Quantifying Intra- and Inter-Observer Variabilities in Manual Contours for Radiotherapy: Evaluation of an MR Tumor Autocontouring Algorithm for Liver, Prostate, and Lung Cancer Patients
Abstract
1. Introduction
2. Materials and Methods
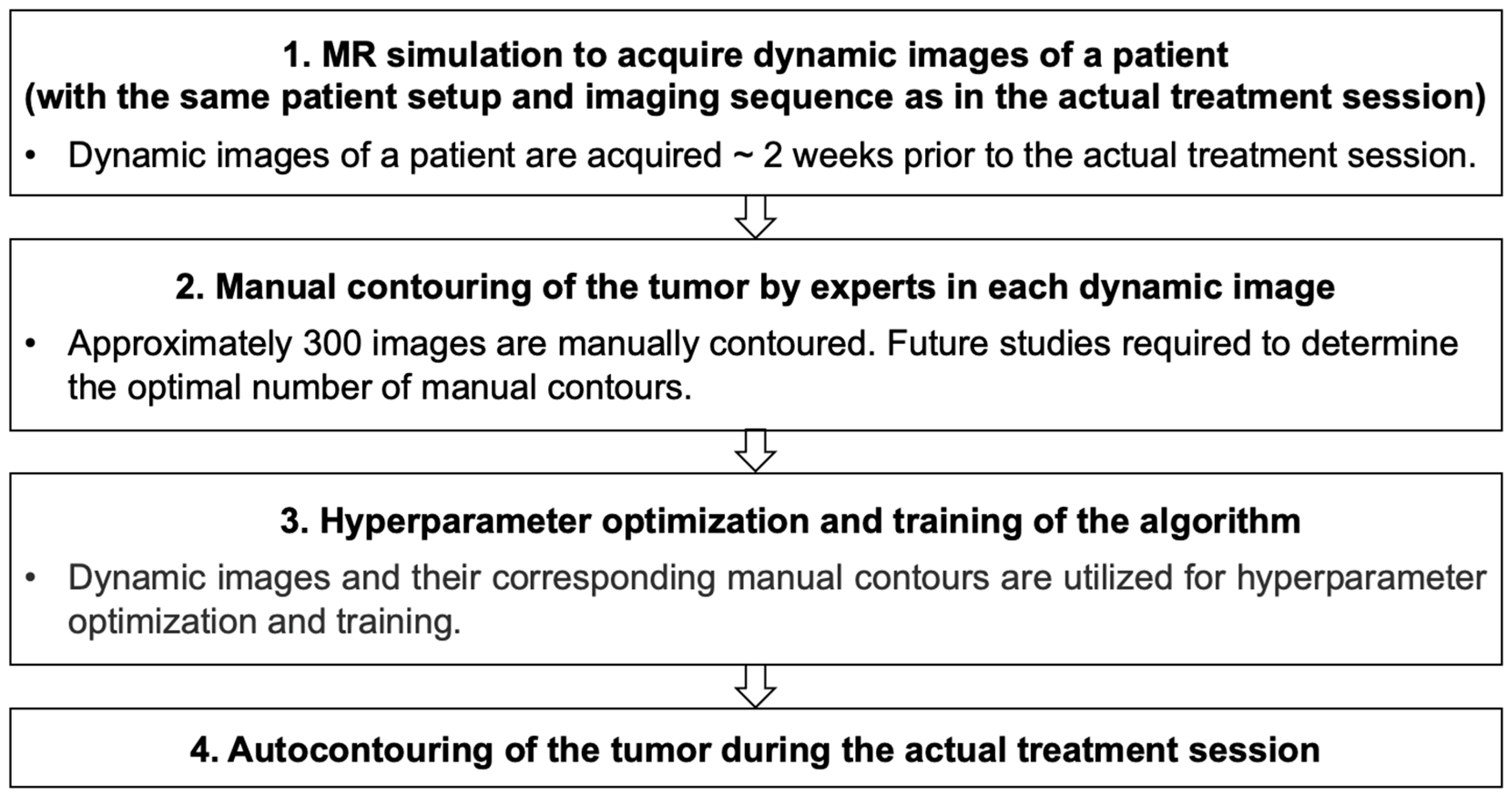
2.1. Patient Imaging and Manual Contouring
2.2. Autocontouring Algorithm
2.3. Evaluation of Contours
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Liver, Automatic vs. Manual | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M11 | M12 | M21 | M22 | M31 | M32 | |||||||||||||
| DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | |
| A11 | 0.92 (0.04) | 1.2 (0.7) | 2.6 (0.8) | 0.80 (0.03) | 2.4 (0.7) | 5.6 (0.9) | 0.76 (0.06) | 3.2 (1.3) | 7.1 (1.7) | 0.73 (0.05) | 3.6 (1.1) | 6.9 (1.4) | 0.67 (0.04) | 2.9 (0.9) | 7.6 (1.1) | 0.65 (0.04) | 3.6 (0.9) | 7.9 (1.1) |
| A12 | 0.78 (0.03) | 2.3 (0.7) | 5.6 (0.9) | 0.93 (0.03) | 1.2 (0.6) | 2.7 (0.8) | 0.79 (0.05) | 3.0 (1.2) | 6.3 (1.5) | 0.78 (0.05) | 3.1 (1.0) | 6.6 (1.3) | 0.60 (0.03) | 3.5 (1.0) | 9.0 (1.3) | 0.66 (0.03) | 3.2 (0.8) | 9.3 (1.1) |
| A21 | 0.75 (0.04) | 3.0 (0.8) | 7.1 (1.1) | 0.80 (0.04) | 2.9 (0.8) | 5.9 (1.1) | 0.89 (0.04) | 1.6 (1.0) | 3.5 (1.2) | 0.85 (0.05) | 2.1 (1.0) | 4.8 (1.5) | 0.64 (0.04) | 3.7 (1.0) | 8.5 (1.1) | 0.68 (0.04) | 3.4 (0.9) | 9.1 (1.1) |
| A22 | 0.75 (0.04) | 2.9 (0.8) | 6.5 (1.1) | 0.80 (0.04) | 2.5 (0.8) | 5.8 (1.2) | 0.86 (0.05) | 2.1 (1.1) | 4.4 (1.4) | 0.89 (0.05) | 1.8 (0.9) | 3.7 (1.2) | 0.63 (0.04) | 4.0 (1.0) | 8.3 (1.3) | 0.71 (0.04) | 3.2 (0.9) | 8.1 (1.2) |
| A31 | 0.66 (0.05) | 2.8 (0.8) | 7.4 (1.1) | 0.61 (0.03) | 3.5 (0.8) | 8.8 (1.2) | 0.65 (0.05) | 3.7 (1.2) | 8.2 (1.7) | 0.64 (0.05) | 3.8 (1.1) | 7.9 (1.4) | 0.87 (0.06) | 1.4 (0.8) | 2.8 (0.9) | 0.73 (0.05) | 2.4 (0.8) | 5.7 (1.0) |
