Evidence for the Widespread Occurrence of Bacteria Implicated in Acute Oak Decline from Incidental Genetic Sampling
Abstract
:1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Quine, C.P.; Atkinson, N.; Denman, S.; Desprez-Loustau, M.-L.; Jackson, R.; Kirby, K. Action Oak Knowledge Review: An Assessment of the Current Evidence on Oak Health in the UK, Identification of Evidence Gaps and Prioritisation of Research Needs; Action Oak: Haslemere, UK, 2019. [Google Scholar]
- Leroy, T.; Plomion, C.; Kremer, A. Oak symbolism in the light of genomics. New Phytol. 2019, 226, 1012–1017. [Google Scholar] [CrossRef]
- Rackham, O. Trees and Woodland in the British Landscape: The Complete History of Britain’s Trees, Woods & Hedgerows; Phoenix Press: London, UK, 2001. [Google Scholar]
- Mitchell, R.; Bellamy, P.; Ellis, C.; Hewison, R.; Hodgetts, N.; Iason, G.; Littlewood, N.; Newey, S.; Stockan, J.; Taylor, A. Collapsing foundations: The ecology of the British oak, implications of its decline and mitigation options. Biol. Conserv. 2019, 233, 316–327. [Google Scholar] [CrossRef]
- Tomlinson, I.; Potter, C.; Bayliss, H. Managing tree pests and diseases in urban settings: The case of Oak Processionary Moth in London, 2006–2012. Urban For. Urban Green. 2015, 14, 286–292. [Google Scholar] [CrossRef] [Green Version]
- Lonsdale, D. Review of oak mildew, with particular reference to mature and veteran trees in Britain. Arboric. J. 2015, 37, 61–84. [Google Scholar] [CrossRef]
- Denman, S.; Brown, N.; Kirk, S.; Jeger, M.; Webber, J. A description of the symptoms of Acute Oak Decline in Britain and a comparative review on causes of similar disorders on oak in Europe. Forestry 2014, 87, 535–551. [Google Scholar] [CrossRef] [Green Version]
- Manion, P.D.; Lachance, D. Forest Decline Concepts; The American Phytopathological Society: St. Paul, MN, USA, 1992. [Google Scholar]
- D Manion, P. Tree Disease Concepts/Paul D. Manion; Prentice-Hall: Englewood Cliffs, NJ, USA, 1981. [Google Scholar]
- Denman, S.; Webber, J. Oak declines—New definitions and new episodes in Britain. Q. J. For. 2009, 103, 285–290. [Google Scholar]
- Brown, N.; Bosch, F.V.D.; Parnell, S.; Denman, S. Integrating regulatory surveys and citizen science to map outbreaks of forest diseases: Acute oak decline in England and Wales. Proc. Biol. Sci. 2017, 284, 20170547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, N.; Vanguelova, E.; Parnell, S.; Broadmeadow, S.; Denman, S. Predisposition of forests to biotic disturbance: Predicting the distribution of Acute Oak Decline using environmental factors. For. Ecol. Manag. 2018, 407, 145–154. [Google Scholar] [CrossRef]
- Pinho, D.; Barroso, C.; Froufe, H.; Brown, N.; Vanguelova, E.; Egas, C.; Denman, S. Linking Tree Health, Rhizosphere Physicochemical Properties, and Microbiome in Acute Oak Decline. Forests 2020, 11, 1153. [Google Scholar] [CrossRef]
- Scarlett, K.; Denman, S.; Clark, D.R.; Forster, J.; Vanguelova, E.; Brown, N.; Whitby, C. Relationships between nitrogen cycling microbial community abundance and composition reveal the indirect effect of soil pH on oak decline. ISME J. 2020, 15, 623–635. [Google Scholar] [CrossRef]
- Denman, S.; Doonan, J.; Ransom-Jones, E.; Broberg, M.; Plummer, S.; Kirk, S.; Scarlett, K.; Griffiths, A.R.; Kaczmarek, M.; Forster, J.; et al. Microbiome and infectivity studies reveal complex polyspecies tree disease in Acute Oak Decline. ISME J. 2018, 12, 386–399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doonan, J.; Denman, S.; Pachebat, J.; McDonald, J.E. Genomic analysis of bacteria in the Acute Oak Decline pathobiome. Microb. Genom. 2019, 5, e000240. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Arnold, D.; McDonald, J.; Denman, S. Taxonomy and identification of bacteria associated with acute oak decline. World J. Microbiol. Biotechnol. 2017, 33, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Cleenwerck, I.; Denman, S.; Venter, S.; Rodriguez-Palenzuela, P.; Coutinho, T.; De Vos, P. Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 2012, 62, 1592–1602. [Google Scholar] [PubMed]
