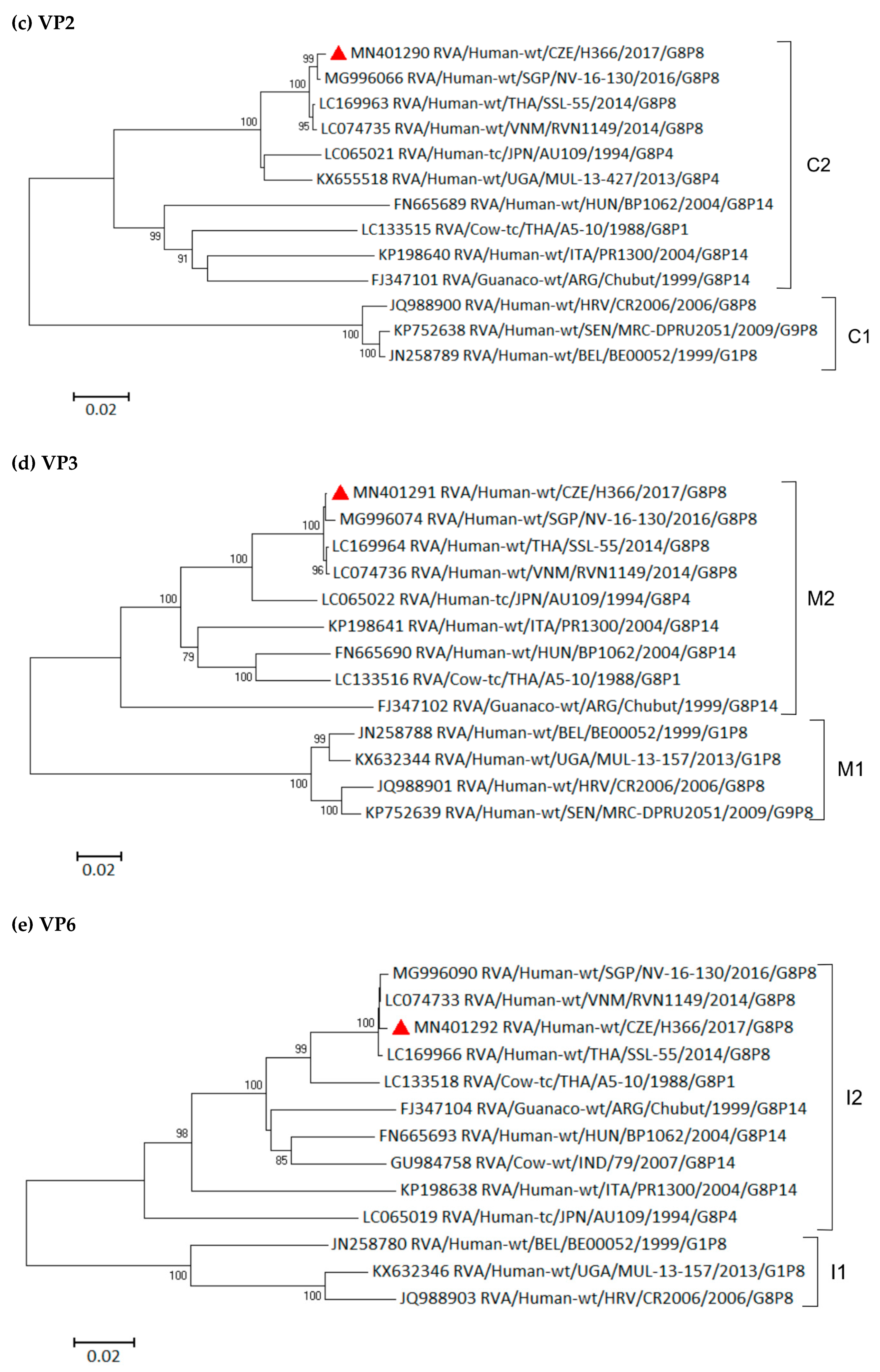

| Strain | 2016–2017 | 2017–2018 | 2018–2019 | Total | |
|---|---|---|---|---|---|
| Fully typed strains | G1P[8] | 24 | 39 | 33 | 96 |
| G2P[4] | 4 | 2 | 4 | 10 | |
| G2P[8] | - | 1 | - | 1 | |
| G3P[8] | - | 10 | 15 | 25 | |
| G3P[9] | 1 | - | 1 | 2 | |
| G4P[8] | - | - | 3 | 3 | |
| G8P[8] | 1 | 12 | 4 | 17 | |
| G9P[4] | - | - | 2 | 2 | |
| G9P[8] | 1 | 16 | 5 | 22 | |
| G12P[8] | - | 2 | 1 | 3 | |
| Partially typed strains | G1P[nd] | - | 5 | - | 5 |
| G2P[nd] | 1 | 2 | 1 | 4 | |
| G3P[nd] | - | - | 2 | 2 | |
| G8P[nd] | - | 4 | - | 4 | |
| G9P[nd] | - | 9 | - | 9 | |
| GndP[4] | - | - | 2 | 2 | |
| GndP[8] | 1 | 3 | 8 | 12 | |
| GndP[nd] | - | 1 | 4 | 5 | |
| Mixed infections | G9P[4][8] | - | 1 | - | 1 |
| Total | 33 | 107 | 85 | 225 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moutelíková, R.; Sauer, P.; Dvořáková Heroldová, M.; Holá, V.; Prodělalová, J. Emergence of Rare Bovine–Human Reassortant DS-1-Like Rotavirus A Strains with G8P[8] Genotype in Human Patients in the Czech Republic. Viruses 2019, 11, 1015. https://doi.org/10.3390/v11111015
Moutelíková R, Sauer P, Dvořáková Heroldová M, Holá V, Prodělalová J. Emergence of Rare Bovine–Human Reassortant DS-1-Like Rotavirus A Strains with G8P[8] Genotype in Human Patients in the Czech Republic. Viruses. 2019; 11(11):1015. https://doi.org/10.3390/v11111015
Chicago/Turabian StyleMoutelíková, Romana, Pavel Sauer, Monika Dvořáková Heroldová, Veronika Holá, and Jana Prodělalová. 2019. "Emergence of Rare Bovine–Human Reassortant DS-1-Like Rotavirus A Strains with G8P[8] Genotype in Human Patients in the Czech Republic" Viruses 11, no. 11: 1015. https://doi.org/10.3390/v11111015
APA StyleMoutelíková, R., Sauer, P., Dvořáková Heroldová, M., Holá, V., & Prodělalová, J. (2019). Emergence of Rare Bovine–Human Reassortant DS-1-Like Rotavirus A Strains with G8P[8] Genotype in Human Patients in the Czech Republic. Viruses, 11(11), 1015. https://doi.org/10.3390/v11111015