| A32 | 0.66 (0.04) | 3.3 (0.8) | 7.8 (1.1) | 0.67 (0.04) | 2.9 (0.8) | 9.2 (1.1) | 0.70 (0.05) | 3.6 (1.2) | 8.9 (1.7) | 0.72 (0.05) | 3.2 (1.1) | 7.6 (1.6) | 0.74 (0.05) | 2.7 (1.0) | 5.6 (1.1) | 0.87 (0.05) | 1.4 (0.7) | 2.7 (0.7) |
| Liver, Manual vs. Manual | ||||||||||||||||||
| M11 | M12 | M21 | M22 | M31 | M32 | |||||||||||||
| DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | |
| M11 | - | - | - | 0.79 (0.04) | 2.6 (0.8) | 5.8 (1.0) | 0.75 (0.06) | 3.4 (1.3) | 7.4 (1.7) | 0.72 (0.05) | 3.7 (1.2) | 7.0 (1.5) | 0.66 (0.05) | 3.0 (1.0) | 6.7 (1.4) | 0.65 (0.05) | 3.6 (1.0) | 7.9 (1.3) |
| M12 | 0.79 (0.04) | 2.6 (0.8) | 5.8 (1.0) | - | - | - | 0.78 (0.06) | 3.1 (1.3) | 6.4 (1.7) | 0.78 (0.06) | 3.0 (1.1) | 6.6 (1.5) | 0.60 (0.04) | 3.6 (1.1) | 9.0 (1.5) | 0.66 (0.04) | 3.1 (1.1) | 9.3 (1.3) |
| M21 | 0.75 (0.06) | 3.4 (1.3) | 7.4 (1.7) | 0.78 (0.06) | 3.1 (1.3) | 6.4 (1.7) | - | - | - | 0.84 (0.06) | 2.5 (1.3) | 5.2 (1.9) | 0.64 (0.06) | 3.9 (1.4) | 8.5 (1.8) | 0.68 (0.05) | 3.7 (1.3) | 9.1 (1.8) |
| M22 | 0.72 (0.05) | 3.7 (1.2) | 7.0 (1.5) | 0.78 (0.06) | 3.0 (1.1) | 6.6 (1.5) | 0.84 (0.06) | 2.5 (1.3) | 5.2 (1.9) | - | - | - | 0.63 (0.05) | 4.1 (1.3) | 8.2 (1.6) | 0.71 (0.05) | 3.3 (1.2) | 7.8 (1.6) |
| M31 | 0.66 (0.05) | 3.0 (1.0) | 6.7 (1.4) | 0.60 (0.04) | 3.6 (1.1) | 9.0 (1.5) | 0.64 (0.06) | 3.9 (1.4) | 8.5 (1.8) | 0.63 (0.05) | 4.1 (1.3) | 8.2 (1.6) | - | - | - | 0.72 (0.06) | 2.9 (1.1) | 6.1 (1.4) |
| M32 | 0.65 (0.05) | 3.6 (1.0) | 7.9 (1.3) | 0.66 (0.04) | 3.1 (1.1) | 9.3 (1.3) | 0.68 (0.05) | 3.7 (1.3) | 9.1 (1.8) | 0.71 (0.05) | 3.3 (1.2) | 7.8 (1.6) | 0.72 (0.06) | 2.9 (1.1) | 6.1 (1.4) | - | - | - |