- Brady, C.; Denman, S.; Kirk, S.; Venter, S.; Rodríguez-Palenzuela, P.; Coutinho, T. Description of Gibbsiella quercinecans gen. nov., sp. nov., associated with Acute Oak Decline. Syst. Appl. Microbiol. 2010, 33, 444–450. [Google Scholar] [CrossRef]
- Brady, C.; Hunter, G.; Kirk, S.; Arnold, D.; Denman, S. Rahnella victoriana sp. nov., Rahnella bruchi sp. nov., Rahnella woolbedingensis sp. nov., classification of Rahnella genomospecies 2 and 3 as Rahnella variigena sp. nov. and Rahnella inusitata sp. nov., respectively and emended description of the genus Rahnella. Syst. Appl. Microbiol. 2014, 37, 545–552. [Google Scholar]
- Li, Y.; Xue, H.; Guo, L.-M.; Koltay, A.; Palacio-Bielsa, A.; Chang, J.; Xie, S.; Yang, X. Elevation of three subspecies of Lonsdalea quercina to species level: Lonsdalea britannica sp. nov., Lonsdalea iberica sp. nov. and Lonsdalea populi sp. nov. Int. J. Syst. Evol. Microbiol. 2017, 67, 4680–4684. [Google Scholar] [CrossRef] [PubMed]
- Soutar, C.D.; Stavrinides, J. Phylogenetic analysis supporting the taxonomic revision of eight genera within the bacterial order Enterobacterales. Int. J. Syst. Evol. Microbiol. 2020, 70, 6524–6530. [Google Scholar] [CrossRef] [PubMed]
- Denman, S.; Brady, C.; Kirk, S.; Cleenwerck, I.; Venter, S.; Coutinho, T.; De Vos, P. Brenneria goodwinii sp. nov., associated with acute oak decline in the UK. Int. J. Syst. Evol. Microbiol. 2012, 62, 2451–2456. [Google Scholar] [CrossRef]
- Vansteenkiste, D.; Tirry, L.; Van Acker, J.; Stevens, M. Predispositions and symptoms of Agrilus borer attack in declining oak trees. Ann. For. Sci. 2004, 61, 815–823. [Google Scholar] [CrossRef] [Green Version]
- Brown, N.; Inward, D.J.; Jeger, M.; Denman, S. A review of Agrilus biguttatus in UK forests and its relationship with acute oak decline. Forestry 2014, 88, 53–63. [Google Scholar] [CrossRef]
- Reed, K.; Denman, S.; Leather, S.R.; Forster, J.; Inward, D.J.G. The lifecycle of Agrilus biguttatus: The role of temperature in its development and distribution, and implications for Acute Oak Decline. Agric. For. Èntomol. 2017, 20, 334–346. [Google Scholar] [CrossRef] [Green Version]
- Doonan, J.M.; Broberg, M.; Denman, S.; McDonald, J.E. Host–microbiota–insect interactions drive emergent virulence in a complex tree disease. Proc. R. Soc. B: Boil. Sci. 2020, 287, 20200956. [Google Scholar] [CrossRef]
- Denman, S.; Plummer, S.; Kirk, S.; Peace, A.; McDonald, J.E. Isolation studies reveal a shift in the cultivable microbiome of oak affected with Acute Oak Decline. Syst. Appl. Microbiol. 2016, 39, 484–490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, A.J.; Ciordia, M. Brenneria goodwinii and Gibbsiella quercinecans isolated from weeping cankers on Quercus robur L. in Spain. Eur. J. Plant Pathol. 2019, 156, 965–969. [Google Scholar] [CrossRef]
- Moradi-Amirabad, Y.; Rahimian, H.; Babaeizad, V.; Denman, S. Brenneria spp. and Rahnella victoriana associated with acute oak decline symptoms on oak and hornbeam in Iran. For. Pathol. 2019, 49, e12535. [Google Scholar] [CrossRef]
- Meaden, S.; Metcalf, C.J.E.; Koskella, B. The effects of host age and spatial location on bacterial community composition in the English Oak tree (Quercus robur). Environ. Microbiol. Rep. 2016, 8, 649–658. [Google Scholar] [CrossRef] [Green Version]
- Stone, B.W.; Weingarten, E.A.; Jackson, C.R. The Role of the Phyllosphere Microbiome in Plant Health and Function. Annu. Plant Rev. Online 2018, 1, 533–556. [Google Scholar]
- Leveau, J. Microbial Communities in the Phyllosphere. In Biology of the Plant Cuticle; Riederer, M.M.C., Ed.; Blackwell: Oxford, UK, 2006; pp. 334–367. [Google Scholar]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828. [Google Scholar] [CrossRef] [PubMed]
