Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication
Abstract
1. Introduction
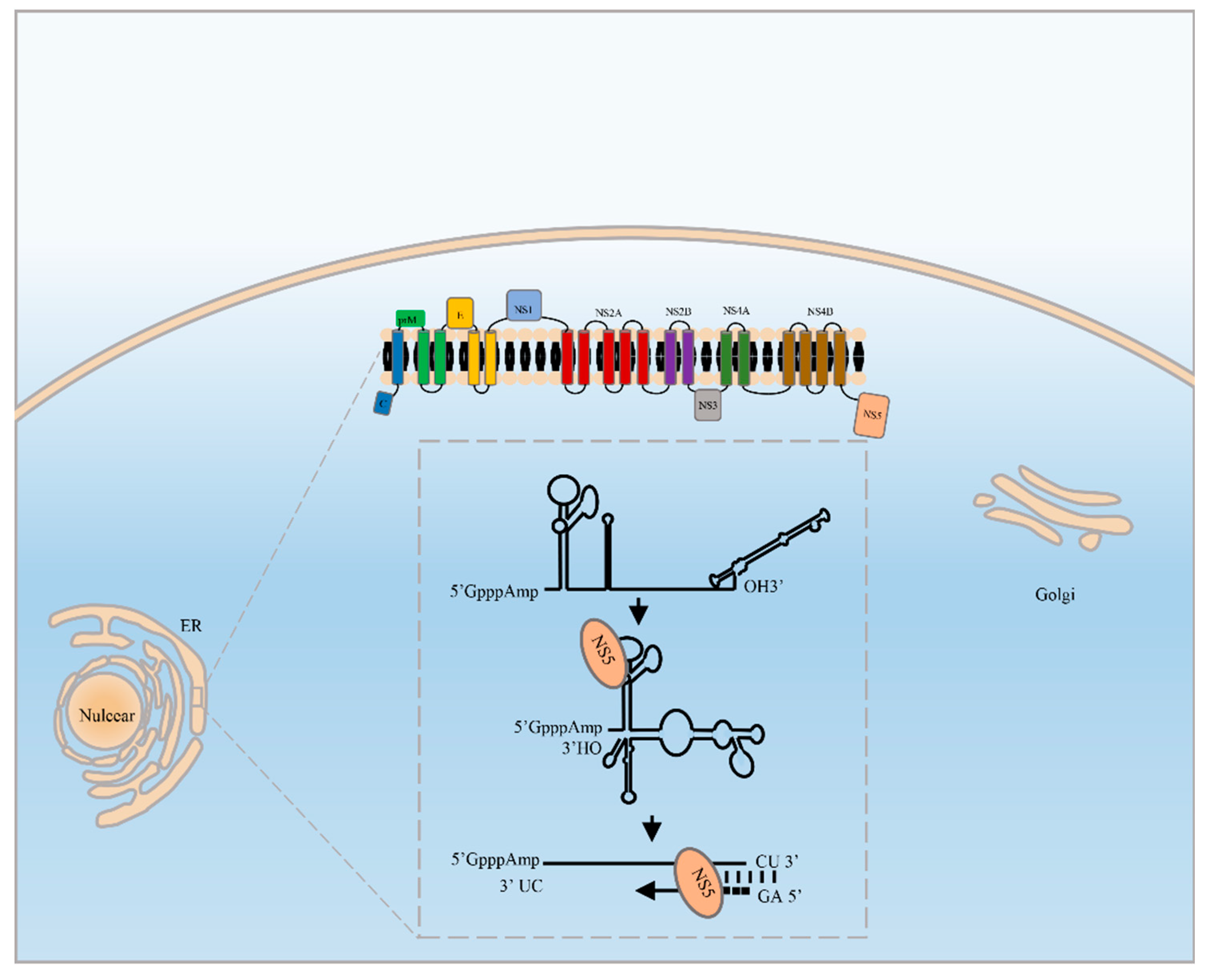
2. The RdRp Manipulates Viral RNA Replication
3. The Functional Motifs of the RdRp
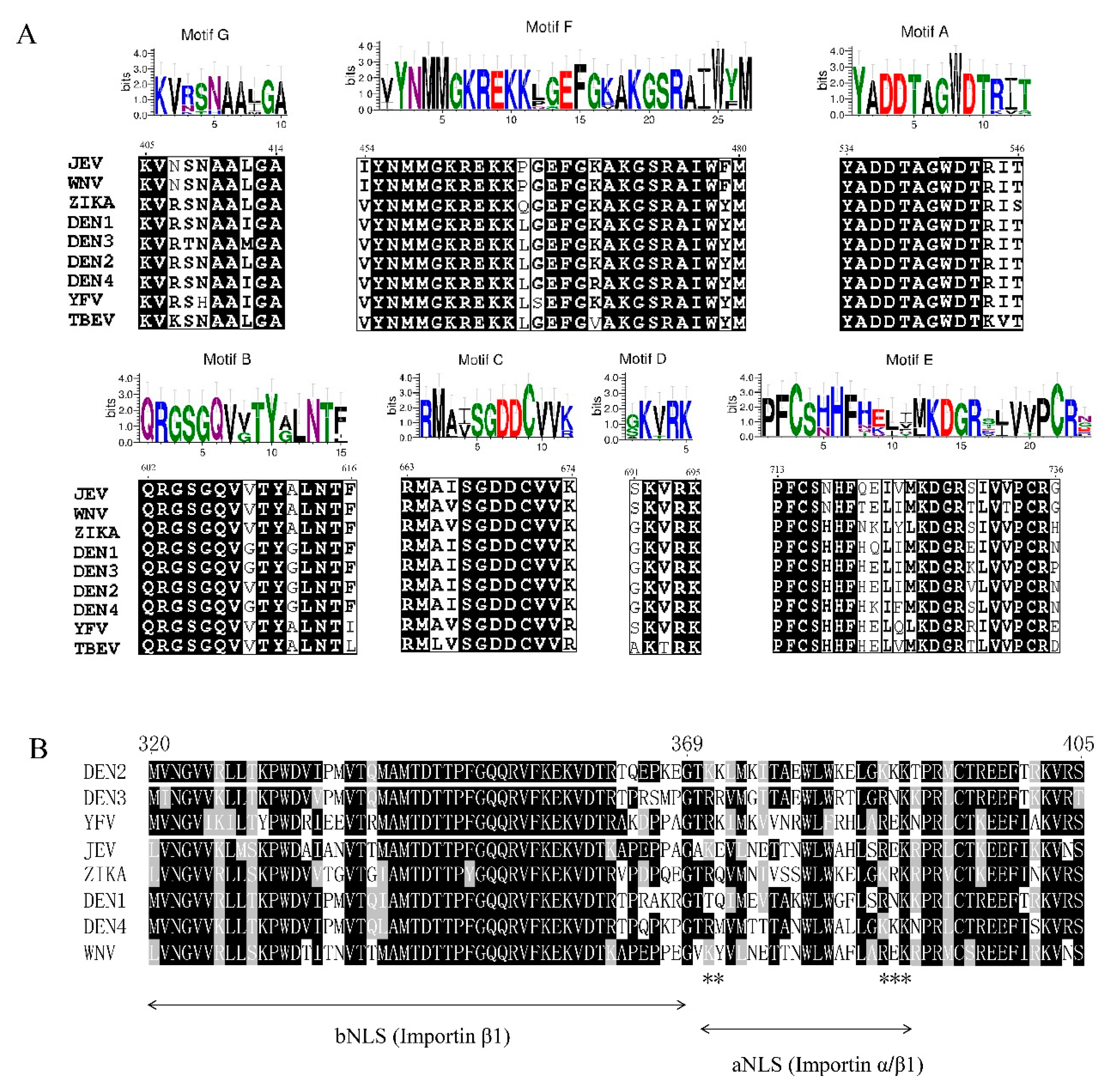
3.1. Motifs A-E of the RdRp
3.2. Nuclear Localization Signal of the RdRp
4. The Interactions Between the RdRp and Functional Viral Components
4.1. RdRp Binds to the 5′UTR and 3′UTR to Initiate RNA Synthesis
4.2. RdRp Cooperates with NS3 to Facilitate RNA Synthesis
4.3. The MTase Interacts with the RdRp to Regulate RNA Synthesis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bayry, J. Emerging and Re-Emerging Infectious Diseases of Livestock; Springer: Cham, Switzerland, 2017. [Google Scholar]
- Malet, H.; Egloff, M.P.; Selisko, B.; Butcher, R.E.; Wright, P.J.; Roberts, M.; Gruez, A.; Sulzenbacher, G.; Vonrhein, C.; Bricogne, G.; et al. Crystal structure of the RNA polymerase domain of the West Nile virus non-structural protein 5. J. Biol. Chem. 2007, 282, 10678–10689. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Rice, C.M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 1999, 73, 4611–4621. [Google Scholar] [PubMed]
- Youn, S.; Li, T.; McCune, B.T.; Edeling, M.A.; Fremont, D.H.; Cristea, I.M.; Diamond, M.S. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 2012, 86, 7360–7371. [Google Scholar] [CrossRef] [PubMed]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Takeda, K.; Markoff, L. Protein-protein interactions among West Nile non-structural proteins and transmembrane complex formation in mammalian cells. Virology 2013, 446, 365–377. [Google Scholar] [CrossRef]
- Xie, X.; Gayen, S.; Kang, C.; Yuan, Z.; Shi, P.Y. Membrane topology and function of dengue virus NS2A protein. J. Virol. 2013, 87, 4609–4622. [Google Scholar] [CrossRef]
- Zou, J.; Lee le, T.; Wang, Q.Y.; Xie, X.; Lu, S.; Yau, Y.H.; Yuan, Z.; Geifman Shochat, S.; Kang, C.; Lescar, J.; et al. Mapping the Interactions between the NS4B and NS3 proteins of dengue virus. J. Virol. 2015, 89, 3471–3483. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Wong, Y.L.; Liew, L.S.; Kang, C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim. Biophys. Acta 2015, 1848, 2244–2252. [Google Scholar] [CrossRef]
- Umareddy, I.; Chao, A.; Sampath, A.; Gu, F.; Vasudevan, S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006, 87, 2605–2614. [Google Scholar] [CrossRef]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antivir. Res. 2015, 118, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Marceau, C.D.; Puschnik, A.S.; Majzoub, K.; Ooi, Y.S.; Brewer, S.M.; Fuchs, G.; Swaminathan, K.; Mata, M.A.; Elias, J.E.; Sarnow, P.; et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Puschnik, A.S.; Majzoub, K.; Ooi, Y.S.; Carette, J.E. A CRISPR toolbox to study virus-host interactions. Nat. Rev. Microbiol. 