Mycoviral Population Dynamics in Spanish Isolates of the Entomopathogenic Fungus Beauveria bassiana
Abstract
1. Introduction
2. Materials and Methods
2.1. Screening of Fungal Isolates and Molecular Cloning
2.2. Computational and Phylogenetic Analysis
2.3. Insect Pathogenicity Experiments
3. Results and Discussion
3.1. Presence of dsRNA Elements in B. bassiana Isolates from the Iberian Peninsula
3.2. Mixed Infections of B. bassiana Isolates with Up to Three Different Mycoviruses
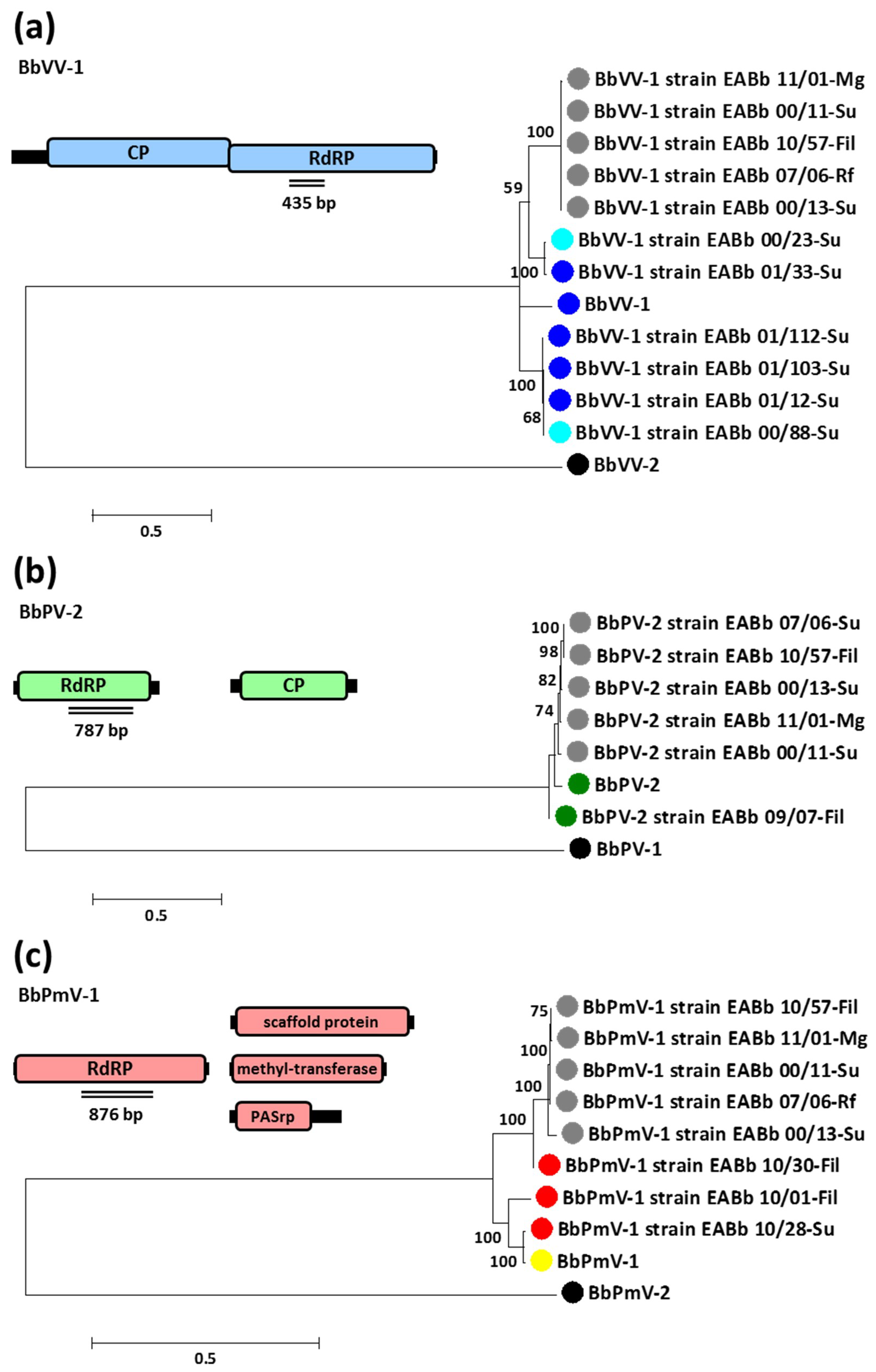
3.3. Selection Pressures and Recombination Events within the Viral Genome
3.4. Evidence of Vertical and Horizontal Transmission of Mycoviruses
3.5. Pathogenicity of Virus-Infected B. Bassiana against the Mediterranean Fruit Fly.
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Chandler, D.; Bailey, A.S.; Tatchell, G.M.; Davidson, G.; Greaves, J.; Grant, W.P. The development, regulation and use of biopesticides for integrated pest management. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 1987–1998. [Google Scholar] [CrossRef] [PubMed]
- De Faria, M.R.; Wraight, S.P. Mycoinsecticides and Mycoacaricides: A comprehensive list with worldwide coverage and international classification of formulation types. Biol. Control 2007, 43, 237–256. [Google Scholar] [CrossRef]
- Lacey, L.A.; Grzywacz, D.; Shapiro-Ilan, D.I.; Frutos, R.; Brownbridge, M.; Goettel, M.S. Insect pathogens as biological control agents: Back to the future. J. Invertebr. Pathol. 2015, 132, 1–41. [Google Scholar] [CrossRef] [PubMed]
- St. Leger, R.J.; Wang, C. Genetic engineering of fungal biocontrol agents to achieve greater efficacy against insect pests. Appl. Microbiol. Biotechnol. 2010, 85, 901–907. [Google Scholar] [CrossRef] [PubMed]
- Kanhayuwa, L.; Kotta-Loizou, I.; Özkan, S.; Gunning, A.P.; Coutts, R.H.A. A novel mycovirus from Aspergillus fumigatus contains four unique dsRNAs as its genome and is infectious as dsRNA. Proc. Natl. Acad. Sci. USA 2015, 112, 9100–9105. [Google Scholar] [CrossRef] [PubMed]
- Kotta-Loizou, I.; Coutts, R.H.A. Studies on the virome of the entomopathogenic fungus Beauveria bassiana reveal novel dsRNA elements and mild hypervirulence. PLoS Pathog. 2017, 13, e1006183. [Google Scholar] [CrossRef] [PubMed]