- Pettifor, B.J.; Doonan, J.; Denman, S.; McDonald, J.E. Survival of Brenneria goodwinii and Gibbsiella quercinecans, associated with acute oak decline, in rainwater and forest soil. Syst. Appl. Microbiol. 2020, 43, 126052. [Google Scholar] [CrossRef] [PubMed]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef] [Green Version]
- Müller, T.; Ruppel, S. Progress in cultivation-independent phyllosphere microbiology. FEMS Microbiol. Ecol. 2014, 87, 2–17. [Google Scholar] [CrossRef]
- Zolfo, M.; Asnicar, F.; Manghi, P.; Pasolli, E.; Tett, A.; Segata, N. Profiling microbial strains in urban environments using metagenomic sequencing data. Biol. Direct 2018, 13, 9. [Google Scholar] [CrossRef] [Green Version]
- Roumpeka, D.D.; Wallace, R.J.; Escalettes, F.; Fotheringham, I.; Watson, M. A Review of Bioinformatics Tools for Bio-Prospecting from Metagenomic Sequence Data. Front. Genet. 2017, 8, 23. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Nocchi, G.; Brown, N.; Coker, T.; Plumb, W.; Stocks, J.; Denman, S.; Buggs, R.J.A. Genomic structure and diversity of oak populations in British parklands. Plants People Planet 2021, 1–15. [Google Scholar] [CrossRef]
- Brown, N.; Jeger, M.; Kirk, S.; Xu, X.; Denman, S. Spatial and temporal patterns in symptom expression within eight woodlands affected by Acute Oak Decline. For. Ecol. Manag. 2016, 360, 97–109. [Google Scholar] [CrossRef]
- Raimbault, P. Physiological Diagnosis; Societe Francaise d’Arboriculture: Versailles, France, 1995. [Google Scholar]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Genet. 2018, 16, 410–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herrmann, M.; Geesink, P.; Richter, R.; Küsel, K. Canopy Position Has a Stronger Effect than Tree Species Identity on Phyllosphere Bacterial Diversity in a Floodplain Hardwood Forest. Microb. Ecol. 2020, 81, 157–168. [Google Scholar] [CrossRef] [PubMed]
- King, T.; Butcher, S.; Zalewski, L. Apocrita—High Performance Computing Cluster for Queen Mary University of London; Zenodo: London, UK, 2017. [Google Scholar]
- Plomion, C.; Aury, J.-M.; Amselem, J.; Leroy, T.; Murat, F.; Duplessis, S.; Faye, S.; Francillonne, N.; Labadie, K.; Le Provost, G.; et al. Oak genome reveals facets of long lifespan. Nat. Plants 2018, 4, 440–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- NCBI. The NCBI Handbook; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013. [Google Scholar]
- McDonald, D.; Clemente, J.C.; Kuczynski, J.; Rideout, J.R.; Stombaugh, J.; Wendel, D.; Wilke, A.; Huse, S.; Hufnagle, J.; Meyer, F.; et al. The Biological Observation Matrix (BIOM) format or: How I learned to stop worrying and love the ome-ome. GigaScience 2012, 1, 7. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- RStudio Team. RStudio: Integrated Development for R; RStudio, Inc.: Boston, MA, USA, 2019. [Google Scholar]
- Oksanen, J.; Guillaume Blanchet, F.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package; R package version 2.5-7. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 30 November 2021).
- Hill, R.; Llewellyn, T.; Downes, E.; Oddy, J.; MacIntosh, C.; Kallow, S.; Panis, B.; Dickie, J.B.; Gaya, E. Seed Banks as Incidental Fungi Banks: Fungal Endophyte Diversity in Stored Seeds of Banana Wild Relatives. Front. Microbiol. 2021, 12, 508. [Google Scholar] [CrossRef]
- Lu, J.; Breitwieser, F.P.; Thielen, P.; Salzberg, S.L. Bracken: Estimating species abundance in metagenomics data. PeerJ Comput. Sci. 2017, 3, e104. [Google Scholar] [CrossRef]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; R Core Team. _ nlme: Linear and Nonlinear Mixed Effects Models; R package version 3.1-148. 2020. Available online: https://CRAN.R-project.org/package=nlme (accessed on 30 November 2021).
