Benchmarking Multiple Large Language Models for Automated Clinical Trial Data Extraction in Aging Research
Abstract
1. Introduction
2. Literature Review
3. Materials and Methods
3.1. Natural Language Processing and Analysis
- openai/o1-mini;
- x-ai/grok-2-1212;
- meta-llama/llama-3.3-70b-instruct;
- google/gemini-flash-1.5-8b;
- deepseek/deepseek-r1-distill-llama-70b.
Prompt Architecture
3.2. Statistical Analysis of Multi-LLM Reliability
- Majority voting for categorical variables (e.g., “Yes” vs. “No” for brain_stimulation_used).
- The mean or median for numeric fields (e.g., stimulation intensity or session duration). These consensus fields serve as the “best estimates” in downstream analyses, including systematic reviews and meta-analyses.
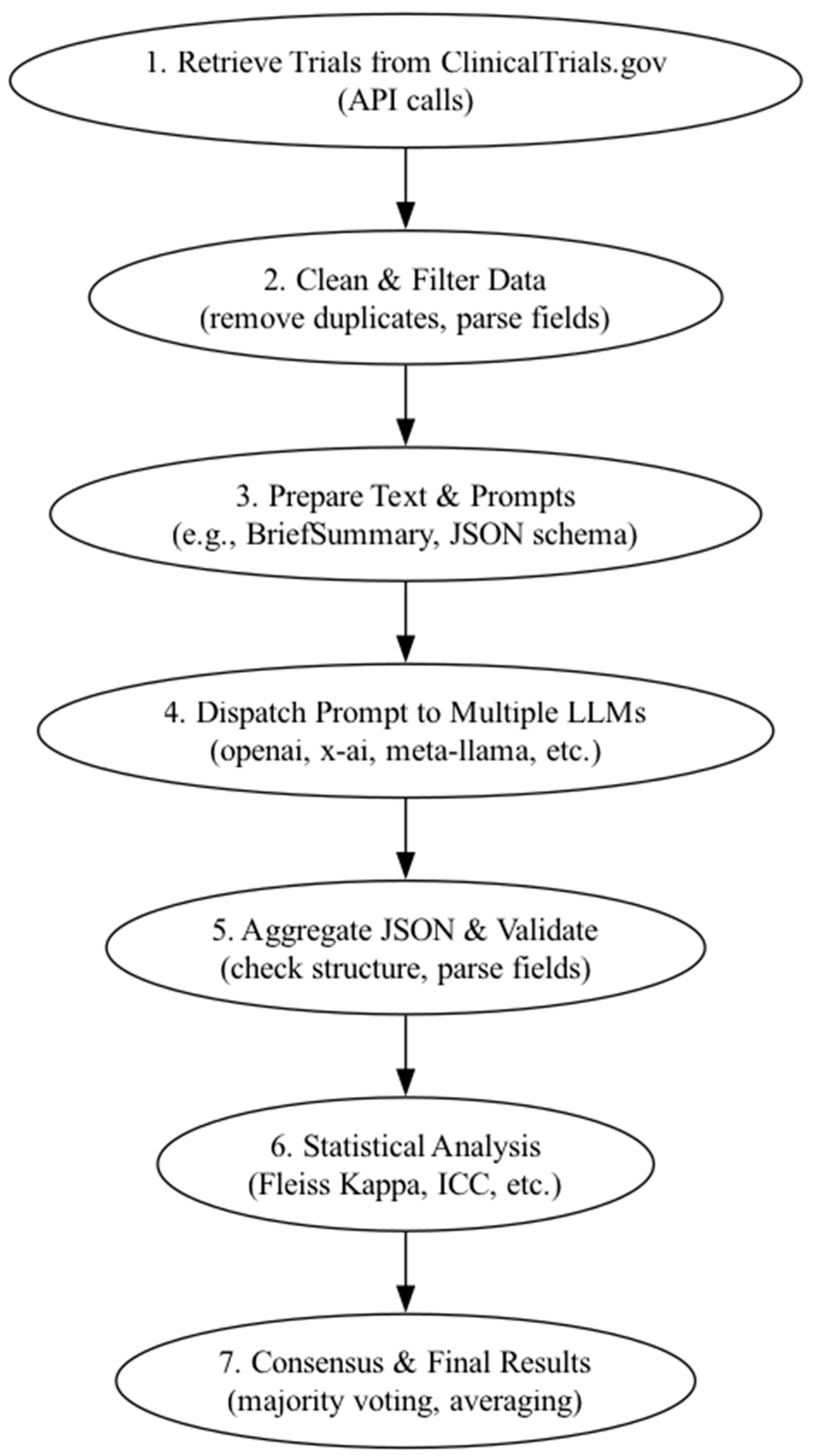
3.3. Analysis Pipeline
- Parses the returned JSON outputs.
- Validates the JSON structure (e.g., required fields and data types).
- Flags malformed outputs for potential manual review.
- Stores validated results in standardized columns, allowing for direct comparison across models.