- Nuss, D.L. Biological control of chestnut blight: An example of virus-mediated attenuation of fungal pathogenesis. Microbiol. Rev. 1992, 56, 561–576. [Google Scholar] [PubMed]
- Bhatti, M.F.; Jamal, A.; Petrou, M.A.; Cairns, T.C.; Bignell, E.M.; Coutts, R.H.A. The effects of dsRNA mycoviruses on growth and murine virulence of Aspergillus fumigatus. Fungal Genet. Biol. 2011, 48, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Xie, J.; Cheng, J.; Fu, Y.; Li, G.; Yi, X.; Jiang, D. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc. Natl. Acad. Sci. USA 2014, 111, 12205–12210. [Google Scholar] [PubMed]
- Özkan, S.; Coutts, R.H.A. Aspergillus fumigatus mycovirus causes mild hypervirulent effect on pathogenicity when tested on Galleria mellonella. Fungal Genet. Biol. 2015, 76, 20–26. [Google Scholar] [CrossRef] [PubMed]
- Okada, R.; Ichinose, S.; Takeshita, K.; Urayama, S.I.; Fukuhara, T.; Komatsu, K.; Arie, T.; Ishihara, A.; Egusa, M.; Kodama, M.; et al. Molecular characterization of a novel mycovirus in Alternaria alternata manifesting two-sided effects: Down-regulation of host growth and up-regulation of host plant pathogenicity. Virology 2018, 519, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Froussard, P. A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res. 1992, 20, 2900. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Raeder, U.; Broda, P. Rapid preparation of DNA from filamentous fungi. Lett. Appl. Microbiol. 1985, 1, 17–20. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes—Application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar] [PubMed]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Jurado, I.; Torrent, J.; Barrón, V.; Corpas, A.; Quesada-Moraga, E. Soil properties affect the availability, movement, and virulence of entomopathogenic fungi conidia against puparia of Ceratitis capitata (Diptera: Tephritidae). Biol. Control 2011, 58, 277–285. [Google Scholar] [CrossRef]
- Garrido-Jurado, I.; Valverde-García, P.; Quesada-Moraga, E. Use of a multiple logistic regression model to determine the effects of soil moisture and temperature on the virulence of entomopathogenic fungi against pre-imaginal Mediterranean fruit fly Ceratitis capitata. Biol. Control 2011, 59, 366–372. [Google Scholar] [CrossRef]