- Boratyn, G.M.; Thierry-Mieg, J.; Thierry-Mieg, D.; Busby, B.; Madden, T.L. Magic-BLAST, an accurate RNA-seq aligner for long and short reads. BMC Bioinform. 2019, 20, 405. [Google Scholar] [CrossRef] [PubMed]
- Uroz, S.; Buée, M.; Murat, C.; Frey-Klett, P.; Martin, F. Pyrosequencing reveals a contrasted bacterial diversity between oak rhizosphere and surrounding soil. Environ. Microbiol. Rep. 2010, 2, 281–288. [Google Scholar] [CrossRef] [PubMed]
- Leff, J.W.; Del Tredici, P.; Friedman, W.E.; Fierer, N. Spatial structuring of bacterial communities within individual Ginkgo biloba trees. Environ. Microbiol. 2015, 17, 2352–2361. [Google Scholar] [CrossRef] [PubMed]
- Jakuschkin, B. Genetic architecture of the interactions between English oak (Quercus robur L.) and the microbial community of its phyllosphere. Ph.D. Thesis, Université de Bordeaux, Bordeaux, France, 2015. [Google Scholar]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bálint, M.; Tiffin, P.; Hallström, B.; O’Hara, R.B.; Olson, M.S.; Fankhauser, J.D.; Piepenbring, M.; Schmitt, I. Host Genotype Shapes the Foliar Fungal Microbiome of Balsam Poplar (Populus balsamifera). PLoS ONE 2013, 8, e53987. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cregger, M.A.; Veach, A.; Yang, Z.K.; Crouch, M.J.; Vilgalys, R.; Tuskan, G.A.; Schadt, C.W. The Populus holobiont: Dissecting the effects of plant niches and genotype on the microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014, 15, R46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kibegwa, F.M.; Bett, R.C.; Gachuiri, C.K.; Stomeo, F.; Mujibi, F.D. A Comparison of Two DNA Metagenomic Bioinformatic Pipelines While Evaluating the Microbial Diversity in Feces of Tanzanian Small Holder Dairy Cattle. BioMed Res. Int. 2020, 2020, 2348560. [Google Scholar] [CrossRef] [PubMed]
- Menzel, P.; Ng, K.; Krogh, A. Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat. Commun. 2016, 7, 11257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crampton, B.G.; Plummer, S.J.; Kaczmarek, M.; McDonald, J.E.; Denman, S. A multiplex real-time PCR assay enables simultaneous rapid detection and quantification of bacteria associated with acute oak decline. Plant Pathol. 2020, 69, 1301–1310. [Google Scholar] [CrossRef]
- Arnold, J.B. ggthemes: Extra Themes, Scales and Geoms for ‘ggplot2’; R package version 4.2.0. 2019. Available online: https://CRAN.R-project.org/package=ggthemes (accessed on 30 November 2021).
- Auguie, B. GridExtra: Miscellaneous Functions for “Grid” Graphics; R package version 2.3. 2017. Available online: https://CRAN.R-project.org/package=gridExtra (accessed on 30 November 2021).
- Wilke, C.O. Cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’; R package version 1.1.1. 2020. Available online: https://CRAN.R-project.org/package=cowplot (accessed on 30 November 2021).
- Henry, L.; Wickham, H. Purrr: Functional Programming Tools; R package version 0.3.4. 2020. Available online: https://CRAN.R-project.org/package=purrr (accessed on 30 November 2021).
- Müller, K.; Wickham, H. Tibble: Simple Data Frames; R package version 3.1.0; 2021. Available online: https://CRAN.R-project.org/package=tibble (accessed on 30 November 2021).
- Wickham, H. The split-apply-combine strategy for data analysis. J. Stat. Softw. 2011, 40, 1–29. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Forcats: Tools for Working with Categorical Variables (Factors); R package verion 0.5.1; 2021. Available online: https://CRAN.R-project.org/package=forcats (accessed on 30 November 2021).
- Wickham, H. Stringr: Simple, Consistent Wrappers for Common String Operations; R package version 1.4.0; 2019. Available online: https://CRAN.R-project.org/package=stringr. (accessed on 30 November 2021).
- Wickham, H. Tidyr: Tidy Messy Data; R package version 1.1.3. 2021. Available online: https://CRAN.R-project.org/package=tidyr. (accessed on 30 November 2021).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. 2020. Available online: https://ggplot2.tidyverse.org/ (accessed on 30 November 2021).
- Wickham, H.; François, R.; Henry, L.; Müller, K. dplyr: A Grammar of Data Manipulation; R package version 1.0.5; 2021. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 30 November 2021).
- Wickham, H.; Hester, J. Readr: Read Rectangular Text Data; R package version 1.4.0; 2020. Available online: https://CRAN.R-project.org/package=readr (accessed on 30 November 2021).