| Prostate, Automatic vs. Manual | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M11 | M12 | M21 | M22 | M31 | M32 | |||||||||||||
| DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | |
| A11 | 0.96 (0.01) | 0.7 (0.4) | 2.7 (0.7) | 0.95 (0.01) | 1.2 (0.4) | 3.8 (0.6) | 0.88 (0.02) | 2.4 (0.8) | 7.3 (1.3) | 0.85 (0.02) | 3.2 (0.9) | 9.0 (1.3) | 0.89 (0.02) | 2.5 (0.7) | 6.4 (1.1) | 0.89 (0.02) | 2.9 (0.5) | 6.8 (0.8) |
| A12 | 0.94 (0.01) | 1.5 (0.4) | 4.1 (0.7) | 0.96 (0.01) | 0.8 (0.3) | 2.7 (0.7) | 0.87 (0.02) | 2.4 (0.8) | 7.7 (1.3) | 0.85 (0.02) | 2.9 (1.0) | 9.0 (1.3) | 0.89 (0.02) | 2.9 (0.8) | 6.4 (0.9) | 0.88 (0.01) | 3.1 (0.5) | 6.6 (0.9) |
| A21 | 0.88 (0.01) | 2.2 (0.5) | 7.3 (0.7) | 0.88 (0.01) | 2.1 (0.6) | 7.4 (0.8) | 0.94 (0.02) | 1.7 (0.8) | 4.6 (1.3) | 0.92 (0.02) | 2.6 (1.1) | 6.3 (1.4) | 0.84 (0.02) | 2.7 (0.8) | 9.2 (1.3) | 0.88 (0.01) | 2.9 (0.7) | 8.4 (1.2) |
| A22 | 0.85 (0.01) | 2.2 (0.5) | 8.3 (0.7) | 0.86 (0.01) | 2.0 (0.4) | 8.2 (0.7) | 0.92 (0.02) | 2.1 (0.9) | 6.1 (1.4) | 0.94 (0.02) | 2.0 (0.9) | 4.9 (1.4) | 0.82 (0.02) | 3.1 (0.8) | 10.5 (1.3) | 0.86 (0.01) | 3.3 (0.6) | 9.8 (0.9) |
| A31 | 0.89 (0.01) | 2.5 (0.5) | 6.2 (0.7) | 0.89 (0.01) | 2.8 (0.5) | 6.0 (0.7) | 0.86 (0.02) | 2.4 (0.8) | 8.4 (1.4) | 0.83 (0.02) | 3.3 (0.9) | 10.4 (1.4) | 0.95 (0.01) | 1.1 (0.6) | 3.3 (0.8) | 0.93 (0.01) | 1.3 (0.5) | 5.1 (0.7) |
| A32 | 0.88 (0.01) | 3.2 (0.4) | 7.3 (0.6) | 0.88 (0.01) | 3.2 (0.5) | 6.8 (0.7) | 0.89 (0.02) | 2.7 (0.8) | 7.5 (1.2) | 0.87 (0.02) | 3.4 (0.8) | 9.7 (1.3) | 0.92 (0.01) | 1.5 (0.7) | 5.4 (0.8) | 0.95 (0.01) | 1.1 (0.5) | 3.3 (0.6) |
| Prostate, Manual vs. Manual | ||||||||||||||||||