- Quesada-Moraga, E.; Valverde-García, P.; Garrido-Jurado, I. The effect of temperature and soil moisture on the development of the preimaginal Mediterranean fruit fly (Diptera: Tephritidae). Environ. Entomol. 2012, 41, 966–970. [Google Scholar] [CrossRef]
- Yousef, M.; Garrido-Jurado, I.; Quesada-Moraga, E. One Metarhizium brunneum strain, two uses to control Ceratitis capitata (Diptera: Tephritidae). J. Econ. Entomol. 2014, 107, 1736–1744. [Google Scholar] [CrossRef] [PubMed]
- Yousef, M.; Aranda-Valera, E.; Quesada-Moraga, E. Lure-and-infect and lure-and-kill devices based on Metarhizium brunneum for spotted wing Drosophila control. J. Pest Sci. 2018, 91, 227–235. [Google Scholar] [CrossRef]
- Yousef, M.; Alba-Ramírez, C.; Garrido Jurado, I.; Mateu, J.; Raya Díaz, S.; Valverde-García, P.; Quesada-Moraga, E. Metarhizium brunneum (Ascomycota; Hypocreales) Treatments Targeting Olive Fly in the Soil for Sustainable Crop Production. Front. Plant Sci. 2018, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Quesada-Moraga, E.; Ruiz-García, A.; Santiago-Alvarez, C. Laboratory evaluation of entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae against puparia and adults of Ceratitis capitata (Diptera: Tephritidae). J. Econ. Entomol. 2006, 99, 1955–1966. [Google Scholar] [CrossRef] [PubMed]
- Steinwender, B.M.; Enkerli, J.; Widmer, F.; Eilenberg, J.; Thorup-Kristensen, K.; Meyling, N.V. Molecular diversity of the entomopathogenic fungal Metarhizium community within an agroecosystem. J. Invertebr. Pathol. 2014, 123, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Meyling, N.V.; Lübeck, M.; Buckley, E.P.; Eilenberg, J.; Rehner, S.A. Community composition, host-range and genetic structure of the fungal entomopathogen Beauveria in adjoining agricultural and semi-natural habitats. Mol. Ecol. 2009, 18, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Garrido-Jurado, I.; Fernández-Bravo, M.; Campos, C.; Quesada-Moraga, E. Diversity of entomopathogenic Hypocreales in soil and phylloplanes of five Mediterranean cropping systems. J. Invertebr. Pathol. 2015, 130, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Herrero, N.; Dueñas, E.; Quesada-Moraga, E.; Zabalgogeazcoa, I. Prevalence and diversity of viruses in the entomopathogenic fungus Beauveria bassiana. Appl. Environ. Microb. 2012, 78, 8523–8530. [Google Scholar] [CrossRef] [PubMed]