3.4. Reproducibility
3.5. Alignment of Research Questions with Analytical Approaches
Code Availability
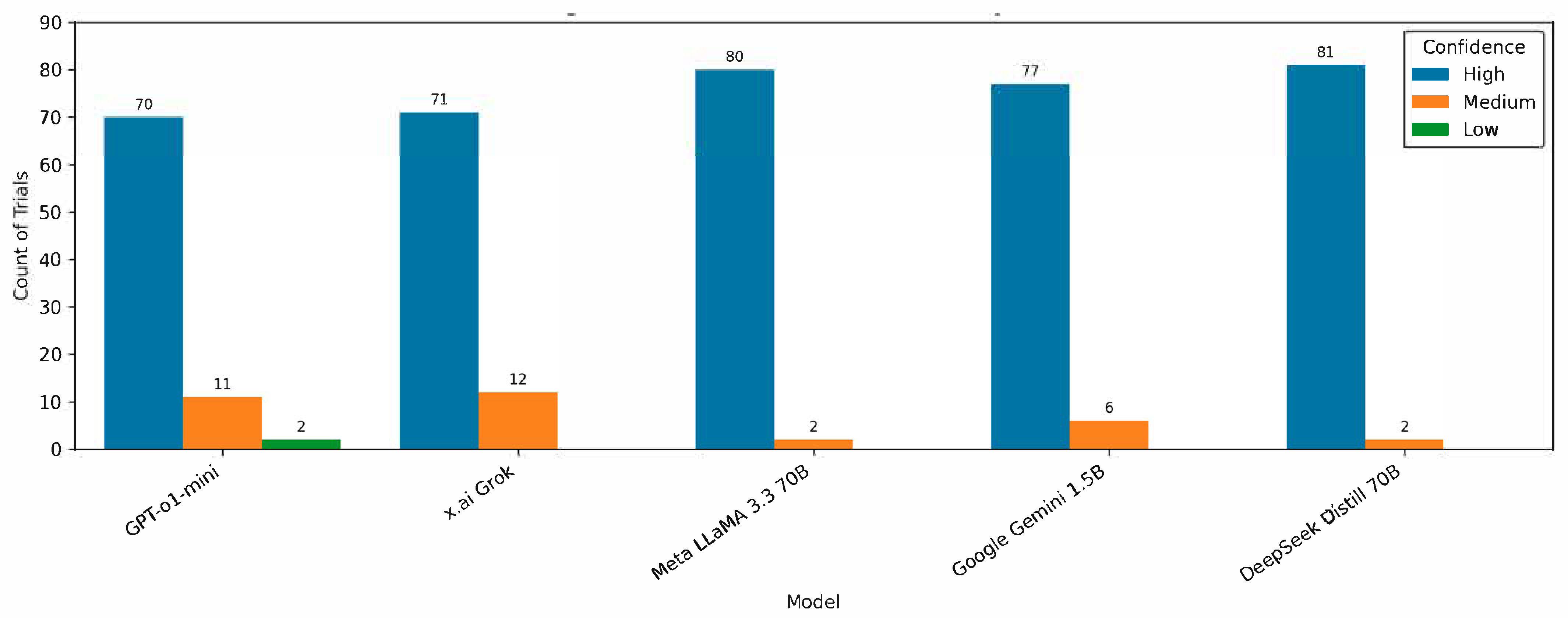
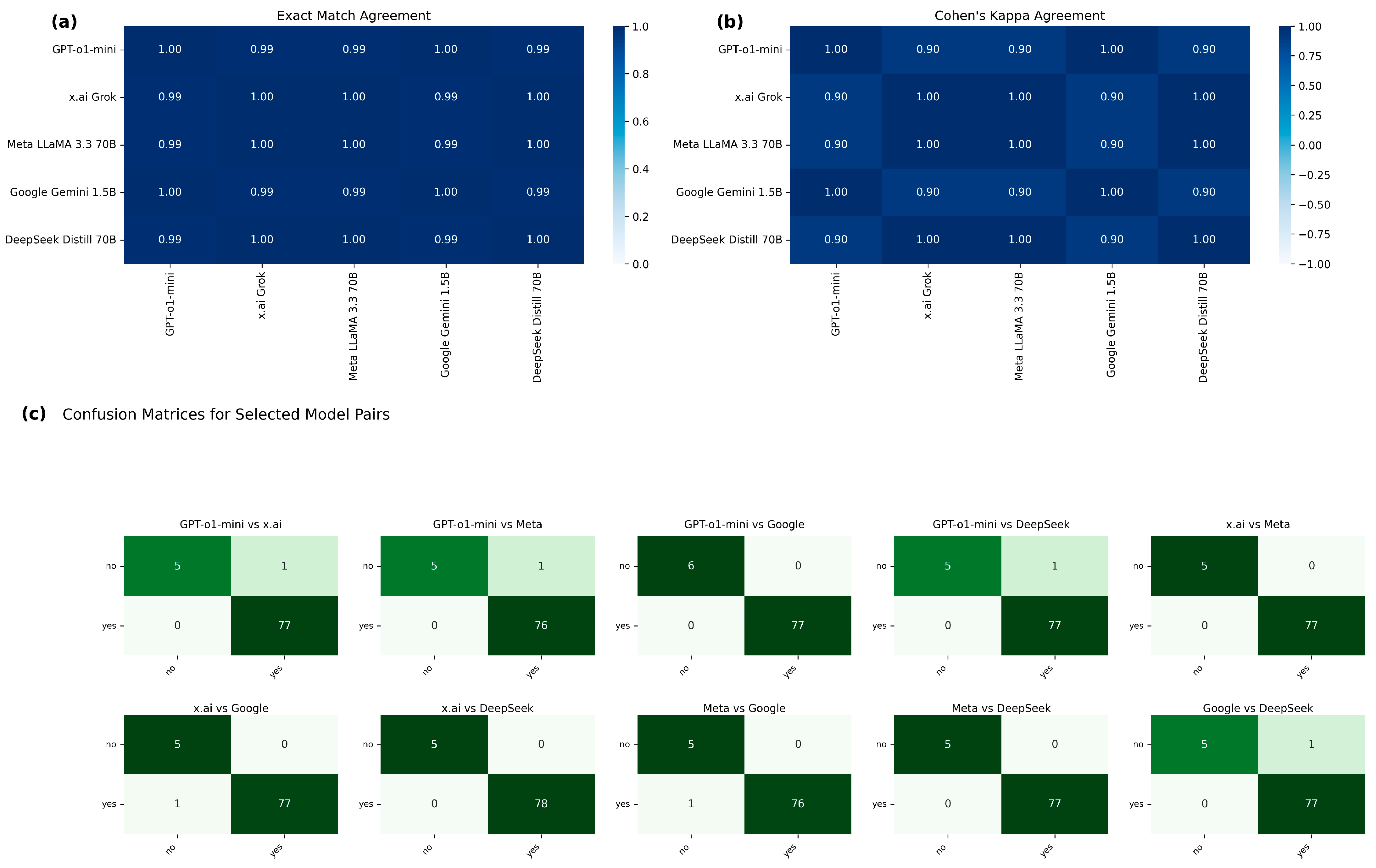
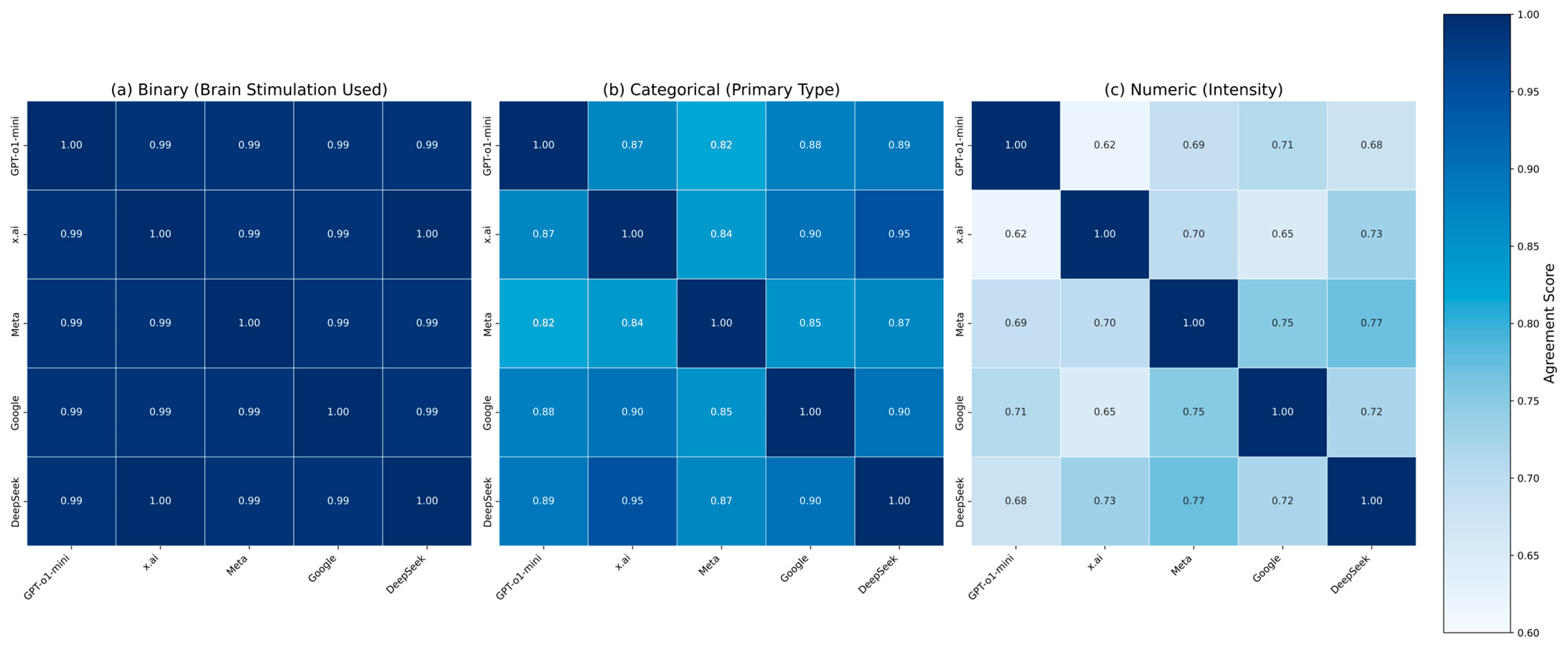
4. Results
5. Discussion
6. Conclusions
Future Research Directions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| API | Application Programming Interface |
| CI | Confidence Interval |
| CSV | Comma-Separated Values |
| DLPFC | Dorsolateral Prefrontal Cortex |
| EMR | Electronic Medical Records |
| ICC | Intraclass Correlation Coefficient |
| JSON | JavaScript Object Notation |
| LLM | Large Language Model |
| MAS | Multi-Agent System |
| NCT | National Clinical Trial |
| NIH | National Institutes of Health |
| NLP | Natural Language Processing |
| Parquet | A columnar storage format for efficient data handling |
| PubMed | A free resource for biomedical and life sciences literature |
| RAG | Retrieval-Augmented Generation |
| tDCS | Transcranial Direct Current Stimulation |
| TMS | Transcranial Magnetic Stimulation |
Appendix A
Appendix B
References
- Ricco, J.B.; Guetarni, F.; Kolh, P. Learning from artificial intelligence and big data in health care. Eur. J. Vasc. Endovasc. Surg. 2020, 59, 868–869. [Google Scholar] [CrossRef] [PubMed]
- Zarin, D.A.; Tse, T.; Williams, R.J.; Carr, S. Trial reporting in clinicaltrials.Gov—The final rule. N. Engl. J. Med. 2016, 375, 1998–2004. [Google Scholar] [CrossRef]
- Zarin, D.A.; Fain, K.M.; Dobbins, H.D.; Tse, T.; Williams, R.J. 10-year update on study results submitted to clinicaltrials.Gov. N. Engl. J. Med. 2019, 381, 1966–1974. [Google Scholar] [CrossRef]
- Chaturvedi, N.; Mehrotra, B.; Kumari, S.; Gupta, S.; Subramanya, H.S.; Saberwal, G. Some data quality issues at clinicaltrials.Gov. Trials 2019, 20, 378. [Google Scholar] [CrossRef]
- Pradhan, R.; Hoaglin, D.C.; Cornell, M.; Liu, W.; Wang, V.; Yu, H. Automatic extraction of quantitative data from clinicaltrials.Gov to conduct meta-analyses. J. Clin. Epidemiol. 2019, 105, 92–100. [Google Scholar] [CrossRef] [PubMed]
- Tasneem, A.; Aberle, L.; Ananth, H.; Chakraborty, S.; Chiswell, K.; McCourt, B.J.; Pietrobon, R. The database for aggregate analysis of clinicaltrials.Gov (aact) and subsequent regrouping by clinical specialty. PLoS ONE 2012, 7, e33677. [Google Scholar] [CrossRef]
- Nye, B.; Jessy Li, J.; Patel, R.; Yang, Y.; Marshall, I.J.; Nenkova, A.; Wallace, B.C. A Corpus with multi-level annotations of patients, interventions and outcomes to support language processing for medical literature. Proc. Conf. Assoc. Comput. Linguist. Meet. 2018, 2018, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Jonnalagadda, S.; Petitti, D. A new iterative method to reduce workload in systematic review process. Int. J. Comput. Biol. Drug Des. 2013, 6, 5–17. [Google Scholar] [CrossRef]
- Contributors, F.; El-Kishky, A.; Selsam, D.; Song, F.; Parascandolo, G.; Ren, H.; Lightman, H.; Won, H.; Akkaya, I.; Sutskever, I.; et al. Openai o1 system card. arXiv 2024, arXiv:2412.16720. [Google Scholar] [CrossRef]
- Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Yang, A.; Fan, A.; et al. The llama 3 herd of models. arXiv 2024, arXiv:abs/2407.21783. [Google Scholar]
- Reid, M.; Savinov, N.; Teplyashin, D.; Lepikhin, D.; Lillicrap, T.; Alayrac, J.-B.; Soricut, R.; Lazaridou, A.; Firat, O.; Schrittwieser, J.; et al. Gemini 1.5: Unlocking multimodal understanding across millions of tokens of context. arXiv 2024, arXiv:2403.05530. [Google Scholar] [CrossRef]
- Guo, D.; Yang, D.; Zhang, H.; Song, J.-M.; Zhang, R.; Xu, R.; Zhu, Q.; Ma, S.; Wang, P.; Bi, X.; et al. DeepSeek-r1: Incentivizing reasoning capability in llms via reinforcement learning. arXiv 2025, arXiv:2501.12948. [Google Scholar] [CrossRef]
- Alkaissi, H.; McFarlane, S.I. Artificial hallucinations in chatgpt: Implications in scientific writing. Cureus 2023, 15, e35179. [Google Scholar] [CrossRef]
- Azamfirei, R.; Kudchadkar, S.R.; Fackler, J. Large language models and the perils of their hallucinations. Crit. Care 2023, 27, 120. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Liu, Y.; Shi, L.; Huang, H.; Wang, R.; Yang, Z.; Zhang, L. Exploring and evaluating hallucinations in llm-powered code generation. arXiv 2024, arXiv:2404.00971. [Google Scholar] [CrossRef]
