Novel Polyomaviruses in Mammals from Multiple Orders and Reassessment of Polyomavirus Evolution and Taxonomy
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Samples and DNA Extraction
2.2. PCR Methods
2.3. Synthesis of Polyomavirus Early Regions and Transfection in Cell Culture
2.4. RNA Extraction and cDNA Synthesis
2.5. Purification of PCR Products and Sequencing
2.6. Prediction of Splice Sites in Early Regions of the Novel Polyomavirus Genomes
2.7. Check of Splice Sites and Coding Sequences (CDS) in Published, Annotated Polyomavirus Genomes
2.8. Sequence Datasets
2.9. Phylogenetic Analyses
2.10. Cophylogenetic Analyses
3. Results
3.1. Identification and Characterization of Polyomaviruses in Non-Hominine Mammals
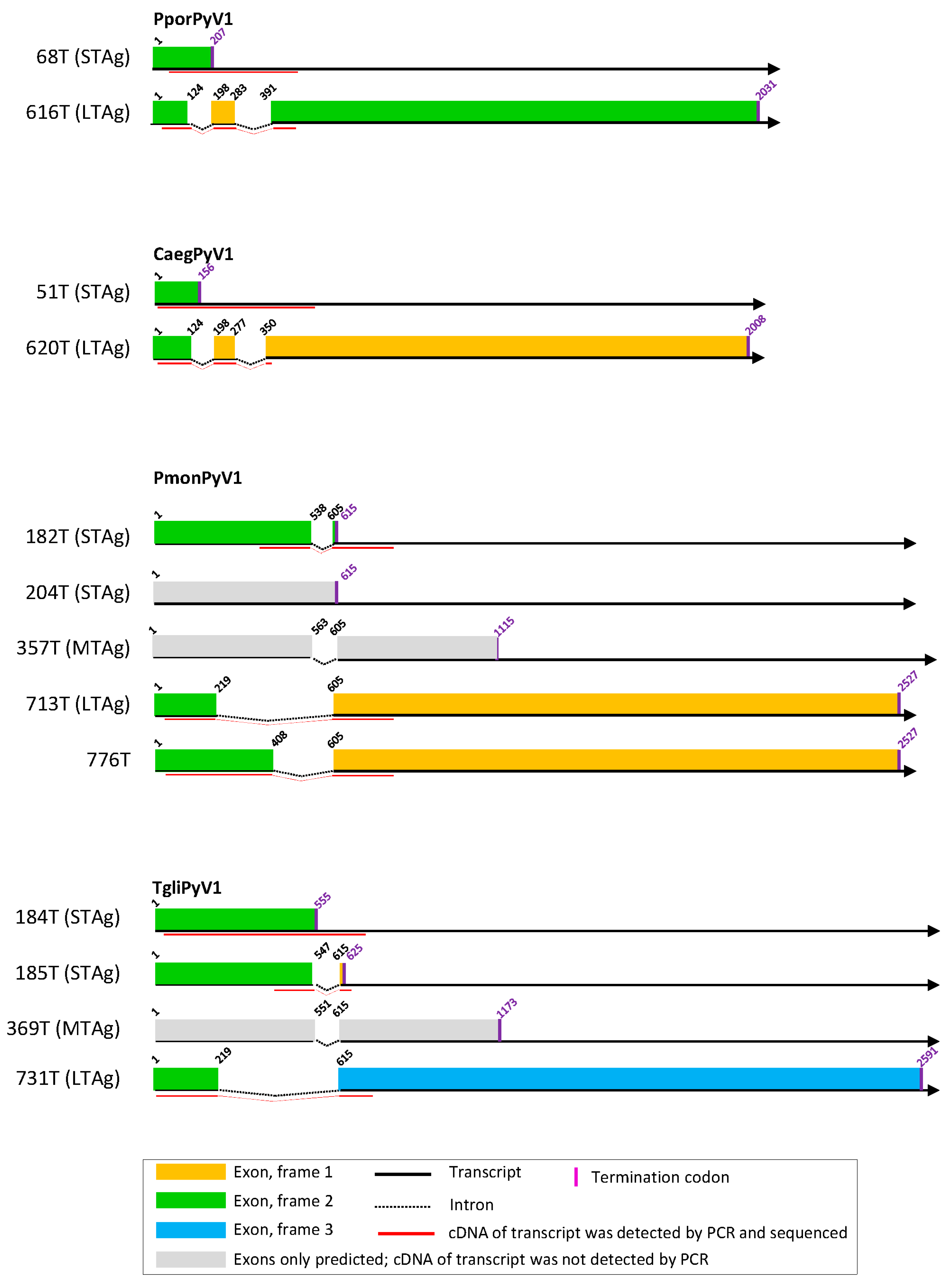
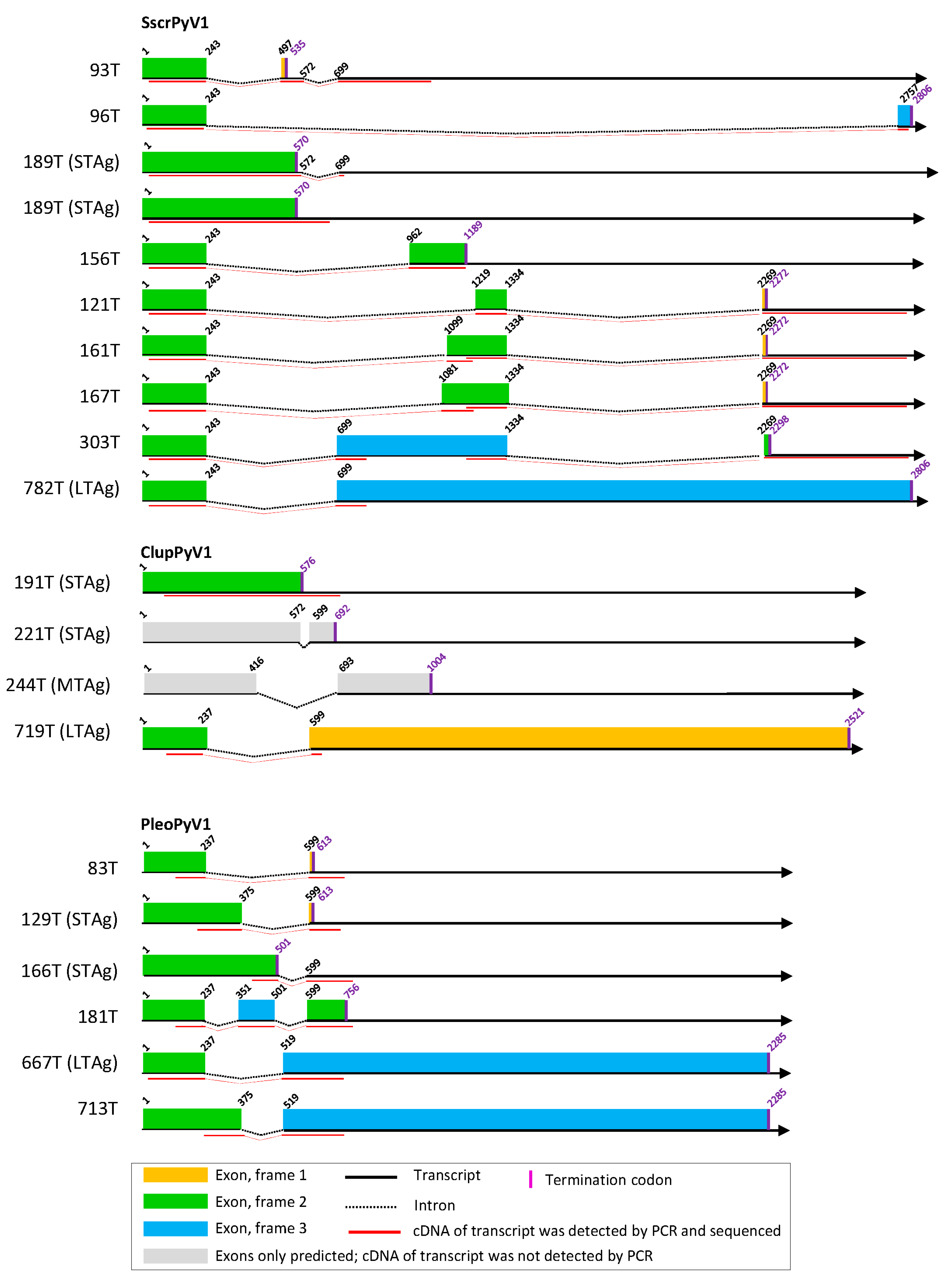
3.2. Cell Culture-Based Identification of Splice Sites in Early Regions of the Novel Polyomavirus Genomes
3.3. Cell Culture-Based Identification of Splice Sites in Early Regions of Selected Published Polyomavirus Genomes
3.4. Phylogeny-Guided Identification of Splice Sites and CDS in Early Regions of Published, Annotated Polyomavirus Genomes
3.5. Other Splice Sites and Coding Sequences in Novel Polyomavirus Genomes
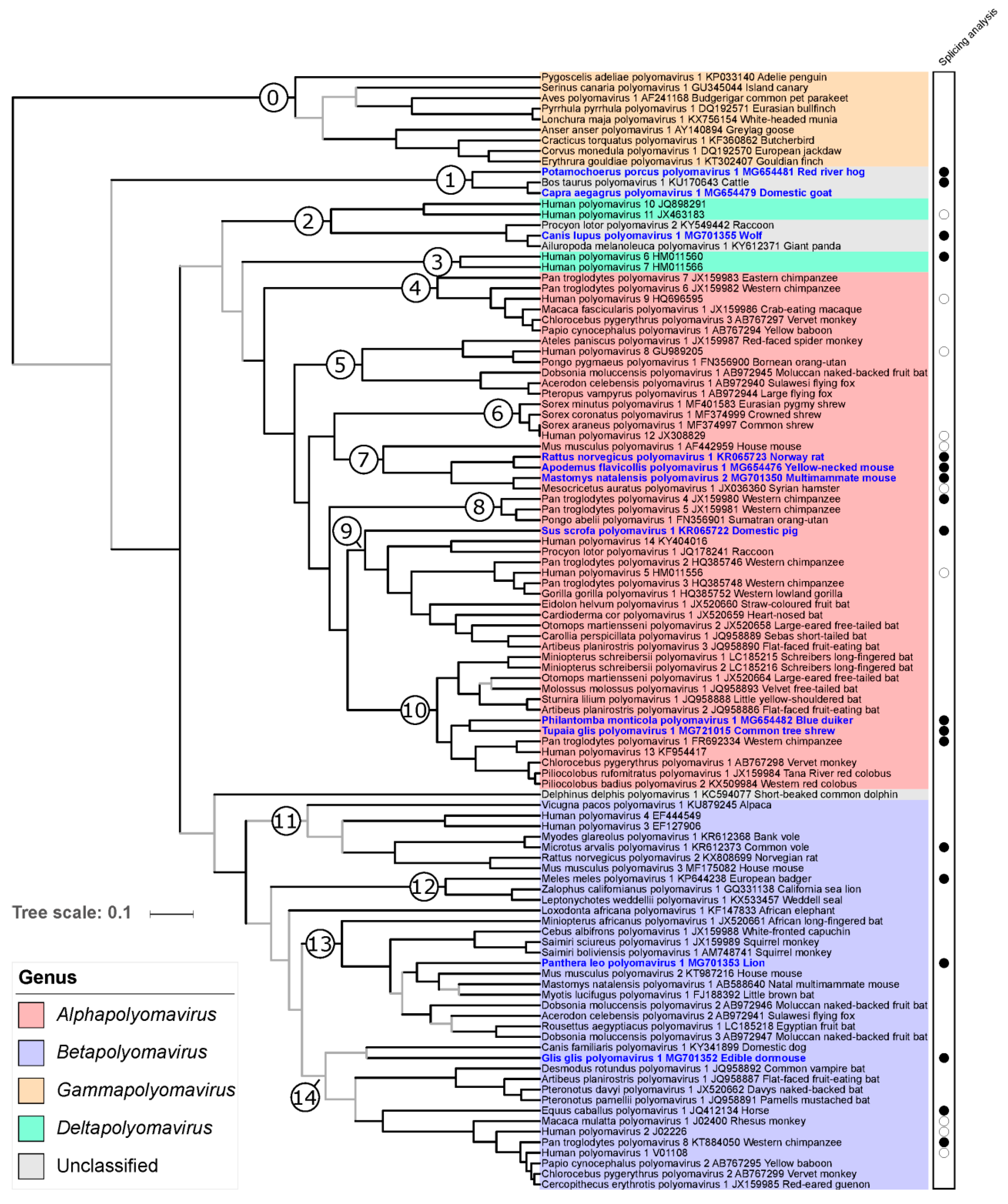
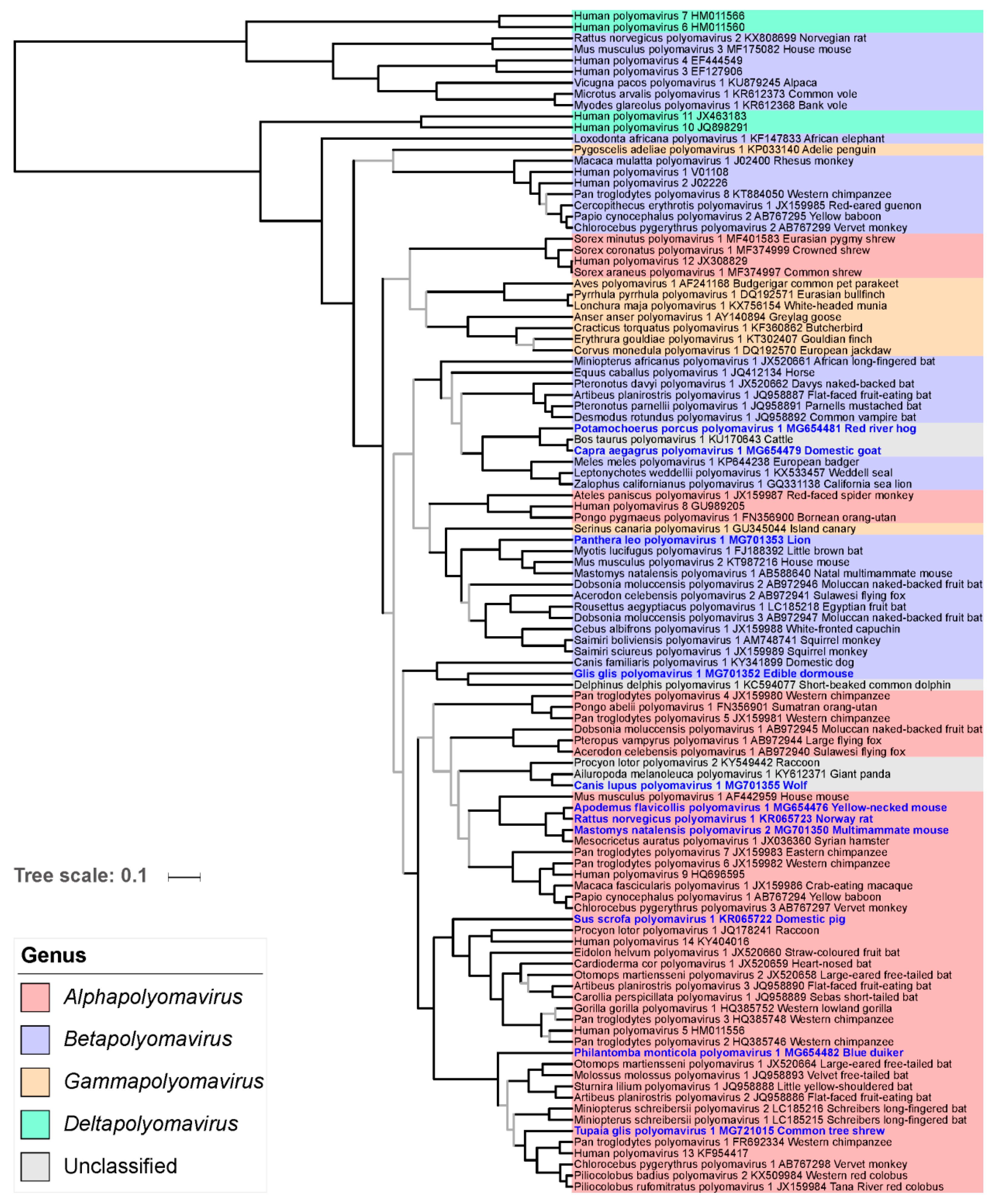
3.6. Phylogenetic Placement of the Novel Polyomaviruses
3.7. Cophylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moens, U.; Krumbholz, A.; Ehlers, B.; Zell, R.; Johne, R.; Calvignac-Spencer, S.; Lauber, C. Biology, evolution, and medical importance of polyomaviruses: An update. Infect. Genet. Evol. 2017, 54, 18–38. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Van Doorslaer, K.; Peretti, A.; Geoghegan, E.M.; Tisza, M.J.; An, P.; Katz, J.P.; Pipas, J.M.; McBride, A.A.; Camus, A.C.; et al. The ancient evolutionary history of polyomaviruses. PLoS Pathog. 2016, 12, e1005574. [Google Scholar] [CrossRef]
- Warden, C.D.; Lacey, S.F. Updated phylogenetic analysis of polyomavirus-host co-evolution. J. Bioinform. Res. 2012, 1, 46–49. [Google Scholar]
- Madinda, N.F.; Ehlers, B.; Wertheim, J.O.; Akoua-Koffi, C.; Bergl, R.A.; Boesch, C.; Akonkwa, D.B.M.; Eckardt, W.; Fruth, B.; Gillespie, T.R.; et al. Assessing host-virus codivergence for close relatives of Merkel cell polyomavirus infecting African great apes. J. Virol. 2016, 90, 8531–8541. [Google Scholar] [CrossRef] [PubMed]
- Geoghegan, J.L.; Duchêne, S.; Holmes, E.C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 2017, 13, e1006215. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.; Gonzalez, G.; Sasaki, M.; Ito, K.; Ishii, A.; Hang’ombe, B.M.; Mweene, A.S.; Orba, Y.; Sawa, H. Discovery of African bat polyomaviruses and infrequent recombination in the large T antigen in the Polyomaviridae. J. Gen. Virol. 2017, 98, 726–738. [Google Scholar] [CrossRef]
- Lim, E.S.; Reyes, A.; Antonio, M.; Saha, D.; Ikumapayi, U.N.; Adeyemi, M.; Stine, O.C.; Skelton, R.; Brennan, D.C.; Mkakosya, R.S.; et al. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 2013, 436, 295–303. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Conrardy, C.; Kuzmin, I.V.; Recuenco, S.; Agwanda, B.; Alvarez, D.A.; Ellison, J.A.; Gilbert, A.T.; Moran, D.; et al. Discovery of diverse polyomaviruses in bats and the evolutionary history of the Polyomaviridae. J. Gen. Virol. 2013, 94, 738–748. [Google Scholar] [CrossRef]
- Hill, S.C.; Murphy, A.A.; Cotten, M.; Palser, A.L.; Benson, P.; Lesellier, S.; Gormley, E.; Richomme, C.; Grierson, S.; Bhuachalla, D.N.; et al. Discovery of a polyomavirus in European badgers (Meles meles) and the evolution of host range in the family Polyomaviridae. J. Gen. Virol. 2015, 96, 1411–1422. [Google Scholar] [CrossRef]