- Leal, S.C.M.; Bertioli, D.J.; Ball, B.V.; Butt, T.M. Presence of double-stranded RNAs and virus-like particles in the entomopathogenic fungus Metarhizium anisopliae. Biocontrol Sci. Technol. 1994, 4, 89–94. [Google Scholar] [CrossRef]
- Bogo, M.R.; Queiroz, M.V.; Silva, D.M.; Giménez, M.P.; Azevedo, J.L.; Schrank, A. Double-stranded RNA and isometric virus-like particles in the entomopathogenic fungus Metarhizium anisopliae. Mycol. Res. 1996, 100, 1468–1472. [Google Scholar] [CrossRef]
- Melzer, M.J.; Bidochka, M.J. Diversity of double-stranded RNA viruses within populations of entomopathogenic fungi and potential implications for fungal growth and virulence. Mycologia 1998, 90, 586–594. [Google Scholar] [CrossRef]
- Martins, M.K.; Furlaneto, M.C.; Sosa-Gomes, D.R.; Faria, M.R.; Fungaro, M.H.P. Double-stranded RNA in the entomopathogenic fungus Metarhizium flavoviride. Curr. Genet. 1999, 36, 94–97. [Google Scholar] [PubMed]
- De la Paz Giménez-Pecci, M.; Bogo, M.; Santi, L.; De Moraes, C.K.; Corrêa, C.T.; Vainstein, M.H.; Schrank, A. Characterization of mycoviruses and analyses of chitinase secretion in the biocontrol fungus Metarhizium anisopliae. Curr. Microbiol. 2002, 45, 334. [Google Scholar] [CrossRef] [PubMed]
- Tiago, P.V.; Fungaro, M.H.; de Faria, M.R.; Furlaneto, M.C. Effects of double-stranded RNA in Metarhizium anisopliae var. acridum and Paecilomyces fumosoroseus on protease activities, conidia production, and virulence. Can. J. Microbiol. 2004, 50, 335–339. [Google Scholar] [PubMed]
- Perinotto, W.M.S.; Golo, P.S.; Coutinho Rodrigues, C.J.B.; Sá, F.A.; Santi, L.; Beys da Silva, W.O.; Junges, A.; Vainstein, M.H.; Schrank, A.; Salles, C.M.C.; et al. Enzymatic activities and effects of mycovirus infection on the virulence of Metarhizium anisopliae in Rhipicephalus microplus. Vet. Parasitol. 2014, 203, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Santos, V.; Mascarin, G.M.; da Silva Lopes, M.; Alves, M.C.D.F.; Rezende, J.M.; Gatti, M.S.V.; Dunlap, C.A.; Júnior, Í.D. Identification of double-stranded RNA viruses in Brazilian strains of Metarhizium anisopliae and their effects on fungal biology and virulence. Plant Gene 2017, 11, 49–58. [Google Scholar] [CrossRef]