2017, 15, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Swarbrick, C.M.D.; Basavannacharya, C.; Chan, K.W.K.; Chan, S.A.; Singh, D.; Wei, N.; Phoo, W.W.; Luo, D.; Lescar, J.; Vasudevan, S.G. NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Res. 2017, 45, 12904–12920. [Google Scholar] [CrossRef] [PubMed]
- Chu, P.W.; Westaway, E.G. Replication strategy of Kunjin virus: Evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology 1985, 140, 68–79. [Google Scholar] [CrossRef]
- Dong, H.; Zhang, B.; Shi, P.Y. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 2008, 381, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Lodeiro, M.F.; Filomatori, C.V.; Gamarnik, A.V. Structural and functional studies of the promoter element for dengue virus RNA replication. J. Virol. 2009, 83, 993–1008. [Google Scholar] [CrossRef]
- You, S.; Falgout, B.; Markoff, L.; Padmanabhan, R. In vitro RNA synthesis from exogenous dengue viral RNA templates requires long range interactions between 5′- and 3′-terminal regions that influence RNA structure. J. Biol. Chem. 2001, 276, 15581–15591. [Google Scholar] [CrossRef]
- Selisko, B.; Potisopon, S.; Agred, R.; Priet, S.; Varlet, I.; Thillier, Y.; Sallamand, C.; Debart, F.; Vasseur, J.J.; Canard, B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012, 8, e1002912. [Google Scholar] [CrossRef]
- Kim, Y.G.; Yoo, J.S.; Kim, J.H.; Kim, C.M.; Oh, J.W. Biochemical characterization of a recombinant Japanese encephalitis virus RNA-dependent RNA polymerase. BMC Mol. Biol. 2007, 8, 59. [Google Scholar] [CrossRef]
- Choi, K.H.; Rossmann, M.G. RNA-dependent RNA polymerases from Flaviviridae. Curr. Opin. Struct. Biol. 2009, 19, 746–751. [Google Scholar] [CrossRef] [PubMed]
- Appleby, T.C.; Perry, J.K.; Murakami, E.; Barauskas, O.; Feng, J.; Cho, A.; Fox, D., 3rd; Wetmore, D.R.; McGrath, M.E.; Ray, A.S.; et al. Viral replication. Structural basis for RNA replication by the hepatitis C virus polymerase. Science 2015, 347, 771–775. [Google Scholar] [CrossRef] [PubMed]
- Godoy, A.S.; Lima, G.M.; Oliveira, K.I.; Torres, N.U.; Maluf, F.V.; Guido, R.V.; Oliva, G. Crystal structure of Zika virus NS5 RNA-dependent RNA polymerase. Nat. Commun. 2017, 8, 14764. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Lim, S.P.; Chen, Y.L.; Liew, C.W.; Yap, L.; Lescar, J.; Shi, P.Y. Conformational flexibility of the Dengue virus RNA-dependent RNA polymerase revealed by a complex with an inhibitor. J. Virol. 2013, 87, 5291–5295. [Google Scholar] [CrossRef] [PubMed]
- Surana, P.; Satchidanandam, V.; Nair, D.T. RNA-dependent RNA polymerase of Japanese encephalitis virus binds the initiator nucleotide GTP to form a mechanistically important pre-initiation state. Nucleic Acids Res. 2014, 42, 2758–2773. [Google Scholar] [CrossRef] [PubMed]