- Abdelghafour, M.A.M.; Mabrouk, M.; Taha, Z. Hallucination mitigation techniques in large language models. Int. J. Intell. Comput. Inf. Sci. 2024, 24, 73–81. [Google Scholar] [CrossRef]
- Ji, Z.; Lee, N.; Frieske, R.; Yu, T.; Su, D.; Xu, Y.; Ishii, E.; Bang, Y.J.; Madotto, A.; Fung, P. Survey of hallucination in natural language generation. ACM Comput. Surv. 2023, 55, 1–38. [Google Scholar] [CrossRef]
- Gartlehner, G.; Kahwati, L.; Hilscher, R.; Thomas, I.; Kugley, S.; Crotty, K.; Viswanathan, M.; Nussbaumer-Streit, B.; Booth, G.; Erskine, N.; et al. Data extraction for evidence synthesis using a large language model: A proof-of-concept study. Res. Synth. Methods 2023, 15, 576–589. [Google Scholar] [CrossRef]
- Valentina, P.; Aneta, H. Breaking down the metrics: A comparative analysis of llm benchmarks. Int. J. Sci. Res. Arch. 2024, 13, 777–788. [Google Scholar] [CrossRef]
- Erdengasileng, A.; Han, Q.; Zhao, T.; Tian, S.; Sui, X.; Li, K.; Wang, W.; Wang, J.; Hu, T.; Pan, F.; et al. Pre-trained models, data augmentation, and ensemble learning for biomedical information extraction and document classification. Database 2022, 2022, baac066. [Google Scholar] [CrossRef]
- Jia, Y.; Wang, H.; Yuan, Z.; Zhu, L.; Xiang, Z.L. Biomedical relation extraction method based on ensemble learning and attention mechanism. BMC Bioinform. 2024, 25, 333. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wei, Q.; Huang, L.C.; Li, J.; Hu, Y.; Chuang, Y.S.; He, J.; Das, A.; Keloth, V.K.; Yang, Y.; et al. Ensemble pretrained language models to extract biomedical knowledge from literature. J. Am. Med. Inform. Assoc. 2024, 31, 1904–1911. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Agarwal, A.; Singh, E. The Vulnerability of language model benchmarks: Do they accurately reflect true llm performance? arXiv 2024, arXiv:2412.03597. [Google Scholar] [CrossRef]
- Bowman, S.R.; Dahl, G.E. What will it take to fix benchmarking in natural language understanding? arXiv 2021, arXiv:abs/2104.02145. [Google Scholar]
- Yosefi, M.H.; Yagedi, Z.; Ahmadizadeh, Z.; Ehsani, F. Effect of transcranial direct current stimulation on learning and motor skill in healthy older adults: A systematic review. J. Maz. Univ. Med. Sci. 2017, 26, 221–231. [Google Scholar]
- Meek, A.W.; Greenwell, D.R.; Nishio, H.; Poston, B.; Riley, Z.A. Anodal m1 tdcs enhances online learning of rhythmic timing videogame skill. PLoS ONE 2024, 19, e0295373. [Google Scholar] [CrossRef]
- Pantovic, M.; Albuquerque, L.L.; Mastrantonio, S.; Pomerantz, A.S.; Wilkins, E.W.; Riley, Z.A.; Guadagnoli, M.A.; Poston, B. transcranial direct current stimulation of primary motor cortex over multiple days improves motor learning of a complex overhand throwing Task. Brain Sci. 2023, 13, 1441. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.A.; Greenwell, D.; Meek, A.W.; Poston, B.; Riley, Z.A. neuroenhancement of a dexterous motor task with anodal tdcs. Brain Res. 2022, 1790, 147993. [Google Scholar] [CrossRef]
- Buch, E.R.; Santarnecchi, E.; Antal, A.; Born, J.; Celnik, P.A.; Classen, J.; Gerloff, C.; Hallett, M.; Hummel, F.C.; Nitsche, M.A.; et al. Effects of tdcs on motor learning and memory formation: A consensus and critical position paper. Clin. Neurophysiol. 2017, 128, 589–603. [Google Scholar] [CrossRef]
- Meek, A.W.; Greenwell, D.; Poston, B.; Riley, Z.A. Anodal tdcs accelerates on-line learning of dart throwing. Neurosci. Lett. 2021, 764, 136211. [Google Scholar] [CrossRef]
- Hardwick, R.M.; Celnik, P.A. Cerebellar direct current stimulation enhances motor learning in older adults. Neurobiol. Aging 2014, 35, 2217–2221. [Google Scholar] [CrossRef]
- Nomura, T.; Kirimoto, H. Anodal transcranial direct current stimulation over the supplementary motor area improves anticipatory postural adjustments in older adults. Front. Hum. Neurosci. 2018, 12, 317. [Google Scholar] [CrossRef]