- Wickham, H.; Seidel, D. Scales: Scale Functions for Visualization; R package version 1.1.0; 2019. Available online: https://CRAN.R-project.org/package=scales. (accessed on 30 November 2021).
- Hope, R.M. Rmisc: Ryan Miscellaneous; R package version 1.5; 2013. Available online: https://CRAN.R-project.org/package=RMisc (accessed on 30 November 2021).
- Sarkar, D. Lattice: Multivariate Data Visualization with R; Springer: New York, NY, USA, 2008. [Google Scholar]
- Kassambara, A. ggpubr: ’ggplot2’ Based Publication Ready Plots; R package version 0.4.0. 2020. Available online: https://CRAN.R-project.org/package=ggpubr. (accessed on 30 November 2021).
- Simpson, G.L. Permut: Functions for Generating Restricted Permutations of Data: R package version 0.9-5. Available online: https://CRAN.R-project.org/package=permute. (accessed on 30 November 2021).
- Wolfe, B.E. Signs: Insert proper Minus Signs; R package version 0.1.2. Available online: https://CRAN.R-project.org/package=signs. (accessed on 30 November 2021).


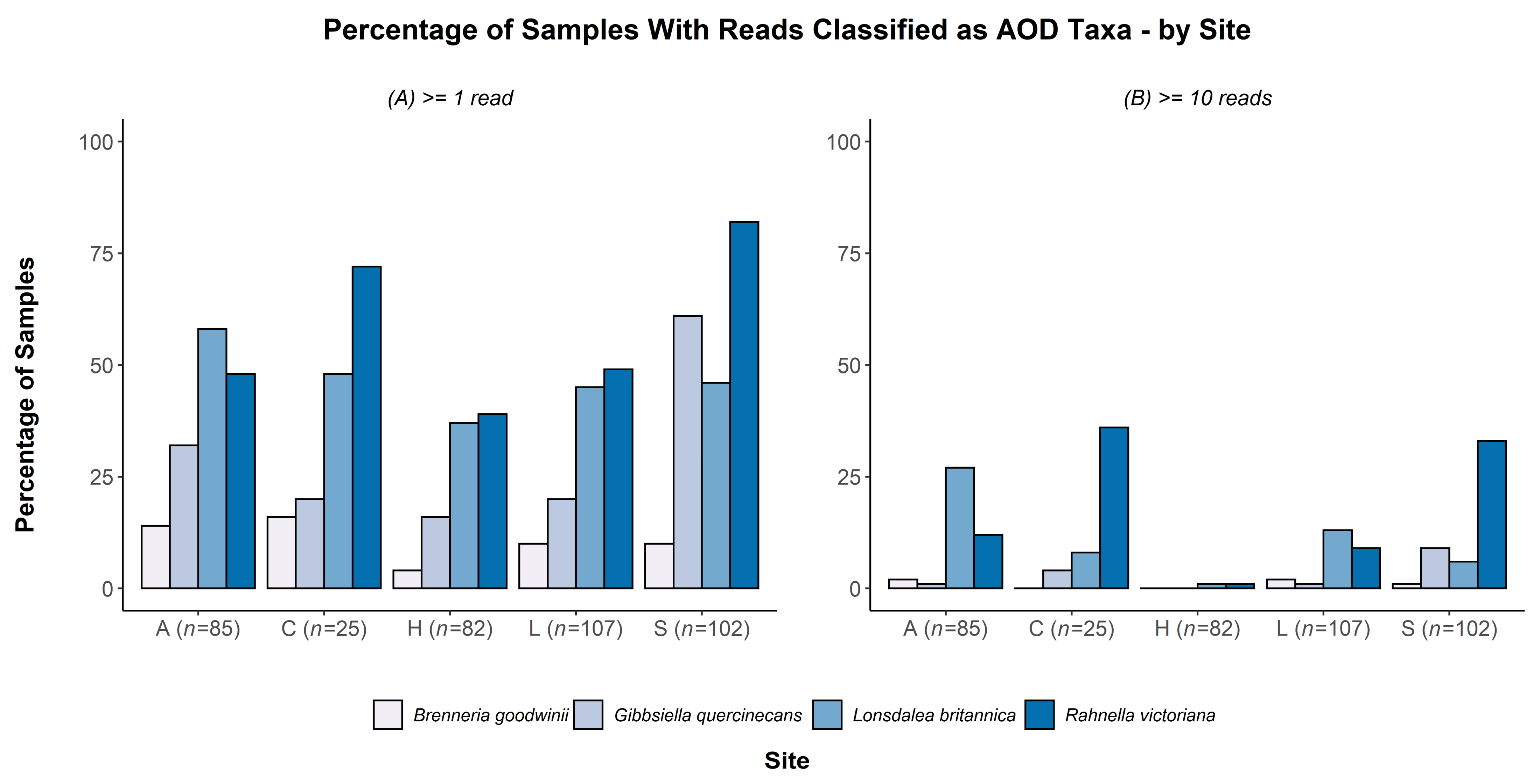
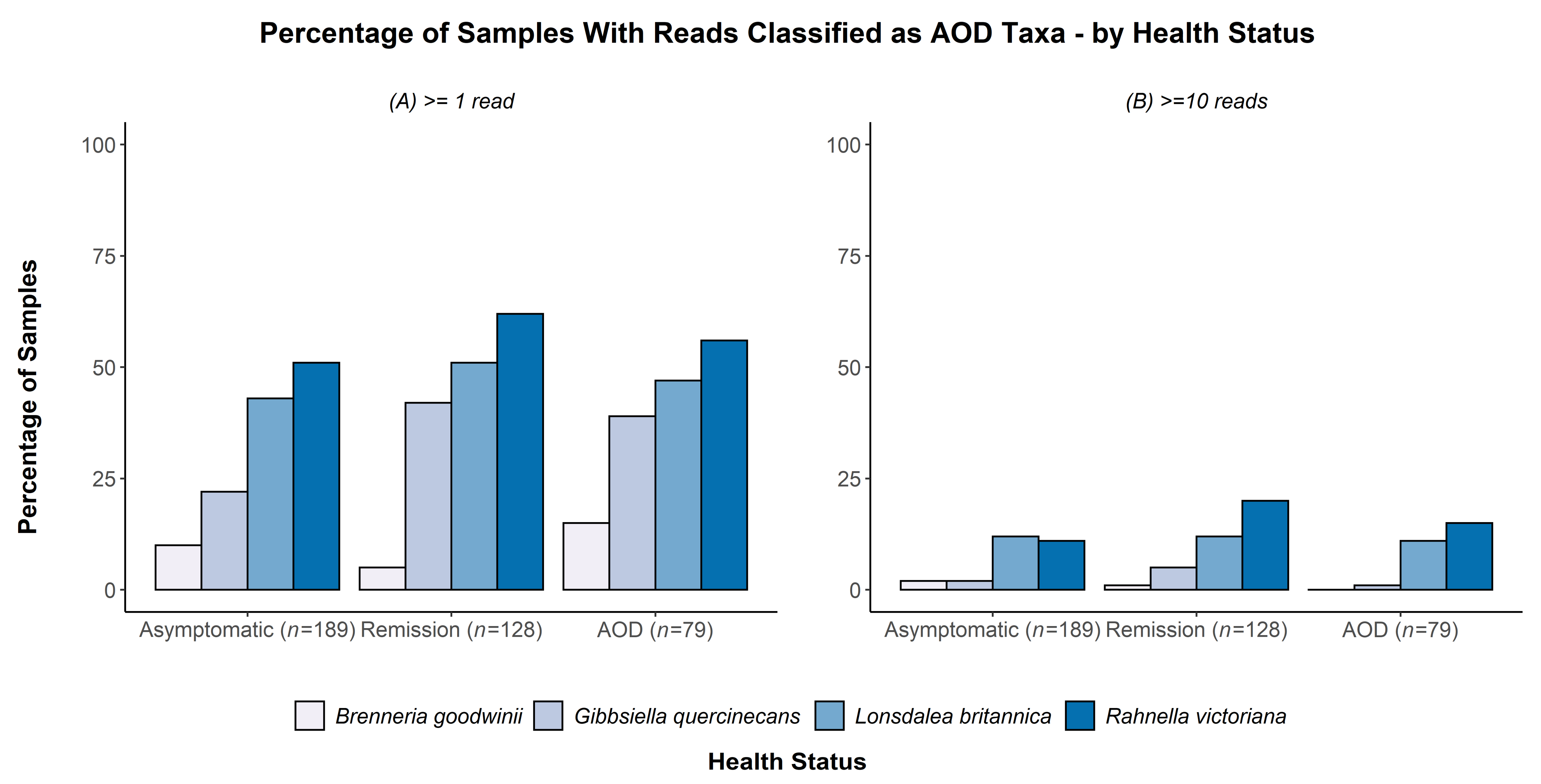
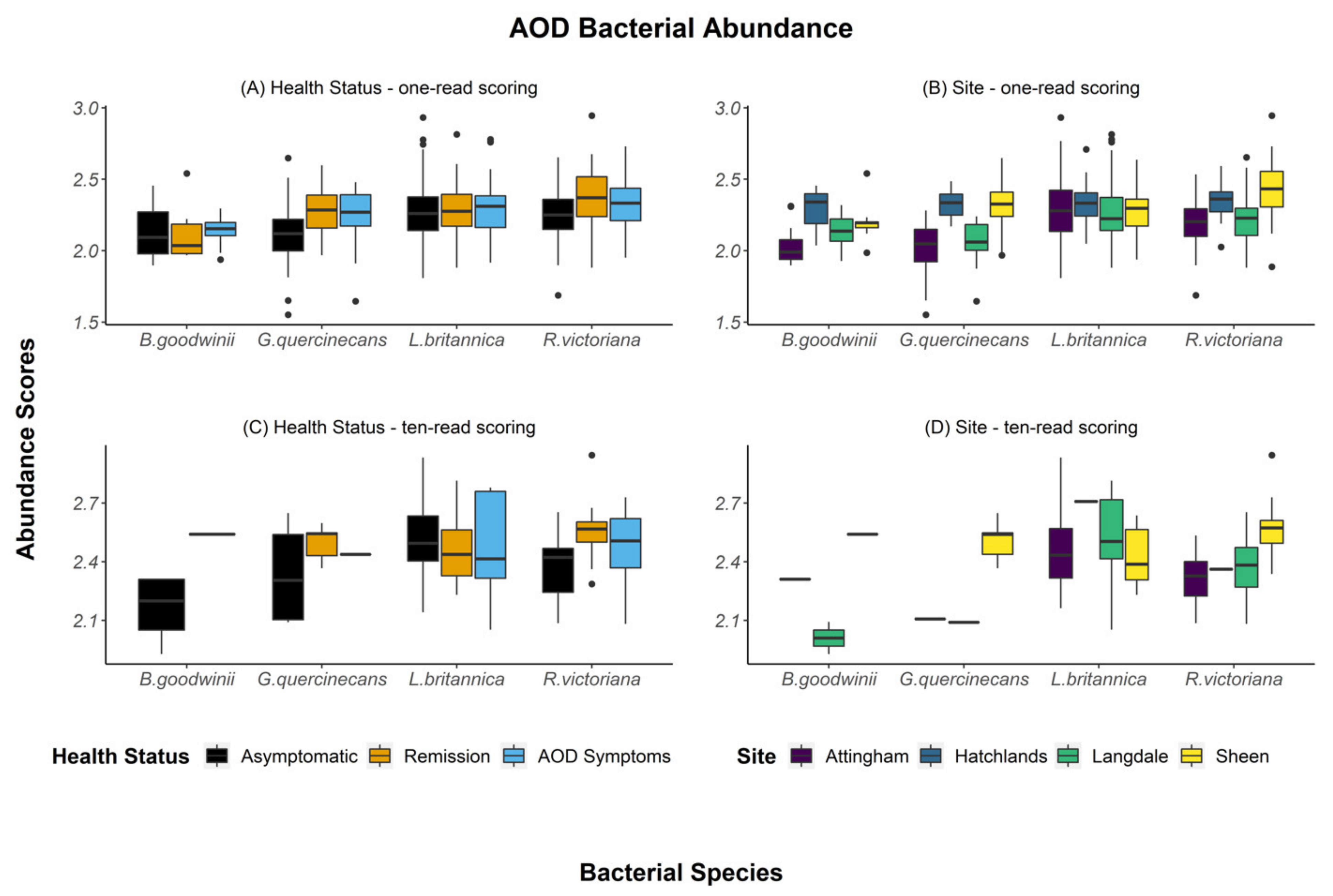
| Health Groups Sampled at Each Site (Asymptomatic 1, Remission 2, Acute Oak Decline 3 or Chronic Oak Decline 4 Symptoms)-Number of Trees in Each Health Category | |||||||
|---|---|---|---|---|---|---|---|