- Shu, B.; Gong, P. The uncoupling of catalysis and translocation in the viral RNA-dependent RNA polymerase. RNA Biol. 2017, 14, 1314–1319. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Smidansky, E.D.; Maksimchuk, K.R.; Lum, D.; Welch, J.L.; Arnold, J.J.; Cameron, C.E.; Boehr, D.D. Motif D of viral RNA-dependent RNA polymerases determines efficiency and fidelity of nucleotide addition. Structure 2012, 20, 1519–1527. [Google Scholar] [CrossRef]
- Dubankova, A.; Boura, E. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antivir. Res. 2019, 169, 104536. [Google Scholar] [CrossRef]
- Potapova, U.; Feranchuk, S.; Leonova, G.; Belikov, S. The rearrangement of motif F in the flavivirus RNA-directed RNA polymerase. Int. J. Biol. Macromol. 2018, 108, 990–998. [Google Scholar] [CrossRef]
- Eyer, L.; Nougairede, A.; Uhlirova, M.; Driouich, J.S.; Zouharova, D.; Valdes, J.J.; Haviernik, J.; Gould, E.A.; De Clercq, E.; de Lamballerie, X.; et al. An E460D Substitution in the NS5 Protein of Tick-Borne Encephalitis Virus Confers Resistance to the Inhibitor Galidesivir (BCX4430) and Also Attenuates the Virus for Mice. J. Virol. 2019, 93, e00367-19. [Google Scholar] [CrossRef]
- Lu, G.; Gong, P. Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013, 9, e1003549. [Google Scholar] [CrossRef] [PubMed]
- Zou, G.; Chen, Y.L.; Dong, H.; Lim, C.C.; Yap, L.J.; Yau, Y.H.; Shochat, S.G.; Lescar, J.; Shi, P.Y. Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 2011, 286, 14362–14372. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Song, H.; Wang, H.; Chai, Y.; Su, C.; Qi, J.; Shi, Y.; Gao, G.F. The crystal structure of Zika virus NS5 reveals conserved drug targets. EMBO J. 2017, 36, 919–933. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Yang, C.; Zhang, W.; Mahalingam, S.; Wang, M.; Cheng, A. Flaviviridae virus nonstructural proteins 5 and 5A mediate viral immune evasion and are promising targets in drug development. Pharmacol. Ther. 2018, 190, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Noble, C.G.; Lim, S.P.; Arora, R.; Yokokawa, F.; Nilar, S.; Seh, C.C.; Wright, S.K.; Benson, T.E.; Smith, P.W.; Shi, P.Y. A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-based Screening. J. Biol. Chem. 2016, 291, 8541–8548. [Google Scholar] [CrossRef] [PubMed]
- Cannalire, R.; Tarantino, D.; Astolfi, A.; Barreca, M.L.; Sabatini, S.; Massari, S.; Tabarrini, O.; Milani, M.; Querat, G.; Mastrangelo, E.; et al. Functionalized 2,1-benzothiazine 2,2-dioxides as new inhibitors of Dengue NS5 RNA-dependent RNA polymerase. Eur. J. Med. Chem. 2018, 143, 1667–1676. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.P.; Noble, C.G.; Seh, C.C.; Soh, T.S.; El Sahili, A.; Chan, G.K.; Lescar, J.; Arora, R.; Benson, T.; Nilar, S.; et al. Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling. PLoS Pathog. 2016, 12, e1005737. [Google Scholar] [CrossRef]