- Carr, M.; Gonzalez, G.; Sasaki, M.; Dool, S.E.; Ito, K.; Ishii, A.; Hang’ombe, B.M.; Mweene, A.S.; Teeling, E.C.; Hall, W.W.; et al. Identification of the same polyomavirus species in different African horseshoe bat species is indicative of short-range host-switching events. J. Gen. Virol. 2017, 98, 2771–2785. [Google Scholar] [CrossRef]
- Gedvilaite, A.; Tryland, M.; Ulrich, R.G.; Schneider, J.; Kurmauskaite, V.; Moens, U.; Preugschas, H.; Calvignac-Spencer, S.; Ehlers, B. Novel polyomaviruses in shrews (Soricidae) with close similarity to human polyomavirus 12. J. Gen. Virol. 2017, 98, 3060–3067. [Google Scholar] [CrossRef] [PubMed]
- Leendertz, F.H.; Scuda, N.; Cameron, K.N.; Kidega, T.; Zuberbuhler, K.; Leendertz, S.A.J.; Couacy-Hymann, E.; Boesch, C.; Calvignac, S.; Ehlers, B. African Great Apes Are Naturally Infected with Polyomaviruses Closely Related to Merkel Cell Polyomavirus. J. Virol. 2011, 85, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Scuda, N.; Hofmann, J.; Calvignac-Spencer, S.; Ruprecht, K.; Liman, P.; Kühn, J.; Hengel, H.; Ehlers, B. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 2011, 85, 4586–4590. [Google Scholar] [CrossRef] [PubMed]
- Scuda, N.; Madinda, N.F.; Akoua-Koffi, C.; Adjogoua, E.V.; Wevers, D.; Hofmann, J.; Cameron, K.N.; Leendertz, S.A.J.; Couacy-Hymann, E.; Robbins, M.; et al. Novel polyomaviruses of nonhuman primates: Genetic and serological predictors for the existence of multiple unknown polyomaviruses within the human population. PLoS Pathog. 2013, 9, e1003429. [Google Scholar] [CrossRef]
- Korup, S.; Rietscher, J.; Calvignac-Spencer, S.; Trusch, F.; Hofmann, J.; Moens, U.; Sauer, I.; Voigt, S.; Schmuck, R.; Ehlers, B. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS ONE 2013, 8, e58021. [Google Scholar] [CrossRef]
- Millán, J.; Chirife, A.D.; Kalema-Zikusoka, G.; Cabezón, O.; Muro, J.; Marco, I.; Cliquet, F.; León-Vizcaíno, L.; Wasniewski, M.; Almería, S.; et al. Serosurvey of dogs for human, livestock, and wildlife pathogens, Uganda. Emerg. Infect. Dis. 2013, 19, 680. [Google Scholar] [CrossRef]
- Hoppe, E.; Pauly, M.; Gillespie, T.R.; Akoua-Koffi, C.; Hohmann, G.; Fruth, B.; Karhemere, S.; Madinda, N.F.; Mugisha, L.; Muyembe, J.-J.; et al. Multiple cross-species transmission events of human adenoviruses (HAdV) during hominine evolution. Mol. Biol. Evol. 2015, 32, 2072–2084. [Google Scholar] [CrossRef]
- Pauly, M.; Akoua-Koffi, C.; Buchwald, N.; Schubert, G.; Weiss, S.; Couacy-Hymann, E.; Anoh, A.E.; Mossoun, A.; Calvignac-Spencer, S.; Leendertz, S.A.; et al. Adenovirus in Rural Côte DIvoire: High Diversity and Cross-Species Detection. Ecohealth 2015, 12, 441–452. [Google Scholar] [CrossRef]
- Ehlers, B.; Dural, G.; Yasmum, N.; Lembo, T.; de Thoisy, B.; Ryser-Degiorgis, M.-P.; Ulrich, R.G.; McGeoch, D.J. Novel mammalian herpesviruses and lineages within the Gammaherpesvirinae: Cospeciation and interspecies transfer. J. Virol. 2008, 82, 3509–3516. [Google Scholar] [CrossRef]
- Ehlers, B.; Borchers, K.; Grund, C.; Fro, K.; Ludwig, H. Detection of new DNA polymerase genes of known and potentially novel herpesviruses by PCR with degenerate and deoxyinosine-substituted primers. Virus Genes 1999, 18, 211–220. [Google Scholar] [CrossRef]
- Ehlers, B.; Küchler, J.; Yasmum, N.; Dural, G.; Voigt, S.; Schmidt-Chanasit, J.; Jäkel, T.; Matuschka, F.-R.; Richter, D.; Essbauer, S.; et al. Identification of novel rodent herpesviruses, including the first gammaherpesvirus of Mus musculus. J. Virol. 2007, 81, 8091–8100. [Google Scholar] [CrossRef] [PubMed]