- Ljubisavljevic, M.R.; Oommen, J.; Filipovic, S.; Bjekic, J.; Szolics, M.; Nagelkerke, N. Effects of tdcs of dorsolateral prefrontal cortex on dual-task performance involving manual dexterity and cognitive task in healthy older adults. Front. Aging Neurosci. 2019, 11, 144. [Google Scholar] [CrossRef]
- Jiang, Y.; Ramasawmy, P.; Antal, A. Uncorking the limitation-improving dual tasking using transcranial electrical stimulation and task training in the elderly: A systematic review. Front. Aging Neurosci. 2024, 16, 1267307. [Google Scholar] [CrossRef] [PubMed]
- Siew-Pin Leuk, J.; Yow, K.E.; Zi-Xin Tan, C.; Hendy, A.M.; Kar-Wing Tan, M.; Hock-Beng Ng, T.; Teo, W.P. A meta-analytical review of transcranial direct current stimulation parameters on upper limb motor learning in healthy older adults and people with parkinson’s disease. Rev. Neurosci. 2023, 34, 325–348. [Google Scholar] [CrossRef]
- Pantovic, M.; Macak, D.; Cokorilo, N.; Moonie, S.; Riley, Z.A.; Madic, D.M.; Poston, B. The influence of transcranial direct current stimulation on shooting performance in elite deaflympic athletes: A case series. J. Funct. Morphol. Kinesiol. 2022, 7, 42. [Google Scholar] [CrossRef]
- Pantovic, M.; Lidstone, D.E.; de Albuquerque, L.L.; Wilkins, E.W.; Munoz, I.A.; Aynlender, D.G.; Morris, D.; Dufek, J.S.; Poston, B. cerebellar transcranial direct current stimulation applied over multiple days does not enhance motor learning of a complex overhand throwing task in young adults. Bioengineering 2023, 10, 1265. [Google Scholar] [CrossRef]
- Pino-Esteban, A.; Megía-García, Á.; Álvarez, D.M.-C.; Beltran-Alacreu, H.; Avendaño-Coy, J.; Gómez-Soriano, J.; Serrano-Muñoz, D. Can transcranial direct current stimulation enhance functionality in older adults? A systematic review. J. Clin. Med. 2021, 10, 2981. [Google Scholar] [CrossRef] [PubMed]
- Marshall, I.J.; Noel-Storr, A.; Kuiper, J.; Thomas, J.; Wallace, B.C. machine learning for identifying randomized controlled trials: An evaluation and practitioner’s guide. Res. Synth. Methods 2018, 9, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Tsafnat, G.; Glasziou, P.; Choong, M.K.; Dunn, A.; Galgani, F.; Coiera, E. Systematic review automation technologies. Syst. Rev. 2014, 3, 74. [Google Scholar] [CrossRef]
- He, C.; Zou, B.; Li, X.; Chen, J.; Xing, J.; Ma, H. Enhancing llm reasoning with multi-path collaborative reactive and reflection agents. arXiv 2024, arXiv:2501.00430. [Google Scholar] [CrossRef]
- Marshall, I.J.; Wallace, B.C. Toward systematic review automation: A practical guide to using machine learning tools in research synthesis. Syst. Rev. 2019, 8, 163. [Google Scholar] [CrossRef] [PubMed]
- O’Mara-Eves, A.; Thomas, J.; McNaught, J.; Miwa, M.; Ananiadou, S. Using text mining for study identification in systematic reviews: A systematic review of current approaches. Syst. Rev. 2015, 4, 5. [Google Scholar] [CrossRef] [PubMed]
- Riaz, I.B.; Naqvi, S.A.A.; Hasan, B.; Murad, M.H. Future of evidence synthesis: Automated, living, and interactive systematic reviews and meta-analyses. Mayo Clin. Proc. Digit. Health 2024, 2, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Sheikhalishahi, S.; Miotto, R.; Dudley, J.T.; Lavelli, A.; Rinaldi, F.; Osmani, V. Natural language processing of clinical notes on chronic diseases: Systematic review. JMIR Med. Inform. 2019, 7, e12239. [Google Scholar] [CrossRef]
- Bernard, N.; Sagawa, Y., Jr.; Bier, N.; Lihoreau, T.; Pazart, L.; Tannou, T. Using artificial intelligence for systematic review: The example of elicit. BMC Med. Res. Methodol. 2025, 25, 75. [Google Scholar] [CrossRef]
- Lai, H.; Liu, J.; Bai, C.; Liu, H.; Pan, B.; Luo, X.; Hou, L.; Zhao, W.; Xia, D.; Tian, J.; et al. Language models for data extraction and risk of bias assessment in complementary medicine. NPJ Digit. Med. 2025, 8, 74. [Google Scholar] [CrossRef]
- Motzfeldt Jensen, M.; Brix Danielsen, M.; Riis, J.; Assifuah Kristjansen, K.; Andersen, S.; Okubo, Y.; Jorgensen, M.G. Chatgpt-4o can serve as the second rater for data extraction in systematic reviews. PLoS ONE 2025, 20, e0313401. [Google Scholar] [CrossRef]