- Buckley, A.; Gaidamovich, S.; Turchinskaya, A.; Gould, E.A. Monoclonal antibodies identify the NS5 yellow fever virus non-structural protein in the nuclei of infected cells. J. Gen. Virol. 1992, 73 Pt 5, 1125–1130. [Google Scholar] [CrossRef]
- Hou, W.; Cruz-Cosme, R.; Armstrong, N.; Obwolo, L.A.; Wen, F.; Hu, W.; Luo, M.H.; Tang, Q. Molecular cloning and characterization of the genes encoding the proteins of Zika virus. Gene 2017, 628, 117–128. [Google Scholar] [CrossRef]
- Hannemann, H.; Sung, P.Y.; Chiu, H.C.; Yousuf, A.; Bird, J.; Lim, S.P.; Davidson, A.D. Serotype-specific differences in dengue virus non-structural protein 5 nuclear localization. J. Biol. Chem. 2013, 288, 22621–22635. [Google Scholar] [CrossRef]
- Lopez-Denman, A.J.; Russo, A.; Wagstaff, K.M.; White, P.A.; Jans, D.A.; Mackenzie, J.M. Nucleocytoplasmic shuttling of the West Nile virus RNA-dependent RNA polymerase NS5 is critical to infection. Cell. Microbiol. 2018, 20, e12848. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.H.; Liu, M.L.; Tien, C.F.; Chou, S.J.; Chang, R.Y. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) interaction with 3′ ends of Japanese encephalitis virus RNA and colocalization with the viral NS5 protein. J. Biomed. Sci. 2009, 16, 40. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.; Brooks, A.J.; Jans, D.A.; Vasudevan, S.G. A small region of the dengue virus-encoded RNA-dependent RNA polymerase, NS5, confers interaction with both the nuclear transport receptor importin-beta and the viral helicase, NS3. J. Gen. Virol. 2001, 82, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Brooks, A.J.; Johansson, M.; John, A.V.; Xu, Y.; Jans, D.A.; Vasudevan, S.G. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3 contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J. Biol. Chem. 2002, 277, 36399–36407. [Google Scholar] [CrossRef] [PubMed]
- Ng, I.H.W.; Chan, K.W.; Tan, M.J.A.; Gwee, C.P.; Smith, K.M.; Jeffress, S.J.; Saw, W.G.; Swarbrick, C.M.D.; Watanabe, S.; Jans, D.A.; et al. Zika Virus NS5 Forms Supramolecular Nuclear Bodies That Sequester Importin-alpha and Modulate the Host Immune and Pro-Inflammatory Response in Neuronal Cells. ACS Infect. Dis. 2019, 5, 932–948. [Google Scholar] [CrossRef] [PubMed]
- Pryor, M.J.; Rawlinson, S.M.; Butcher, R.E.; Barton, C.L.; Waterhouse, T.A.; Vasudevan, S.G.; Bardin, P.G.; Wright, P.J.; Jans, D.A.; Davidson, A.D. Nuclear localization of dengue virus nonstructural protein 5 through its importin alpha/beta-recognized nuclear localization sequences is integral to viral infection. Traffic 2007, 8, 795–807. [Google Scholar] [CrossRef]