- Ehlers, B.; Richter, D.; Matuschka, F.-R.; Ulrich, R.G. Genome sequences of a rat polyomavirus related to murine polyomavirus, Rattus norvegicus polyomavirus 1. Genome Announc. 2015, 3, e00997-15. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, S.; Goltz, M.; Ehlers, B. Characterization of the DNA polymerase loci of the novel porcine lymphotropic herpesviruses 1 and 2 in domestic and feral pigs. J. Gen. Virol. 1999, 80, 3199–3205. [Google Scholar] [CrossRef] [PubMed]
- García-Bocanegra, I.; Barranco, I.; Rodríguez-Gómez, I.; Pérez, B.; Gómez-Laguna, J.; Rodríguez, S.; Ruiz-Villamayor, E.; Perea, A. Tuberculosis in alpacas (Lama pacos) caused by Mycobacterium bovis. J. Clin. Microbiol. 2010, 48, 1960–1964. [Google Scholar] [CrossRef]
- de Val, B.P.; Grau-Roma, L.; Segalés, J.; Domingo, M.; Vidal, E. Mycobacteriosis outbreak caused by Mycobacterium avium subsp. avium detected through meat inspection in five porcine fattening farms. Vet. Rec. 2013, 174, 96. [Google Scholar]
- Benson, K.A.S.; Manire, C.A.; Ewing, R.Y.; Saliki, J.T.; Townsend, F.I.; Ehlers, B.; Romero, C.H. Identification of novel alpha-and gammaherpesviruses from cutaneous and mucosal lesions of dolphins and whales. J. Virol. Methods 2006, 136, 261–266. [Google Scholar] [CrossRef]
- Schlegel, M.; Ali, H.S.; Stieger, N.; Groschup, M.H.; Wolf, R.; Ulrich, R.G. Molecular identification of small mammal species using novel cytochrome B gene-derived degenerated primers. Biochem. Genet. 2012, 50, 440–447. [Google Scholar] [CrossRef]
- Clercx, C.; Fastrès, A.; Roels, E. Idiopathic pulmonary fibrosis in West Highland white terriers: An update. Vet. J. 2018. [Google Scholar] [CrossRef]
- Fiebig, U.; Hartmann, M.G.; Bannert, N.; Kurth, R.; Denner, J. Transspecies transmission of the endogenous koala retrovirus. J. Virol. 2006, 80, 5651–5654. [Google Scholar] [CrossRef]
- Johne, R.; Enderlein, D.; Nieper, H.; Muller, H. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J. Virol. 2005, 79, 3883–3887. [Google Scholar] [CrossRef]
- Desmet, F.O.; Hamroun, D.; Lalande, M.; Collod-Beroud, G.; Claustres, M.; Beroud, C. Human Splicing Finder: An online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009, 37, e67. [Google Scholar] [CrossRef] [PubMed]
- Hyde-DeRuyscher, R.; Carmichael, G.G. Polyomavirus early-late switch is not regulated at the level of transcription initiation and is associated with changes in RNA processing. Proc. Natl. Acad. Sci. USA 1988, 85, 8993–8997. [Google Scholar] [CrossRef] [PubMed]
- Schuurman, R.; Jacobs, M.; van Strien, A.; van der Noordaa, J.; Sol, C. Analysis of splice sites in the early region of bovine polyomavirus: Evidence for a unique pattern of large T mRNA splicing. J. Gen. Virol. 1992, 73, 2879–2886. [Google Scholar] [CrossRef] [PubMed]
- Zerrahn, J.; Knippschild, U.; Winkler, T.; Deppert, W. Independent expression of the transforming amino-terminal domain of SV40 large I antigen from an alternatively spliced third SV40 early mRNA. EMBO J. 1993, 12, 4739–4746. [Google Scholar] [CrossRef]
- Abend, J.R.; Joseph, A.E.; Das, D.; Campbell-Cecen, D.B.; Imperiale, M.J. A truncated T antigen expressed from an alternatively spliced BK virus early mRNA. J. Gen. Virol. 2009, 90, 1238–1245. [Google Scholar] [CrossRef]
- Huang, Y.; Carmichael, G.G. RNA processing in the polyoma virus life cycle. Front. Biosci. 2009, 14, 4968. [Google Scholar] [CrossRef]
- Riley, M.I.; Yoo, W.; Mda, N.Y.; Folk, W.R. Tiny T antigen: An autonomous polyomavirus T antigen amino-terminal domain. J. Virol. 1997, 71, 6068–6074. [Google Scholar]
- Shuda, M.; Feng, H.; Kwun, H.J.; Rosen, S.T.; Gjoerup, O.; Moore, P.S.; Chang, Y. T antigen mutations are a human tumor-specific signature for Merkel cell polyomavirus. Proc. Natl. Acad. Sci. USA 2008, 105, 16272–16277. [Google Scholar] [CrossRef]