- Liu, J.; Lai, H.; Zhao, W.; Huang, J.; Xia, D.; Liu, H.; Luo, X.; Wang, B.; Pan, B.; Hou, L.; et al. Ai-driven evidence synthesis: Data extraction of randomized controlled trials with large language models. Int. J. Surg. 2025, 111, 2722–2726. [Google Scholar] [CrossRef]
- Stuhlmiller, T.J.; Rabe, A.J.; Rapp, J.; Manasco, P.; Awawda, A.; Kouser, H.; Salamon, H.; Chuyka, D.; Mahoney, W.; Wong, K.K.; et al. A scalable method for validated data extraction from electronic health records with large language models. medRxiv 2025. [Google Scholar] [CrossRef]
- Khan, M.A.; Ayub, U.; Naqvi, S.A.A.; Khakwani, K.Z.R.; Sipra, Z.B.R.; Raina, A.; Zou, S.; He, H.; Hossein, S.A.; Hasan, B.; et al. Collaborative large language models for automated data extraction in living systematic reviews. medRxiv 2024. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Zhang, R.; Doi, S.A.; Furuya-Kanamori, L.; Yu, T.; Lin, L.; Xu, C. How good are large language models for automated data extraction from randomized trials? medRxiv 2024. [Google Scholar] [CrossRef]
- Fan, D.; Che, X.; Jiang, Y.; He, Q.; Yu, J.; Zhao, H. Noninvasive brain stimulations modulated brain modular interactions to ameliorate working memory in community-dwelling older adults. Cereb. Cortex 2024, 34, bhae140. [Google Scholar] [CrossRef] [PubMed]
- Manor, B. Transcranial electrical stimulation as a tool to understand and enhance mobility in older adults. Innov. Aging 2023, 7, 281. [Google Scholar] [CrossRef]
- Manor, B.; Lo, O.; Zhou, J.; Dhami, P.; Farzan, F. Noninvasive brain stimulation to reduce falls in older adults. In Falls and Cognition in Older Persons: Fundamentals, Assessment and Therapeutic Options; Springer: Cham, Switzerland, 2019; pp. 373–398. [Google Scholar] [CrossRef]
- Zhou, J.; Lo, O.; Halko, M.; Harrison, R.; Lipsitz, L.; Manor, B. Noninvasive brain stimulation increases the complexity of resting-state brain network activity in older adults. Innov. Aging 2018, 2, 402. [Google Scholar] [CrossRef]
- Moon, I.H. Performance Comparison of large language models on advanced calculus problems. arXiv 2025, arXiv:2503.03960. [Google Scholar] [CrossRef]
- Shojaee-Mend, H.; Mohebbati, R.; Amiri, M.; Atarodi, A. Evaluating the strengths and weaknesses of large language models in answering neurophysiology questions. Sci. Rep. 2024, 14, 10785. [Google Scholar] [CrossRef]
- Chen, X.; Xiang, J.; Lu, S.; Liu, Y.; He, M.; Shi, D. Evaluating large language models in medical applications: A survey. arXiv 2024, arXiv:2405.07468. [Google Scholar] [CrossRef]
- Guo, Z.; Jin, R.; Liu, C.; Huang, Y.; Shi, D.; Supryadi; Yu, L.; Liu, Y.; Li, J.; Xiong, B.; et al. Evaluating large language models: A comprehensive survey. arXiv 2023, arXiv:2310.19736. [Google Scholar] [CrossRef]
- Mondorf, P.; Plank, B. Beyond Accuracy: Evaluating the reasoning behavior of large language models—A survey. arXiv 2024, arXiv:2404.01869. [Google Scholar] [CrossRef]
- Anthropic. Claude 3 Model Card: October Addendum. PDF, n.d. Available online: https://assets.anthropic.com/m/1cd9d098ac3e6467/original/Claude-3-Model-Card-October-Addendum.pdf (accessed on 2 May 2025).
- Barnett, S.; Brannelly, Z.; Kurniawan, S.; Wong, S. Fine-tuning or fine-failing? debunking performance myths in large language models. arXiv 2024, arXiv:2406.11201. [Google Scholar] [CrossRef]
- Liu, S.; McCoy, A.B.; Wright, A. Improving large language model applications in biomedicine with retrieval-augmented generation: A systematic review, meta-analysis, and clinical development guidelines. J. Am. Med. Inform. Assoc. 2025, 32, 605–615. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, X.; Yu, H. Exploring llm multi-agents for icd coding. arXiv 2024, arXiv:2406.15363. [Google Scholar] [CrossRef]
- Pandey, H.; Amod, A.; Kumar, S. Advancing healthcare automation: Multi-agent system for medical necessity justification. Workshop Biomed. Nat. Lang. Process. 2024, arXiv:2404.17977. [Google Scholar] [CrossRef]