- Tay, M.Y.; Smith, K.; Ng, I.H.; Chan, K.W.; Zhao, Y.; Ooi, E.E.; Lescar, J.; Luo, D.; Jans, D.A.; Forwood, J.K.; et al. The C-terminal 18 Amino Acid Region of Dengue Virus NS5 Regulates its Subcellular Localization and Contains a Conserved Arginine Residue Essential for Infectious Virus Production. PLoS Pathog. 2016, 12, e1005886. [Google Scholar] [CrossRef]
- Rawlinson, S.M.; Pryor, M.J.; Wright, P.J.; Jans, D.A. CRM1-mediated nuclear export of dengue virus RNA polymerase NS5 modulates interleukin-8 induction and virus production. J. Biol. Chem. 2009, 284, 15589–15597. [Google Scholar] [CrossRef]
- De Maio, F.A.; Risso, G.; Iglesias, N.G.; Shah, P.; Pozzi, B.; Gebhard, L.G.; Mammi, P.; Mancini, E.; Yanovsky, M.J.; Andino, R.; et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016, 12, e1005841. [Google Scholar] [CrossRef]
- Grant, A.; Ponia, S.S.; Tripathi, S.; Balasubramaniam, V.; Miorin, L.; Sourisseau, M.; Schwarz, M.C.; Sanchez-Seco, M.P.; Evans, M.J.; Best, S.M.; et al. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 2016, 19, 882–890. [Google Scholar] [CrossRef]
- Morrison, J.; Laurent-Rolle, M.; Maestre, A.M.; Rajsbaum, R.; Pisanelli, G.; Simon, V.; Mulder, L.C.; Fernandez-Sesma, A.; Garcia-Sastre, A. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013, 9, e1003265. [Google Scholar] [CrossRef] [PubMed]
- Ashour, J.; Laurent-Rolle, M.; Shi, P.Y.; Garcia-Sastre, A. NS5 of dengue virus mediates STAT2 binding and degradation. J. Virol. 2009, 83, 5408–5418. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Buhler, S.; Selisko, B.; Davidson, A.; Mulder, K.; Canard, B.; Miller, S.; Bartenschlager, R. Nuclear localization of dengue virus nonstructural protein 5 does not strictly correlate with efficient viral RNA replication and inhibition of type I interferon signaling. J. Virol. 2013, 87, 4545–4557. [Google Scholar] [CrossRef] [PubMed]
- Jans, D.A.; Xiao, C.Y.; Lam, M.H. Nuclear targeting signal recognition: A key control point in nuclear transport? Bioessays 2000, 22, 532–544. [Google Scholar] [CrossRef]
- Tay, M.Y.; Fraser, J.E.; Chan, W.K.; Moreland, N.J.; Rathore, A.P.; Wang, C.; Vasudevan, S.G.; Jans, D.A. Nuclear localization of dengue virus (DENV) 1-4 non-structural protein 5; protection against all 4 DENV serotypes by the inhibitor Ivermectin. Antivir. Res. 2013, 99, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yang, S.N.Y.; Smith, K.; Forwood, J.K.; Jans, D.A. Nuclear import inhibitor N-(4-hydroxyphenyl) retinamide targets Zika virus (ZIKV) nonstructural protein 5 to inhibit ZIKV infection. Biochem. Biophys. Res. Commun. 2017, 493, 1555–1559. [Google Scholar] [CrossRef] [PubMed]