- Trowbridge, P.W.; Frisque, R.J. Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. J. Neurovirol. 1995, 1, 195–206. [Google Scholar] [CrossRef]
- Orba, Y.; Kobayashi, S.; Nakamura, I.; Ishii, A.; Hang’ombe, B.M.; Mweene, A.S.; Thomas, Y.; Kimura, T.; Sawa, H. Detection and characterization of a novel polyomavirus in wild rodents. J. Gen. Virol. 2011, 92, 789–795. [Google Scholar] [CrossRef]
- Theiss, J.M.; Günther, T.; Alawi, M.; Neumann, F.; Tessmer, U.; Fischer, N.; Grundhoff, A. A comprehensive analysis of replicating Merkel cell polyomavirus genomes delineates the viral transcription program and suggests a role for mcv-miR-M1 in episomal persistence. PLoS Pathog. 2015, 11, e1004974. [Google Scholar] [CrossRef] [PubMed]
- van der Meijden, E.; Kazem, S.; Dargel, C.A.; van Vuren, N.; Hensbergen, P.J.; Feltkamp, M.C. Characterization of T Antigens, Including Middle T and Alternative T, Expressed by the Human Polyomavirus Associated with Trichodysplasia Spinulosa. J. Virol. 2015, 89, 9427–9439. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, R.; Sommerfeld, K.; Schröder, A.; Prokoph, H.; Arnold, W.; Krüger, D.H.; Scherneck, S. Hamster polyomavirus-encoded proteins: Gene cloning, heterologous expression and immunoreactivity. Virus Genes 1996, 12, 265–274. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Guindon, S.; Lethiec, F.; Duroux, P.; Gascuel, O. PHYML Online--a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005, 33, W557–W559. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Anisimova, M.; Gil, M.; Dufayard, J.F.; Dessimoz, C.; Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 2011, 60, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Suchard, M.A.; Lemey, P.; Baele, G.; Ayres, D.L.; Drummond, A.J.; Rambaut, A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018, 4, vey016. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.P.; Charleston, M.A. A cophylogenetic perspective of RNA-virus evolution. Mol. Biol. Evol. 2004, 21, 45–57. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gottschling, M.; Goker, M.; Stamatakis, A.; Bininda-Emonds, O.R.; Nindl, I.; Bravo, I.G. Quantifying the phylodynamic forces driving papillomavirus evolution. Mol. Biol. Evol. 2011, 28, 2101–2113. [Google Scholar] [CrossRef]
- Conow, C.; Fielder, D.; Ovadia, Y.; Libeskind-Hadas, R. Jane: A new tool for the cophylogeny reconstruction problem. Algorithms Mol. Biol. 2010, 5, 16. [Google Scholar] [CrossRef]
- Esselstyn, J.A.; Oliveros, C.H.; Swanson, M.T.; Faircloth, B.C. Investigating Difficult Nodes in the Placental Mammal Tree with Expanded Taxon Sampling and Thousands of Ultraconserved Elements. Genome Biol. Evol. 2017, 9, 2308–2321. [Google Scholar] [CrossRef]
- Schuurman, R.; Sol, C.; Van Der Noordaa, J. The complete nucleotide sequence of bovine polyomavirus. J. Gen. Virol. 1990, 71, 1723–1735. [Google Scholar] [CrossRef]
- Kilham, L. Isolation in suckling mice of a virus from C3H mice harboring Bittner milk agent. Science 1952, 116, 391–392. [Google Scholar] [CrossRef]
- Ben Salem, N.; Moens, U.; Ehlers, B. Genome Sequences of Murine Pneumotropic Virus (Polyomaviridae) Detected in Wild House Mice (Mus musculus). Genome Announc. 2016, 4, e01545-15. [Google Scholar] [CrossRef] [PubMed]
- Ben Salem, N.; Perez de Val, B.; Martin, M.; Moens, U.; Ehlers, B. Genome Sequence of Bovine Polyomavirus 1 Detected in a Salers Cow (Bos taurus) from Catalonia, Spain. Genome Announc. 2016, 4, e01658-15. [Google Scholar] [CrossRef] [PubMed]