- Yang, H.; Chen, H.; Guo, H.; Chen, Y.; Lin, C.; Hu, S.; Hu, J.; Wu, X.; Wang, X. Llm-medqa: Enhancing medical question answering through case studies in large language models. arXiv 2024, arXiv:2501.05464. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, Z.; Lei, X.; Wang, N.; Long, Z.; Zhao, J.; Wang, Z.; Yang, P.; Hua, M.; Ma, C.; et al. Quantifying the capability boundary of deepseek models: An application-driven performance analysis. arXiv 2025, arXiv:2502.11164. [Google Scholar] [CrossRef]
- Gao, T.; Jin, J.; Ke, Z.; Moryoussef, G. A comparison of deepseek and other llms. arXiv 2025, arXiv:2502.03688. [Google Scholar] [CrossRef]
- Sapkota, R.; Raza, S.; Karkee, M. Comprehensive analysis of transparency and accessibility of chatgpt, deepseek, and other sota large language models. arXiv 2025, arXiv:2502.18505. [Google Scholar] [CrossRef]
- Yang, J.; Luo, J.; Tian, X.; Zhao, Y.; Li, Y.; Wu, X. Progress in understanding oxidative stress, aging, and aging-related diseases. Antioxidants 2024, 13, 394. [Google Scholar] [CrossRef]
- Lee, J.; Kim, H.J. Normal aging induces changes in the brain and neurodegeneration progress: Review of the structural, biochemical, metabolic, cellular, and molecular changes. Front. Aging Neurosci. 2022, 14, 931536. [Google Scholar] [CrossRef]
- Epel, E.S.; Prather, A.A. Stress, telomeres, and psychopathology: Toward a deeper understanding of a triad of early aging. Annu. Rev. Clin. Psychol. 2018, 14, 371–397. [Google Scholar] [CrossRef]
- Trapp, C.; Schmidt-Hegemann, N.; Keilholz, M.; Brose, S.F.; Marschner, S.N.; Schonecker, S.; Maier, S.H.; Dehelean, D.C.; Rottler, M.; Konnerth, D.; et al. Patient-and clinician-based evaluation of large language models for patient education in prostate cancer radiotherapy. Strahlenther. Onkol. 2025, 201, 333–342. [Google Scholar] [CrossRef]
- Hao, Y.; Holmes, J.; Waddle, M.; Yu, N.; Vickers, K.; Preston, H.; Margolin, D.; Löckenhoff, C.E.; Vashistha, A.; Ghassemi, M.; et al. Outlining the borders for llm applications in patient education: Developing an expert-in-the-loop llm-powered chatbot for prostate cancer patient education. arXiv 2024, arXiv:2409.19100. [Google Scholar] [CrossRef]
- Yang, Y.; Peng, Q.; Wang, J.; Zhang, W. Multi-llm-agent systems: Techniques and business perspectives. arXiv 2024, arXiv:2411.14033. [Google Scholar] [CrossRef]
- Fuellen, G.; Kulaga, A.; Lobentanzer, S.; Unfried, M.; Avelar, R.A.; Palmer, D.; Kennedy, B.K. Validation requirements for ai-based intervention-evaluation in aging and longevity research and practice. Ageing Res. Rev. 2025, 104, 102617. [Google Scholar] [CrossRef] [PubMed]
- Shusterman, R.; Waters, A.C.; O’Neill, S.; Bangs, M.; Luu, P.; Tucker, D.M. An active inference strategy for prompting reliable responses from large language models in medical practice. NPJ Digit. Med. 2025, 8, 119. [Google Scholar] [CrossRef]
- Tonmoy, S.; Zaman, S.M.M.; Jain, V.; Rani, A.; Rawte, V.; Chadha, A.; Das, A. A Comprehensive survey of hallucination mitigation techniques in large language models. arXiv 2024, arXiv:2401.01313. [Google Scholar] [CrossRef]
- Duan, H.; Yang, Y.; Tam, K. Do llms know about hallucination? an empirical investigation of llm’s hidden states. arXiv 2024, arXiv:2402.09733. [Google Scholar] [CrossRef]
- Ciatto, G.; Agiollo, A.; Magnini, M.; Omicini, A. Large language models as oracles for instantiating ontologies with domain-specific knowledge. Knowledge-Based Syst. 2025, 310, 112940. [Google Scholar] [CrossRef]
- Belisle-Pipon, J.C. Why we need to be careful with llms in medicine. Front. Med. 2024, 11, 1495582. [Google Scholar] [CrossRef]
| Description | Details |
|---|---|
| Data Source | ClinicalTrials.gov |
| API Endpoint URL | https://clinicaltrials.gov/data-api/api |
| Query Terms-Keywords | “Aged” |
| Number of Trials Retrieved | 10,030 trials |
| Inclusion Criteria | Trials explicitly mentioning tDCS, targeting older adults (aged ≥ 65), and involving brain stimulation methods |
| Exclusion Criteria | Trials not explicitly mentioning tDCS, animal studies, duplicates, and unrelated interventions |
| Final Number of Trials Analyzed | 83 trials |
| Date of Data Extraction | 1 January 2025 |
| NCTId | # Models Saying Yes | # Models Saying No | Consensus |
|---|---|---|---|
| NCT05511259 | 3 | 2 | Yes |
| NCT06501755 | 4 | 0 | Yes |