- Fraser, J.E.; Watanabe, S.; Wang, C.; Chan, W.K.; Maher, B.; Lopez-Denman, A.; Hick, C.; Wagstaff, K.M.; Mackenzie, J.M.; Sexton, P.M.; et al. A nuclear transport inhibitor that modulates the unfolded protein response and provides in vivo protection against lethal dengue virus infection. J. Infect. Dis. 2014, 210, 1780–1791. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.N.Y.; Atkinson, S.C.; Fraser, J.E.; Wang, C.; Maher, B.; Roman, N.; Forwood, J.K.; Wagstaff, K.M.; Borg, N.A.; Jans, D.A. Novel Flavivirus Antiviral That Targets the Host Nuclear Transport Importin alpha/beta1 Heterodimer. Cells 2019, 8, 281. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Iglesias, N.G.; Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J. Biol. Chem. 2011, 286, 6929–6939. [Google Scholar] [CrossRef] [PubMed]
- Filomatori, C.V.; Lodeiro, M.F.; Alvarez, D.E.; Samsa, M.M.; Pietrasanta, L.; Gamarnik, A.V. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006, 20, 2238–2249. [Google Scholar] [CrossRef]
- Yu, L.; Markoff, L. The topology of bulges in the long stem of the flavivirus 3′ stem-loop is a major determinant of RNA replication competence. J. Virol. 2005, 79, 2309–2324. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, D.E.; Lodeiro, M.F.; Luduena, S.J.; Pietrasanta, L.I.; Gamarnik, A.V. Long-range RNA-RNA interactions circularize the dengue virus genome. J. Virol. 2005, 79, 6631–6643. [Google Scholar] [CrossRef] [PubMed]
- Polacek, C.; Foley, J.E.; Harris, E. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J. Virol. 2009, 83, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Friebe, P.; Harris, E. Interplay of RNA elements in the dengue virus 5′ and 3′ ends required for viral RNA replication. J. Virol. 2010, 84, 6103–6118. [Google Scholar] [CrossRef] [PubMed]
- Bujalowski, P.J.; Bujalowski, W.; Choi, K.H. Interactions between the Dengue Virus Polymerase NS5 and Stem-Loop A. J. Virol. 2017, 91, e00047-17. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Li, X.F.; Jiang, T.; Deng, Y.Q.; Ye, Q.; Zhao, H.; Yu, J.Y.; Qin, C.F. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife 2016, 5, e17636. [Google Scholar] [CrossRef]
- Hodge, K.; Tunghirun, C.; Kamkaew, M.; Limjindaporn, T.; Yenchitsomanus, P.T.; Chimnaronk, S. Identification of a Conserved RNA-dependent RNA Polymerase (RdRp)-RNA Interface Required for Flaviviral Replication. J. Biol. Chem. 2016, 291, 17437–17449. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Sanles, A.; Rios-Marco, P.; Romero-Lopez, C.; Berzal-Herranz, A. Functional Information Stored in the Conserved Structural RNA Domains of Flavivirus Genomes. Front. Microbiol. 2017, 8, 546. [Google Scholar] [CrossRef]