- Dilworth, S.M. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat. Rev. Cancer 2002, 2, 951. [Google Scholar] [CrossRef] [PubMed]
- Carter, J.J.; Daugherty, M.D.; Qi, X.; Bheda-Malge, A.; Wipf, G.C.; Robinson, K.; Roman, A.; Malik, H.S.; Galloway, D.A. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proc. Natl. Acad. Sci. USA 2013, 110, 12744–12749. [Google Scholar] [CrossRef] [PubMed]
- Renshaw, R.W.; Wise, A.G.; Maes, R.K.; Dubovi, E.J. Complete genome sequence of a polyomavirus isolated from horses. J. Virol. 2012, 86, 8903. [Google Scholar] [CrossRef]
- Schowalter, R.M.; Pastrana, D.V.; Pumphrey, K.A.; Moyer, A.L.; Buck, C.B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 2010, 7, 509–515. [Google Scholar] [CrossRef]
- Nainys, J.; Timinskas, A.; Schneider, J.; Ulrich, R.G.; Gedvilaite, A. Identification of two novel members of the tentative genus Wukipolyomavirus in wild rodents. PLoS ONE 2015, 10, e0140916. [Google Scholar] [CrossRef]
- Deuzing, I.; Fagrouch, Z.; Groenewoud, M.J.; Niphuis, H.; Kondova, I.; Bogers, W.; Verschoor, E.J. Detection and characterization of two chimpanzee polyomavirus genotypes from different subspecies. Virol J. 2010, 7, 347. [Google Scholar] [CrossRef]
- Salem, N.B.; Leendertz, F.H.; Ehlers, B. Genome sequences of polyomaviruses from the wild-living red colobus (Piliocolobus badius) and western chimpanzee (Pan troglodytes verus). Genome Announc. 2016, 4, e01101-16. [Google Scholar]
- Anthony, S.J.; St. Leger, J.A.; Navarrete-Macias, I.; Nilson, E.; Sanchez-Leon, M.; Liang, E.; Seimon, T.; Jain, K.; Karesh, W.; Daszak, P.; et al. Identification of a Novel Cetacean Polyomavirus from a Common Dolphin (Delphinus delphis) with Tracheobronchitis. PLoS ONE 2013, 8, e68239. [Google Scholar] [CrossRef]
- Polyomaviridae Study Group of the International Committee on Taxonomy of Viruses; Calvignac-Spencer, S.; Feltkamp, M.C.; Daugherty, M.D.; Moens, U.; Ramqvist, T.; Johne, R.; Ehlers, B. A taxonomy update for the family Polyomaviridae. Arch. Virol. 2016, 161, 1739–1750. [Google Scholar] [CrossRef] [PubMed]
- Reddy, V.; Thimmappaya, B.; Dhar, R.; Subramanian, K.N.; Zain, B.; Pan, J.; Ghosh, P.; Celma, M.; Weissman, S. The genome of simian virus 40. Science 1978, 200, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, K.A.; Villarreal, L.P. Natural biology of polyomavirus middle T antigen. Microbiol. Mol. Biol. Rev. 2001, 65, 288–318. [Google Scholar] [CrossRef] [PubMed]
- Demetriou, S.K.; Ona-Vu, K.; Sullivan, E.M.; Dong, T.K.; Hsu, S.W.; Oh, D.H. Defective DNA repair and cell cycle arrest in cells expressing Merkel cell polyomavirus T antigen. Int. J. Cancer 2012, 131, 1818–1827. [Google Scholar] [CrossRef]
- Korup-Schulz, S.-V.; Lucke, C.; Moens, U.; Schmuck, R.; Ehlers, B. Large T antigen variants of human polyomaviruses 9 and 12 and seroreactivity against their N terminus. J. Gen. Virol. 2017, 98, 704–714. [Google Scholar] [CrossRef]
- Aiewsakun, P.; Katzourakis, A. Time-Dependent Rate Phenomenon in Viruses. J. Virol. 2016, 90, 7184–7195. [Google Scholar] [CrossRef]
- Membrebe, J.V.; Suchard, M.A.; Rambaut, A.; Baele, G.; Lemey, P. Bayesian inference of evolutionary histories under time-dependent substitution rates. Mol. Biol. Evol. 2019, 36, 1793–1803. [Google Scholar] [CrossRef]
- Wertheim, J.O.; Kosakovsky Pond, S.L. Purifying selection can obscure the ancient age of viral lineages. Mol. Biol. Evol. 2011, 28, 3355–3365. [Google Scholar] [CrossRef]
- Siddell, S.G.; Walker, P.J.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; et al. Additional changes to taxonomy ratified in a special vote by the International Committee on Taxonomy of Viruses (October 2018). Arch. Virol. 2019, 164, 943–946. [Google Scholar] [CrossRef]