| NCTId | Model | Primary Type | Intensity | Confidence |
|---|---|---|---|---|
| NCT06658795 | OpenAI_o1_mini | tDCS | None | High |
| NCT06658795 | x_ai_grok_2_1212 | tDCS | None | High |
| NCT06658795 | Meta_LLaMA_3.3_70B | tDCS | None | High |
| NCT06658795 | Google_Gemini_Flash_1.5_8b | tDCS | None | High |
| NCT06658795 | DeepSeek_Distill_70b | tDCS | None | High |
| NCT03814304 | OpenAI_o1_mini | tDCS | None | High |
| NCT03814304 | x_ai_grok_2_1212 | tDCS | None | High |
| NCT03814304 | Meta_LLaMA_3.3_70B | tDCS | None | High |
| NCT03814304 | Google_Gemini_Flash_1.5_8b | tDCS | None | High |
| NCT03814304 | DeepSeek_Distill_70b | tDCS | 2mA | High |
| NCT02436915 | OpenAI_o1_mini | tDCS | None | High |
| NCT02436915 | x_ai_grok_2_1212 | tDCS | None | High |
| NCT02436915 | Meta_LLaMA_3.3_70B | tDCS | None | High |
| NCT02436915 | Google_Gemini_Flash_1.5_8b | tDCS | None | High |
| NCT02436915 | DeepSeek_Distill_70b | tDCS | 2mA | High |
| Field | FleissKappa | Mean % Agreement | # Trials | Categories |
|---|---|---|---|---|
| brain_stimulation_used | 0.941 | 0.993 | 83 | yes, no |
| primary_type | 0.709 | 0.938 | 78 | ctdcs, dtms, dtms, tdcs, hd-tdcs, missing, non-invasive brain stimulation, tacs, tcs, tdcs, tdcs and tacs, tdcs, tacs, tms, tps, tps (transcranial pulse stimulation), tps (transcranial pulse stimulation, also known as low-intensity extracorporeal shock wave therapy (li-eswt)) |
| is_noninvasive | −0.003 | 0.978 | 81 | true, false |
| primary_target | 0.53 | 0.747 | 76 | anodal, anodal transcranial direct current stimulation (atdcs), audio-visual associative memory areas, bifrontal, brain regions associated with cognitive function, brain regions associated with sleep spindles, brain regions involved in active cognitive function, brain regions underneath the neocortex, brain underneath the neocortex, center electrode, central nervous system, cerebellum, cerebral cortex, cognitive control network (ccn), cortical and deep brain structures, cortical areas, corticothalamic and corticospinal projections, dlpfc, dorso-lateral prefrontal cortex, dorso-lateral prefrontal cortices, dorsolateral prefrontal cortex, dorsolateral prefrontal cortex (dlpfc), dorsolateral prefrontal cortex (f3), dorsolateral prefrontal cortex, areas of the memory and language network, dorsolateral prefrontal cortex, memory and language network, f3, frontal brain regions, frontal circuits, fronto-central, fronto-central alpha, fronto-central alpha region, fronto-central region, frontopolar cortex, inferior frontal lobe, inferior frontal lobe and superior parietal lobe, knee oa-related areas, language areas, language areas of the brain, lateral prefrontal cortex (lpfc), lateral prefrontal cortex (lpfc) and default mode network (dmn), left dlpfc, left dorsal lateral prefrontal cortex (dlpfc), left dorsolateral prefrontal cortex, left dorsolateral prefrontal cortex (dlpfc), left m1, left prefrontal cortex, m1, m1 (primary motor cortex), memory-related brain regions, mid-cingulate cortex (mcc), missing, motor cortex, neocortex, not explicitly stated, not specified, null, pain-related brain regions, pf areas, pfc, pre-frontal (pf) areas, pre-frontal (pf) brain areas, pre-frontal areas, precuneus, prefrontal circuits, prefrontal cortex, prefrontal cortex and angular gyrus, prefrontal cortex, angular gyrus, prefrontal cortex, specifically the dlpfc, prefrontal cortical activity, prefrontal regions f3/f4, prefrontal right region, primary motor cortex, primary motor cortex (m1), primary motor cortex (m1) contralateral to the moving leg (cm1), primary motor cortex (m1), posterior parietal cortex (ppc), and cerebellar cortex (cbm), right dlpfc and right ppc, right dorsolateral prefrontal cortex (dlpfc), right dorsolateral prefrontal cortex (dlpfc) and right posterior parietal cortex (ppc), right inferior frontal lobe, right temporoparietal junction, right temporoparietal junction (rtpj), right temporoparietal junction (rtpj) or dorsomedial prefrontal cortex (dmpfc), scalp, subcortical areas, theta frequency, unspecified |