- Villordo, S.M.; Carballeda, J.M.; Filomatori, C.V.; Gamarnik, A.V. RNA Structure Duplications and Flavivirus Host Adaptation. Trends Microbiol. 2016, 24, 270–283. [Google Scholar] [CrossRef]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef]
- Tilgner, M.; Deas, T.S.; Shi, P.Y. The flavivirus-conserved penta-nucleotide in the 3′ stem-loop of the West Nile virus genome requires a specific sequence and structure for RNA synthesis, but not for viral translation. Virology 2005, 331, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Mosley, R.T.; Edwards, T.E.; Murakami, E.; Lam, A.M.; Grice, R.L.; Du, J.; Sofia, M.J.; Furman, P.A.; Otto, M.J. Structure of hepatitis C virus polymerase in complex with primer-template RNA. J. Virol. 2012, 86, 6503–6511. [Google Scholar] [CrossRef] [PubMed]
- Nomaguchi, M.; Ackermann, M.; Yon, C.; You, S.; Padmanabhan, R. De novo synthesis of negative-strand RNA by Dengue virus RNA-dependent RNA polymerase in vitro: Nucleotide, primer, and template parameters. J. Virol. 2003, 77, 8831–8842. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, M.; Padmanabhan, R. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 2001, 276, 39926–39937. [Google Scholar] [CrossRef] [PubMed]
- Mayhoub, A.S. Hepatitis C RNA-dependent RNA polymerase inhibitors: A review of structure-activity and resistance relationships; different scaffolds and mutations. Bioorg. Med. Chem. 2012, 20, 3150–3161. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.H.; Fu, J.; Sugrue, R.J.; Yap, E.H.; Chan, Y.C.; Tan, Y.H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 1996, 216, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Xu, T.; Hunke, C.; Gruber, G.; Vasudevan, S.G.; Lescar, J. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008, 82, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Jain, R.; Coloma, J.; Garcia-Sastre, A.; Aggarwal, A.K. Structure of the NS3 helicase from Zika virus. Nat. Struct. Mol. Biol. 2016, 23, 752–754. [Google Scholar] [CrossRef] [PubMed]
- Tay, M.Y.; Saw, W.G.; Zhao, Y.; Chan, K.W.; Singh, D.; Chong, Y.; Forwood, J.K.; Ooi, E.E.; Gruber, G.; Lescar, J.; et al. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J. Biol. Chem. 2015, 290, 2379–2394. [Google Scholar] [CrossRef]
- Yon, C.; Teramoto, T.; Mueller, N.; Phelan, J.; Ganesh, V.K.; Murthy, K.H.; Padmanabhan, R. Modulation of the nucleoside triphosphatase/RNA helicase and 5′-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 2005, 280, 27412–27419. [Google Scholar] [CrossRef]