- Van Doorslaer, K.; Kraberger, S.; Austin, C.; Farkas, K.; Bergeman, M.; Paunil, E.; Davison, W.; Varsani, A. Fish polyomaviruses belong to two distinct evolutionary lineages. J. Gen. Virol. 2018, 99, 567–573. [Google Scholar] [CrossRef]

| Number | Host Common Name | Host Taxonomic Name | Higher Host Taxon | n Animals (Samples) Tested with Generic PCR a | Organs Tested with Generic PCR | Countries of Origin of Animals Tested | Polyomavirus Positive in Generic PCR b |
|---|---|---|---|---|---|---|---|
| 1 | Domestic cattle | Bos taurus | Artiodactyla | 239 (247) | feces, lung, lymph node, spleen | Côte d’Ivoire, Spain, Uganda | + |
| 2 | Blue duiker | Philantomba monticola | Artiodactyla | 19 (19) | feces, intestine, spleen | Democratic Republic of the Congo | + |
| 3 | Black-fronted duiker | Cephalophus nigrifrons | Artiodactyla | 2 (3) | intestine, lung, spleen | Democratic Republic of the Congo | - |
| 4 | Peters´ duiker | Cephalophus callipygus | Artiodactyla | 8 (12) | feces, intestine, lung, spleen | Democratic Republic of the Congo | + |
| 5 | Yellow-backed duiker | Cephalophus silvicultor | Artiodactyla | 1 (1) | spleen | Democratic Republic of the Congo | - |
| 6 | Domestic goat | Capra aegagrus hircus | Artiodactyla | 148 (159) | feces, pooled lymph nodes | Côte d’Ivoire, Spain, Uganda | + |
| 7 | Domestic pig | Sus scrofa domesticus | Artiodactyla | 173 (359) | blood and diverse organs | Belgium, Côte d’Ivoire, Germany, Spain, Sweden, Switzerland, Uganda | + |
| 8 | Wild boar | Sus scrofa | Artiodactyla | 22 (52) | bladder, bone marrow, spleen, tonsil | Germany | - |
| 9 | Red river hog | Potamochoerus porcus | Artiodactyla | 1 (1) | spleen | Democratic Republic of the Congo | + |
| 10 | Sitatunga | Tragelaphus spekii | Artiodactyla | 1 (1) | intestine | Democratic Republic of the Congo | - |
| 11 | Water chevrotein | Hyemoschus aquaticus | Artiodactyla | 4 (8) | feces, intestine, lung, spleen | Democratic Republic of the Congo | - |
| 12 | Alpaca | Vicugna pacos | Artiodactyla | 3 (3) | liver, lung, lymph node | Spain | - |
| 13 | Domestic sheep | Ovis aries | Artiodactyla | 52 (52) | feces | Côte d’Ivoire, Uganda | - |
| 14 | Bottlenose dolphin | Tursiops truncatus | Artiodactyla | 2 (6) | kidney, liver, lung, skin, spleen | Germany | + |
| 15 | Mountain zebra | Equus zebra | Persissodactyla | 12 (12) | blood | Namibia | + |
| 16 | Domestic dog | Canis familiaris | Carnivora | 33 (36) | blood, feces, lung, salivary gland, spleen | Germany, USA, Uganda, Belgium | - |
| 17 | Wolf | Canis lupus | Carnivora | 49 (103) | salivary gland, spleen | Germany | + |
| 18 | Black-backed jackal | Canis mesomelas | Carnivora | 3 (3) | lung | Tanzania | - |
| 19 | Lion | Panthera leo | Carnivora | 28 (28) | blood, lung | Tanzania | + |
| 20 | Bat-eared fox | Otocyon megalotis | Carnivora | 3 (3) | lung | Tanzania | - |
| 21 | Spotted hyena | Crocuta crocuta | Carnivora | 6 (6) | blood, lung | Tanzania | - |
| 22 | Red fox | Vulpes vulpes | Carnivora | 11 (11) | spleen | Germany | - |
| 23 | European polecat | Mustela putorius | Carnivora | 3 (3) | spleen | Germany | - |
| 24 | Raccoon dog | Nyctereutes procyonoides | Carnivora | 20 (20) | spleen | Germany | - |
| 25 | Common tree shrew | Tupaia glis | Scandentia | 4 (12) | blood, lung | Thailand | + |
| 26 | Black rat | Rattus rattus | Rodentia | 5 (5) | spleen | Thailand | - |
| 27 | Norway rat | Rattus norvegicus | Rodentia | 33 (33) | spleen | Germany | + |
| 28 | Malayan field rat | Rattus tiomanicus | Rodentia | 7 (7) | spleen | Germany | - |
| 29 | Greater bandicoot rat | Bandicota indica | Rodentia | 13 (19) | lymph node, spleen | Germany | - |