| Features | AOD First Recorded [42] | A 1 | R 2 | AOD 3 | COD 4 | Total | |
| Attingham Park | Small area of closely planted oak dominated amenity woodland within parkland. Area of new planting with trees approximately 30 years old | 2007 | 58 | 12 | 15 | 0 | 85 |
| Hatchlands Park | Landscaped parkland with shelterbelts | After 2001 | 37 | 26 | 19 | 0 | 82 |
| Langdale Wood | Managed open forest, not closely planted, mowed grass below | Before 2006 | 77 | 30 | 20 | 0 | 127 |
| Sheen Wood | Oak dominated woodland in a Royal Park, trees not closely planted. Location as a boundary belt on the edge of the park in an urban area | 1991 | 17 | 60 | 25 | 0 | 102 |
| Chestnuts Wood | Area of plantation 50–60 years old in the Forest of Dean. Close to natural woodland. COD present. No AOD in the woodland sampled, but AOD is present in the wider area | N/A | 14 | 0 | 0 | 11 | 25 |
| Total | 203 | 128 | 79 | 11 | 421 | ||
| Permanova | Permdisp | |||||
|---|---|---|---|---|---|---|
| Adonis 2 | Adonis | |||||
| Variable | DF | Marginal R2 | p-Value | R2 | p-Value | p |
| Tissue | 2 | 0.018 | 0.001 | 0.084 | 0.001 | 0.172 |
| Site | 4 | 0.1 | <0.001 | 0.118 | 0.001 | <0.001 |
| Health Status | 3 | 0.01 | 0.011 | 0.01 | 0.009 | 0.31 |
| Tree Species | 4 | 0.007 | 0.52 | 0.007 | 0.506 | 0.052 |