- Herrero, N. A novel monopartite dsRNA virus isolated from the entomopathogenic and nematophagous fungus Purpureocillium lilacinum. Arch. Virol. 2016, 161, 3375–3384. [Google Scholar] [CrossRef] [PubMed]
- Inglis, P.W.; Valadares-Inglis, M.C. Rapid isolation of double-stranded RNAs from entomopathogenic species of the fungus Paecilomyces using a commercial minicolumn system. J. Virol. Methods 1997, 67, 113–116. [Google Scholar] [CrossRef]
- Andino, R.; Domingo, E. Viral quasispecies. Virology 2015, 479–480, 46–51. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Perales, C. Quasispecies and virus. Eur. Biophys. J. 2018, 47, 443. [Google Scholar] [CrossRef] [PubMed]
- Briones, C.; Domingo, E. Minority report: Hidden memory genomes in HIV-1 quasispecies and possible clinical implications. AIDS Rev. 2008, 10, 93–109. [Google Scholar] [PubMed]
- Rios, A. Fundamental challenges to the development of a preventive HIV vaccine. Curr. Opin. Virol. 2018, 29, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Lauring, A.S.; Andino, R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010, 6, e1001005. [Google Scholar] [CrossRef] [PubMed]
- Domingo, E.; Sheldon, J.; Perales, C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef] [PubMed]
- Couteaudier, Y.; Viaud, M. New insights into population structure of Beauveria bassiana with regard to vegetative compatibility groups and telomeric restriction fragment length polymorphisms. FEMS Microbiol. Ecol. 1997, 22, 175–182. [Google Scholar] [CrossRef]
- Castrillo, L.A.; Griggs, M.H.; Vandenberg, J.D. Vegetative compatibility groups in indigenous and mass-released strains of the entomopathogenic fungus Beauveria bassiana: Likelihood of recombination in the field. J. Invertebr. Pathol. 2004, 86, 26–37. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Melzer, M.S.; Boland, G.J. Vegetative compatibility and transmission of hypovirulence-associated dsRNA in Sclerotinia homoeocarpa. Can. J. Plant Pathol. 2002, 24, 481–488. [Google Scholar] [CrossRef]
- Xiao, G.; Ying, S.H.; Zheng, P.; Wang, Z.L.; Zhang, S.; Xie, X.Q.; Shang, Y.; St Leger, R.J.; Zhao, G.P.; Wang, C.; et al. Genomic perspectives on the evolution of fungal entomopathogenicity in Beauveria bassiana. Sci. Rep. 2012, 2, 483. [Google Scholar] [CrossRef] [PubMed]