| parameters_intensity | 0.498 | 0.786 | 28 | 1 ma, 1–2 ma, 1–2 ma (tdcs), 1–2 milliampere (ma), 1.5 ma, 1.5 ma, 1 ma, 2 ma, 2 milliampere (ma), 2 ma, 3 ma, 3 ma, anodal stimulation, low-intensity, missing, not specified, null, very low energy, very weak electrical current, very weak electrical current from a 9-volt battery |
| parameters_duration | 0.498 | 0.765 | 40 | 1 h/week, 1 h/week for 10 weeks, 1 min, 1 min, 1 min on, 5 s off, 10 sessions over 2 weeks, 13 min, 13 min, 1 min with 5 s breaks, 2 weeks, 20 min, 20 to 60 min, 20–30 min, 10 days, and 2 weeks, 3 sessions per week for 2–4 weeks, 3 sessions per week for 3 weeks (9 sessions total), 3 sessions per week, 6000 pulses each, for 2–4 weeks, 30 min, 30 min for 5 successive days for temporal cortex stimulation, 30 min per session, 30 min per session, 10 sessions over two weeks, 4 weeks, 4-week (3 times per week) treatment, 4000 pulses each, every 6 months for an average of two to four years; 3 tps sessions (6000 pulses each) per week for 2–4 weeks, 5 consecutive days, 5 consecutive days, 30 min per session, 5 sessions/day over 4 days, 5 sessions/day over 4 days only, 6 sessions over 2 weeks, 8 sessions of anodal tdcs completed twice a week for 4 weeks, 8 sessions, twice a week for 4 weeks, 9 sessions (3 sessions per week for 3 weeks), acute (one-time), applied during training sessions, approximately 20 min, daily over 4 weeks, during slow wave sleep, missing, null, repeated sessions, two weeks, two-week, varied (e.g., 3 times per week for 2–4 weeks) |
| (a) | |||
| Field | FleissKappa | 95% CI | Interpretation |
| brain_stimulation_used | 0.904 | 0.68–1.0 | Almost Perfect |
| primary_type | 0.709 | 0.6–0.82 | Substantial |
| is_noninvasive | 0.59 | 0.4–0.71 | Moderate |
| (b) | |||
| Parameter | ICC(2,1) | 95% CI | Interpretation |
| intensity | 0.95 | 0.9–0.98 | Excellent |
| duration | 0.964 | 0.94–0.98 | Excellent |
| frequency | 1 | 1.0–1.0 | Excellent |
| NCT ID | Brain Stimulation | Stimulation Type | Intensity (mA) | Duration (Minutes) | Primary Target | Confidence | Unanimous? | Comments |
|---|---|---|---|---|---|---|---|---|
| NCT06658795 | Yes | tDCS | lateral prefrontal cortex (LPFC) | High | No | Intensity (mA) note: no numeric parse; duration (minutes) note: no numeric parse | ||
| NCT05511259 | Yes | TMS | None | High | No | Intensity (mA) note: no numeric parse; duration (minutes) note: no numeric parse | ||
| NCT04154397 | Yes | ctDCS | cerebellum | High | No | Intensity (mA) note: no numeric parse; duration (minutes) note: no numeric parse | ||
| NCT03814304 | Yes | tDCS | 2.00 | 20.00 | left dorsolateral prefrontal cortex (dlPFC) | High | No | |
| NCT02436915 | Yes | tDCS | 2.00 | 20.00 | left prefrontal cortex | High | No |
| Model Suffix | Brain Stim. | Primary Type | Is Noninvasive | Intensity | Duration | Primary Target | Avg. Agreement |
|---|---|---|---|---|---|---|---|
| GPT-o1-mini | 98.8 | 90.2 | 95.2 | 77.8 | 68.8 | 87.3 | 86.3 |
| x.ai Grok | 100 | 96.3 | 100 | 77.8 | 90.6 | 75.9 | 90.1 |
| Meta LLaMA 3.3 70B | 98.8 | 92.7 | 98.8 | 77.8 | 90.6 | 86.1 | 90.8 |
| Google Gemini Flash 1.5B | 98.8 | 96.3 | 97.6 | 77.8 | 78.1 | 83.5 | 88.7 |
| DeepSeek Distill 70B | 100 | 98.8 | 97.6 | 94.4 | 90.6 | 50.6 | 88.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Young, R.J.; Matthews, A.M.; Poston, B. Benchmarking Multiple Large Language Models for Automated Clinical Trial Data Extraction in Aging Research. Algorithms 2025, 18, 296. https://doi.org/10.3390/a18050296
Young RJ, Matthews AM, Poston B. Benchmarking Multiple Large Language Models for Automated Clinical Trial Data Extraction in Aging Research. Algorithms. 2025; 18(5):296. https://doi.org/10.3390/a18050296
Chicago/Turabian StyleYoung, Richard J., Alice M. Matthews, and Brach Poston. 2025. "Benchmarking Multiple Large Language Models for Automated Clinical Trial Data Extraction in Aging Research" Algorithms 18, no. 5: 296. https://doi.org/10.3390/a18050296
APA StyleYoung, R. J., Matthews, A. M., & Poston, B. (2025). Benchmarking Multiple Large Language Models for Automated Clinical Trial Data Extraction in Aging Research. Algorithms, 18(5), 296. https://doi.org/10.3390/a18050296