| Drying Method | 2 | 0.002 | 0.84 | 0.002 | 0.84 | 0.37 |
| Presence = One-Read Scoring | Presence = 10-Read Scoring | |||||||
|---|---|---|---|---|---|---|---|---|
| Pearson’s chi-squared test with Hatchlands (df = 3) | Pearson’s chi-squared test with Hatchlands removed (df = 2) | Pearson’s chi-squared test with Hatchlands (df = 3) | Pearson’s chi-squared test with Hatchlands removed (df =2 ) | |||||
| X2 | p-Value | X2 | p-Value | X2 | p-Value | X2 | p-Value | |
| G. quercinecans | 57.448 | <0.001 | 42.359 | <0.001 | 18.818 | <0.001 | 12.673 | 0.002 |
| B. goodwinii | 5.400 | 0.145 | 1.049 | 0.591 | 2.024 | 0.568 | 0.558 | 0.757 |
| L. britannica | 7.619 | 0.054 | 3.745 | 0.154 | 31.762 | <0.001 | 17.456 | <0.001 |
| R. victoriana | 42.861 | <0.001 | 32.608 | <0.001 | 44.24 | <0.001 | 24.952 | <0.001 |
| Fixed Effect | Numerator DF | Denominator DF | F-Value | p-Value | |
|---|---|---|---|---|---|
| (A) One-read scoring(n = 305) | Tissue | 2 | 297 | 28.46 | <0.001 |
| Age | 2 | 297 | 16.95 | <0.001 | |
| Bacterial Species | 3 | 250 | 25.58 | <0.001 | |
| Site | 3 | 297 | 2.98 | 0.0316 | |
| Bacterial Species: Site | 9 | 250 | 7.34 | <0.001 | |
| (B) Ten-read scoring(n = 97) | Tissue | 2 | 89 | 10.274 | <0.001 |
| Age | 2 | 89 | 0.107 | 0.899 | |
| Bacterial Species | 3 | 19 | 3.870 | 0.0257 | |