- Coenen, A.; Kevei, F.; Hoekstra, R.F. Factors affecting the spread of double-stranded RNA viruses in Aspergillus nidulans. Genet. Res. 1997, 69, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Anagnostakis, S.L.; Chen, B.; Geletka, L.M.; Nuss, D.L. Hypovirus transmission to ascospore progeny by field-released transgenic hypovirulent strains of Cryphonectria parasitica. Phytopathology 1998, 88, 598–604. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Rinyu, E.; Kevei, E.; Tóth, B.; Kozakiewicz, Z. Double-stranded RNA mycoviruses in species of Aspergillus sections Circumdati and Fumigati. Can. J. Microbiol. 1998, 44, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Allen, T.D.; Hillman, B.I.; Nuss, D.L. Comparative analysis of alterations in host phenotype and transcript accumulation following hypovirus and mycoreovirus infections of the chestnut blight fungus Cryphonectria parasitica. Eukaryot. Cell 2007, 6, 1286–1298. [Google Scholar] [CrossRef] [PubMed]
- Chun, S.J.; Lee, Y.H. Inheritance of dsRNAs in the rice blast fungus, Magnaporthe grisea. FEMS Microbiol. Lett. 2009, 148, 159–162. [Google Scholar] [CrossRef]
- Kotta-Loizou, I.; Sipkova, J.; Coutts, R.H.A. Identification and sequence determination of a novel double-stranded RNA mycovirus from the entomopathogenic fungus Beauveria bassiana. Arch. Virol. 2015, 160, 873–875. [Google Scholar] [CrossRef] [PubMed]
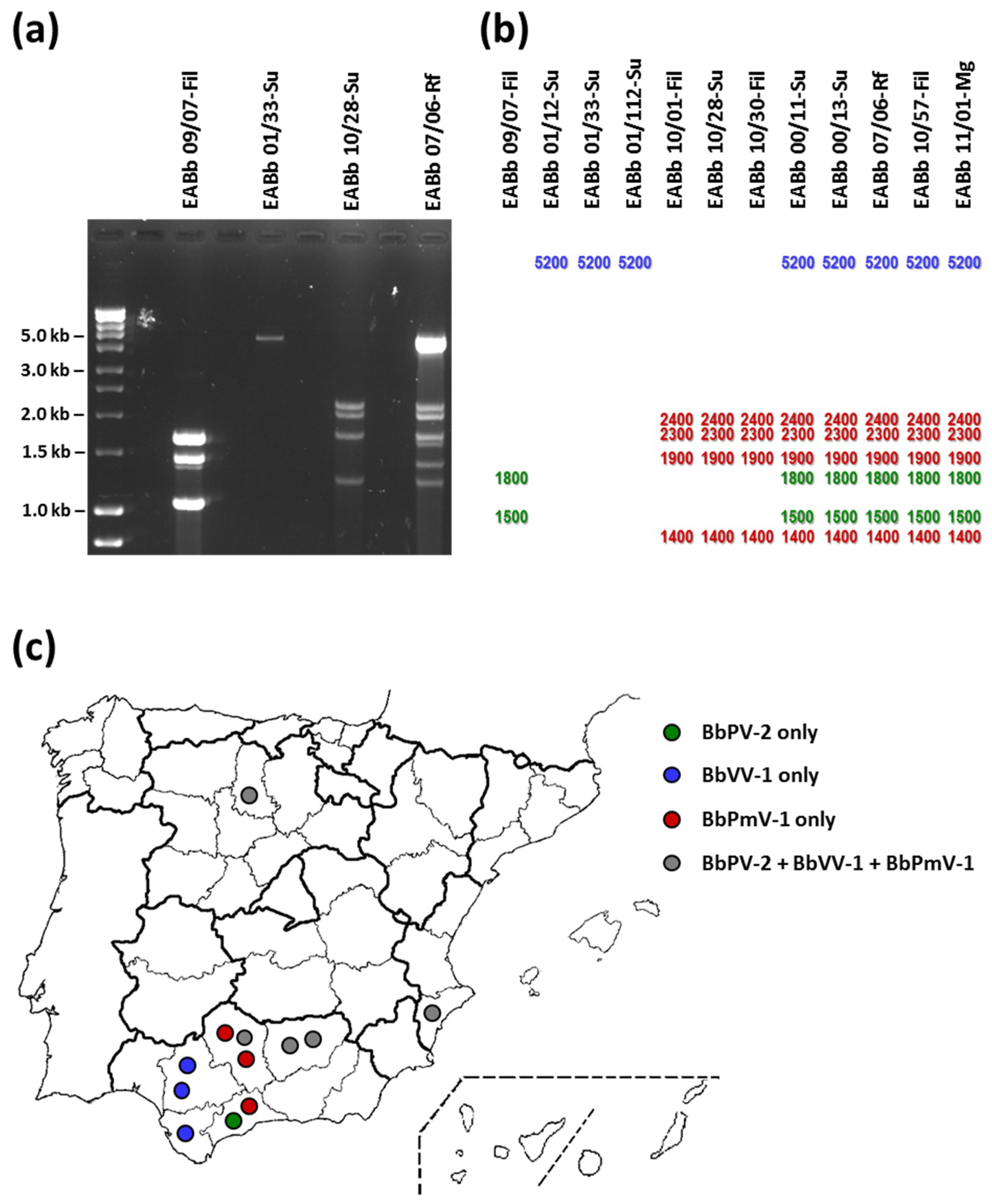
| Species | Isolate | Habitat | Location | Mycovirus * |
|---|---|---|---|---|
| B. bassiana | EABb 00/11-Su | Soil (scrubland) | Jaen | BbVV-1 + BbPV-2 + BbPmV-1 |
| B. bassiana | EABb 00/13-Su | Soil (woodland) | Jaen | BbVV-1 + BbPV-2 + BbPmV-1 |
| B. bassiana | EABb 01/12-Su | Soil (scrubland) | Seville | BbVV-1 |
| B. bassiana | EABb 01/33-Su | Soil (olive grove) | Cadiz | BbVV-1 |
| B. bassiana | EABb 01/112-Su | Soil (wheat field) | Seville | BbVV-1 |
| B. bassiana | EABb 07/06-Rf | Rhynchophorus ferrugineus | Alicante | BbVV-1 + BbPV-2 + BbPmV-1 |
| B. bassiana | EABb 09/07-Fil | Phylloplane (meadow) | Malaga | BbPV-2 |
| B. bassiana | EABb 10/01-Fil | Phylloplane (olive grove) | Malaga | BbPmV-1 |
| B. bassiana | EABb 10/28-Su | Soil (olive grove) | Cordoba | BbPmV-1 |
| B. bassiana | EABb 10/30-Fil | Phylloplane (olive grove) | Cordoba | BbPmV-1 |