| 30 | Savile’s bandicoot rat | Bandicota savilei | Rodentia | 6 (6) | spleen | Germany | - |
| 31 | House mouse | Mus musculus | Rodentia | 57 (76) | spleen, lung | Germany | + |
| 32 | Striped field mouse | Apodemus agrarius | Rodentia | 12 (29) | heart, kidney, liver, lung, lymph node, spleen | Germany | - |
| 33 | Wood mouse | Apodemus sylvaticus | Rodentia | 23 (26) | kidney, lung, spleen | Germany | - |
| 34 | Yellow-necked mouse | Apodemus flavicollis | Rodentia | 77 (81) | chest cavity fluid, lung, spleen | Germany | + |
| 35 | Bank vole | Myodes glareolus | Rodentia | 19 (23) | kidney, lung, lymph node, spleen | Germany | - |
| 36 | Multimammate mouse | Mastomys natalensis | Rodentia | 49 (59) | lung, spleen | Côte d’Ivoire | + |
| 37 | Edible dormouse | Glis glis | Rodentia | 3 (6) | spleen | Germany | + |
| 38 | Garden dormouse | Eliomys quercinus | Rodentia | 3 (5) | kidney, spleen | Germany | - |
| 39 | Hazel dormouse | Muscardinus avellanarius | Rodentia | 3 (4) | kidney, spleen | Germany | - |
| 40 | Common vole | Microtus arvalis | Rodentia | 30 (36) | kidney, lung, lymph node, spleen | Germany | - |
| 41 | Muskrat | Ondatra zibethicus | Rodentia | 19 (19) | lymph node | Germany | - |
| 42 | Syrian hamster | Mesocricetus auratus | Rodentia | 8 (8) | spleen | Germany | - |
| 43 | Koala | Phascolarctos cinereus | Diprotodontia | 6 (6) | blood | Germany | - |
| 44 | Rock hyrax | Procavia capensis | Hyracoidea | 2 (6) | liver, nervus axillaris, esophagus, parotid gland, spleen | Germany | - |
| Number | Host Common Name | Name of Identified Polyomavirus (Abbreviation) | n Samples Positive in Generic PCR (Body Compartment) | n Samples Positive in Specific PCR (Body Compartment) | n Animals Positive in Generic or Specific PCR | Country of Origin of PCR Positive Samples | Known or Novel Polyomavirus |
|---|---|---|---|---|---|---|---|
| 1 | Domestic cattle | Bovine polyomavirus (BoPyV) | 1 (lymph node) | not done | 1 | Spain | known |
| 2 | Blue duiker | Philantomba monticola polyomavirus 1 (PmonPyV1) | 4 (spleen) | not done | 4 | Democratic Republic of the Congo | novel |
| 3 | Peters´ duiker | Cephalophus callipygus polyomavirus 1 (CcalPyV1) | 1 (intestine) | not done | 1 | Democratic Republic of the Congo | novel |
| 4 | Domestic goat | Capra aegagrus polyomavirus 1 (CaegPyV1) | 1 (pooled lymph nodes) | 10 (pooled lymph nodes) | 6 | Spain | novel |
| 5 | Domestic goat | Capra aegagrus polyomavirus 2 (CaegPyV2) | 2 (pooled lymph nodes) | not done | 2 | Spain | novel |
| 6 | Domestic goat | Capra aegagrus polyomavirus 3 (CaegPyV3) | 1 (feces) | not done | 1 | Uganda | novel |
| 7 | Bottlenose dolphin | Tursiops truncatus polyomavirus 1 (TtruPyV1) | 1 (spleen) | not done | 1 | Germany | novel |
| 8 | Mountain zebra | Equus zebra polyomavirus 1 (EzebPyV1) | 1 (blood) | not done | 1 | Namibia | novel |
| 9 | Domestic pig | Sus scrofa polyomavirus 1 (SscrPyV1) | 1 (spleen) | 1 (spleen) | 1 | Germany | novel |
| 10 | Red river hog | Potamochoerus porcus polyomavirus 1 (PporPyV1) | 1 (spleen) | not done | 1 | Democratic Republic of the Congo | novel |
| 11 | Wolf | Canis lupus polyomavirus 1 (ClupPyV1) | 1 (spleen) | 5 (spleen, blood, pancreas) | 4 | Germany | novel |
| 12 | Lion | Panthera leo polyomavirus 1 (PleoPyV1) | 1 (lung) | 2 (lung) | 2 | Tanzania | novel |
| 13 | Common tree shrew | Tupaia glis polyomavirus 1 (TgliPyV1) | 1 (spleen) | 4 (spleen, lymph node) | 2 | Thailand | novel |
| 14 | Norway rat | Rattus norvegicus polyomavirus 1 (RnorPyV1) | 22 (spleen) | not done | 22 | Germany | novel |
| 15 | House mouse | Murine pneumotropic virus (MPtV) | 3 (spleen) | not done | 3 | Germany | known |