| Site | 3 | 89 | 0.835 | 0.478 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gathercole, L.A.P.; Nocchi, G.; Brown, N.; Coker, T.L.R.; Plumb, W.J.; Stocks, J.J.; Nichols, R.A.; Denman, S.; Buggs, R.J.A. Evidence for the Widespread Occurrence of Bacteria Implicated in Acute Oak Decline from Incidental Genetic Sampling. Forests 2021, 12, 1683. https://doi.org/10.3390/f12121683
Gathercole LAP, Nocchi G, Brown N, Coker TLR, Plumb WJ, Stocks JJ, Nichols RA, Denman S, Buggs RJA. Evidence for the Widespread Occurrence of Bacteria Implicated in Acute Oak Decline from Incidental Genetic Sampling. Forests. 2021; 12(12):1683. https://doi.org/10.3390/f12121683
Chicago/Turabian StyleGathercole, Louise A. P., Gabriele Nocchi, Nathan Brown, Timothy L. R. Coker, William J. Plumb, Jonathan J. Stocks, Richard A. Nichols, Sandra Denman, and Richard J. A. Buggs. 2021. "Evidence for the Widespread Occurrence of Bacteria Implicated in Acute Oak Decline from Incidental Genetic Sampling" Forests 12, no. 12: 1683. https://doi.org/10.3390/f12121683
APA StyleGathercole, L. A. P., Nocchi, G., Brown, N., Coker, T. L. R., Plumb, W. J., Stocks, J. J., Nichols, R. A., Denman, S., & Buggs, R. J. A. (2021). Evidence for the Widespread Occurrence of Bacteria Implicated in Acute Oak Decline from Incidental Genetic Sampling. Forests, 12(12), 1683. https://doi.org/10.3390/f12121683