| B. bassiana | EABb 10/57-Fil | Phylloplane (meadow) | Cordoba | BbVV-1 + BbPV-2 + BbPmV-1 |
| B. bassiana | EABb 11/01-Mg | Monochamus galloprovincialis | Palencia | BbVV-1 + BbPV-2 + BbPmV-1 |
| Treatment * | Mortality (Mean ± SE)% ** | Kaplan-Meier Survival Analysis | |
|---|---|---|---|
| Total Mortality | Fungal Outgrowth | AST *** (d, Mean ± SE) | |
| Control | 13.3 ± 3.3 a | 0.0 ± 0.0 a | 7.5 ± 0.3 a |
EABb 10/30-Fil  | 80.0 ± 0.0 b | 53.3 ± 3.3 b | 5.5 ± 0.4 b |
EABb 01/33-Su  | 86.7 ± 3.3 b | 76.7 ± 8.8 c | 5.4 ± 0.3 b |
EABb 00/11-Su  | 90.0 ± 0.0 b | 70.0 ± 10.0 b | 5.2 ± 0.4 bc |
EABb 00/13-Su  | 90.0 ± 5.8 b | 66.6 ± 12.0 b | 5.1 ± 0.3 bc |
EABb 01/12-Su  | 93.3 ± 6.7 bc | 76.7 ± 8.8 c | 4.5 ± 0.4 c |
EABb 10/01-Fil  | 93.3 ± 3.3 bc | 80.0 ± 10.0 c | 5.3 ± 0.3 bc |
EABb 10/28-Su  | 96.7 ± 3.3 c | 63.3 ± 18.6 b | 4.7 ± 0.4 bc |
EABb 10/57-Fil  | 96.7 ± 3.3 c | 70.0 ± 15.2 b | 5.0 ± 0.4 bc |
EABb 01/112-Su  | 100.0 ± 0.0 c | 50.0 ± 10.0 b | 4.2 ± 0.4 c |
EABb 07/06-Rf  | 100.0±0.0 c | 90.0 ± 0.0 c | 4.8 ± 0.3 c |
EABb 09/07-Fil  | 100.0±0.0 c | 83.3 ± 6.7 c | 5.2 ± 0.3 bc |
EABb 11/01-Mg  | 100.0±0.0 c | 70.0 ± 5.8 b | 5.1 ± 0.1 bc |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippou, C.; Garrido-Jurado, I.; Meyling, N.V.; Quesada-Moraga, E.; Coutts, R.H.A.; Kotta-Loizou, I. Mycoviral Population Dynamics in Spanish Isolates of the Entomopathogenic Fungus Beauveria bassiana. Viruses 2018, 10, 665. https://doi.org/10.3390/v10120665
Filippou C, Garrido-Jurado I, Meyling NV, Quesada-Moraga E, Coutts RHA, Kotta-Loizou I. Mycoviral Population Dynamics in Spanish Isolates of the Entomopathogenic Fungus Beauveria bassiana. Viruses. 2018; 10(12):665. https://doi.org/10.3390/v10120665
Chicago/Turabian StyleFilippou, Charalampos, Inmaculada Garrido-Jurado, Nicolai V. Meyling, Enrique Quesada-Moraga, Robert H. A. Coutts, and Ioly Kotta-Loizou. 2018. "Mycoviral Population Dynamics in Spanish Isolates of the Entomopathogenic Fungus Beauveria bassiana" Viruses 10, no. 12: 665. https://doi.org/10.3390/v10120665
APA StyleFilippou, C., Garrido-Jurado, I., Meyling, N. V., Quesada-Moraga, E., Coutts, R. H. A., & Kotta-Loizou, I. (2018). Mycoviral Population Dynamics in Spanish Isolates of the Entomopathogenic Fungus Beauveria bassiana. Viruses, 10(12), 665. https://doi.org/10.3390/v10120665