| 16 | Yellow-necked mouse | Apodemus flavicollis polyomavirus 1 (AflaPyV1) | 9 (lung, chest cavity fluid) | 16 (lung, chest cavity fluid) | 17 | Germany | novel |
| 17 | Edible dormouse | Glis glis polyomavirus 1 (GgliPyV1) | 1 (spleen) | 2 (spleen, kidney) | 1 | Germany | novel |
| 18 | Multimammate mouse | Mastomys natalensis polyomavirus 2 (MnatPyV2) | 2 (lung) | 1 (lung) | 3 | Côte d’Ivoire | novel |
| Number | Name of Identified Polyomavirus | n Genomes (Sample ID) | n Animals with Full Genome Detected | Genome Length (bp) | Early Region mRNA Splice Sites Identified in Cell Culture | GenBank Accession Numbers |
|---|---|---|---|---|---|---|
| 1 | Philantomba monticola polyomavirus 1 | 1 (#9781) | 1 | 5034 | + | MG654482 |
| 2 | Capra aegagrus polyomavirus 1 | 2 (#7515, #9483) | 2 | 4699 | + | MG654479, MG654480 |
| 3 | Sus scrofa polyomavirus 1 | 1 (#0471) | 1 | 5058 | + | KR065722 |
| 4 | Potamochoerus porcus polyomavirus 1 | 1 (#9780) | 1 | 4825 | + | MG654481 |
| 5 | Canis lupus polyomavirus 1 | 2 (#8472; #8476) | 2 | 5215 | + | MG701355, MG701356 |
| 6 | Panthera leo polyomavirus 1 | 2 (#3884; #3887) | 2 | 5018 | + | MG701353, MG701354 |
| 7 | Tupaia glis polyomavirus 1 | 3 (#4373; #4376; #4472) | 3 | 5234 | + | MG721015, MG721016, MG721017 |
| 8 | Rattus norvegicus polyomavirus 1 | 6 (#3671; #3687; #3690; #5700; #5704; #5709) | 6 | 5318 | + | KR065723, KR065724, KR075943, KR075944, KR075945, KR075946 |
| 9 | Apodemus flavicollis polyomavirus 1 | 3 (#3349; #4021; #9779) | 3 | 5327 | + | MG654476, MG654477, MG654478 |
| 10 | Mastomys natalensis polyomavirus 2 | 2 (8173; #8174) | 2 | 5322 | + | MG701350, MG701351 |
| 11 | Glis glis polyomavirus 1 | 1 (#3327) | 1 | 5338 | + a | MG701352 |
| Analysis a | # Cospeciations | # Duplications | # Duplications and Host Switches | # Losses | # Failures to Diverge | Cost | % Samples Best Cost ≤ Original |
|---|---|---|---|---|---|---|---|
| Terrestrial vertebrate poylomaviruses (8679 solutions—all same costs) b | 60 | 15 | 30 | 208 | 0 | −60 | 0 |
| Alphapolyomaviruses (9795 solutions—all same costs) c | 28 | 8 | 12 | 81 | 0 | −28 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ehlers, B.; Anoh, A.E.; Ben Salem, N.; Broll, S.; Couacy-Hymann, E.; Fischer, D.; Gedvilaite, A.; Ingenhütt, N.; Liebmann, S.; Martin, M.; et al. Novel Polyomaviruses in Mammals from Multiple Orders and Reassessment of Polyomavirus Evolution and Taxonomy. Viruses 2019, 11, 930. https://doi.org/10.3390/v11100930
Ehlers B, Anoh AE, Ben Salem N, Broll S, Couacy-Hymann E, Fischer D, Gedvilaite A, Ingenhütt N, Liebmann S, Martin M, et al. Novel Polyomaviruses in Mammals from Multiple Orders and Reassessment of Polyomavirus Evolution and Taxonomy. Viruses. 2019; 11(10):930. https://doi.org/10.3390/v11100930
Chicago/Turabian StyleEhlers, Bernhard, Augustin E. Anoh, Nicole Ben Salem, Sebastian Broll, Emmanuel Couacy-Hymann, Daniela Fischer, Alma Gedvilaite, Nanina Ingenhütt, Sonja Liebmann, Maite Martin, and et al. 2019. "Novel Polyomaviruses in Mammals from Multiple Orders and Reassessment of Polyomavirus Evolution and Taxonomy" Viruses 11, no. 10: 930. https://doi.org/10.3390/v11100930
APA StyleEhlers, B., Anoh, A. E., Ben Salem, N., Broll, S., Couacy-Hymann, E., Fischer, D., Gedvilaite, A., Ingenhütt, N., Liebmann, S., Martin, M., Mossoun, A., Mugisha, L., Muyembe-Tamfum, J.-J., Pauly, M., Pérez de Val, B., Preugschas, H., Richter, D., Schubert, G., Szentiks, C. A., ... Calvignac-Spencer, S. (2019). Novel Polyomaviruses in Mammals from Multiple Orders and Reassessment of Polyomavirus Evolution and Taxonomy. Viruses, 11(10), 930. https://doi.org/10.3390/v11100930