- Xu, S.; Ci, Y.; Wang, L.; Yang, Y.; Zhang, L.; Xu, C.; Qin, C.; Shi, L. Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase. Nucleic Acids Res. 2019, 47, 8693–8707. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Yi, G.; Du, F.; Chuang, Y.C.; Vaughan, R.C.; Sankaran, B.; Kao, C.C.; Li, P. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017, 8, 14762. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.K.; Cyr, M.; Longenecker, K.; Tripathi, R.; Sun, C.; Kempf, D.J. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Klema, V.J.; Ye, M.; Hindupur, A.; Teramoto, T.; Gottipati, K.; Padmanabhan, R.; Choi, K.H. Dengue Virus Nonstructural Protein 5 (NS5) Assembles into a Dimer with a Unique Methyltransferase and Polymerase Interface. PLoS Pathog. 2016, 12, e1005451. [Google Scholar] [CrossRef] [PubMed]
- Ferrero, D.S.; Ruiz-Arroyo, V.M.; Soler, N.; Uson, I.; Guarne, A.; Verdaguer, N. Supramolecular arrangement of the full-length Zika virus NS5. PLoS Pathog. 2019, 15, e1007656. [Google Scholar] [CrossRef] [PubMed]
- Potisopon, S.; Priet, S.; Collet, A.; Decroly, E.; Canard, B.; Selisko, B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014, 42, 11642–11656. [Google Scholar] [CrossRef]
- Teramoto, T.; Balasubramanian, A.; Choi, K.H.; Padmanabhan, R. Serotype-specific interactions among functional domains of dengue virus 2 nonstructural proteins (NS) 5 and NS3 are crucial for viral RNA replication. J. Biol. Chem. 2017, 292, 9465–9479. [Google Scholar] [CrossRef]
- Li, X.D.; Shan, C.; Deng, C.L.; Ye, H.Q.; Shi, P.Y.; Yuan, Z.M.; Gong, P.; Zhang, B. The interface between methyltransferase and polymerase of NS5 is essential for flavivirus replication. PLoS Negl. Trop. Dis. 2014, 8, e2891. [Google Scholar] [CrossRef]
- Saw, W.G.; Chan, K.W.; Vasudevan, S.G.; Gruber, G. Zika virus nonstructural protein 5 residue R681 is critical for dimer formation and enzymatic activity. FEBS Lett. 2019, 593, 1272–1291. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duan, Y.; Zeng, M.; Jiang, B.; Zhang, W.; Wang, M.; Jia, R.; Zhu, D.; Liu, M.; Zhao, X.; Yang, Q.; et al. Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication. Viruses 2019, 11, 929. https://doi.org/10.3390/v11100929
Duan Y, Zeng M, Jiang B, Zhang W, Wang M, Jia R, Zhu D, Liu M, Zhao X, Yang Q, et al. Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication. Viruses. 2019; 11(10):929. https://doi.org/10.3390/v11100929
Chicago/Turabian StyleDuan, YanPing, Miao Zeng, Bowen Jiang, Wei Zhang, Mingshu Wang, Renyong Jia, Dekang Zhu, Mafeng Liu, Xinxin Zhao, Qiao Yang, and et al. 2019. "Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication" Viruses 11, no. 10: 929. https://doi.org/10.3390/v11100929
APA StyleDuan, Y., Zeng, M., Jiang, B., Zhang, W., Wang, M., Jia, R., Zhu, D., Liu, M., Zhao, X., Yang, Q., Wu, Y., Zhang, S., Liu, Y., Zhang, L., Yu, Y., Pan, L., Chen, S., & Cheng, A. (2019). Flavivirus RNA-Dependent RNA Polymerase Interacts with Genome UTRs and Viral Proteins to Facilitate Flavivirus RNA Replication. Viruses, 11(10), 929. https://doi.org/10.3390/v11100929