| M11 | M12 | M21 | M22 | M31 | M32 | |||||||||||||
| DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | |
| M11 | - | - | - | 0.94 (0.01) | 1.4 (0.5) | 4.0 (0.7) | 0.88 (0.02) | 2.3 (0.8) | 7.3 (1.3) | 0.85 (0.02) | 3.1 (0.9) | 9.1 (1.3) | 0.89 (0.02) | 2.5 (0.7) | 6.4 (0.9) | 0.88 (0.01) | 2.9 (0.6) | 6.7 (0.8) |
| M12 | 0.94 (0.01) | 1.4 (0.5) | 4.0 (0.7) | - | - | - | 0.88 (0.02) | 2.4 (0.9) | 7.5 (1.3) | 0.86 (0.02) | 2.9 (1.0) | 8.8 (1.3) | 0.89 (0.02) | 2.8 (0.8) | 6.2 (0.9) | 0.89 (0.01) | 3.1 (0.6) | 6.4 (0.9) |
| M21 | 0.88 (0.02) | 2.3 (0.8) | 7.3 (1.3) | 0.88 (0.02) | 2.4 (0.9) | 7.5 (1.3) | - | - | - | 0.91 (0.02) | 2.5 (1.2) | 6.9 (1.9) | 0.85 (0.02) | 2.4 (0.8) | 8.4 (1.4) | 0.89 (0.02) | 2.5 (0.9) | 7.4 (1.5) |
| M22 | 0.85 (0.02) | 3.1 (0.9) | 9.1 (1.3) | 0.86 (0.02) | 2.9 (1.0) | 8.8 (1.3) | 0.91 (0.02) | 2.5 (1.2) | 6.9 (1.9) | - | - | - | 0.83 (0.02) | 3.4 (1.1) | 10.6 (1.6) | 0.87 (0.02) | 3.3 (0.9) | 9.9 (1.5) |
| M31 | 0.89 (0.02) | 2.5 (0.7) | 6.4 (0.9) | 0.89 (0.02) | 2.8 (0.8) | 6.2 (0.9) | 0.85 (0.02) | 2.4 (0.8) | 8.4 (1.4) | 0.83 (0.02) | 3.4 (1.1) | 10.6 (1.6) | - | - | - | 0.92 (0.01) | 1.3 (0.6) | 5.1 (0.8) |
| M32 | 0.88(0.01) | 2.9 (0.6) | 6.7 (0.8) | 0.89 (0.01) | 3.1 (0.6) | 6.4 (0.9) | 0.89 (0.02) | 2.5 (0.9) | 7.4 (1.5) | 0.87 (0.02) | 3.3 (0.9) | 9.9 (1.5) | 0.92 (0.01) | 1.3 (0.6) | 5.1 (0.8) | - | - | - |
| Lung, Automatic vs. Manual | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M11 | M12 | M21 | M22 | M31 | M32 | |||||||||||||
| DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | |
| A11 | 0.92 (0.03) | 1.0 (0.6) | 2.3 (0.7) | 0.91 (0.04) | 1.1 (0.6) | 2.5 (0.8) | 0.82 (0.05) | 2.2 (1.0) | 5.0 (1.4) | 0.82 (0.04) | 2.0 (1.0) | 5.0 (1.3) | 0.77 (0.05) | 2.0 (0.9) | 5.4 (1.1) | 0.73 (0.04) | 1.6 (0.7) | 5.8 (1.0) |
| A12 | 0.90 (0.03) | 1.1 (0.6) | 2.5 (0.7) | 0.92 (0.03) | 1.0 (0.6) | 2.3 (0.7) | 0.82 (0.05) | 2.4 (1.0) | 5.1 (1.4) | 0.82 (0.04) | 2.2 (1.0) | 5.2 (1.4) | 0.75 (0.05) | 2.0 (0.9) | 5.7 (1.1) | 0.70 (0.04) | 1.8 (0.7) | 6.1 (0.9) |
| A21 | 0.86 (0.04) | 1.9 (0.9) | 4.2 (1.2) | 0.85 (0.04) | 2.0 (0.8) | 4.5 (1.1) | 0.87 (0.05) | 1.6 (0.8) | 3.1 (1.1) | 0.84 (0.04) | 1.8 (0.9) | 3.6 (1.1) | 0.76 (0.05) | 2.0 (1.0) | 5.0 (1.1) | 0.71 (0.04) | 1.6 (0.8) | 5.1 (1.1) |
| A22 | 0.83 (0.04) | 1.9 (0.8) | 4.6 (1.1) | 0.83 (0.04) | 2.0 (0.8) | 4.9 (1.1) | 0.82 (0.05) | 2.0 (1.0) | 4.1 (1.2) | 0.89 (0.05) | 1.5 (0.9) | 2.9 (1.1) | 0.72 (0.04) | 2.4 (1.0) | 5.7 (1.0) | 0.68 (0.04) | 1.9 (0.7) | 5.8 (0.9) |
| A31 | 0.77 (0.05) | 1.7 (0.7) | 5.6 (1.1) | 0.76 (0.04) | 1.8 (0.7) | 5.9 (1.0) | 0.75 (0.05) | 2.1 (1.0) | 5.2 (1.3) | 0.72 (0.04) | 2.2 (1.0) | 5.8 (1.2) | 0.87 (0.06) | 1.4 (0.8) | 2.7 (0.9) | 0.83 (0.05) | 1.5 (0.7) | 3.1 (0.9) |
| A32 | 0.74 (0.04) | 1.6 (0.7) | 6.1 (1.1) | 0.72 (0.04) | 1.7 (0.8) | 6.4 (1.0) | 0.71 (0.05) | 1.8 (0.9) | 5.5 (1.3) | 0.68 (0.04) | 2.1 (1.0) | 6.0 (1.2) | 0.81 (0.06) | 1.7 (0.9) | 3.6 (1.1) | 0.89 (0.05) | 1.1 (0.6) | 2.1 (0.7) |
| Lung, Manual vs. Manual | ||||||||||||||||||
| M11 | M12 | M21 | M22 | M31 | M32 | |||||||||||||
| DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | DC | CD | HD | |
| M11 | - | - | - | 0.90 (0.04) | 1.3 (0.7) | 2.8 (0.8) | 0.81 (0.05) | 2.4 (1.1) | 5.2 (1.5) | 0.81 (0.05) | 2.2 (1.2) | 5.3 (1.5) | 0.76 (0.05) | 2.1 (0.9) | 5.8 (1.3) | 0.72 (0.05) | 1.8 (0.9) | 6.2 (1.3) |
| M12 | 0.90 (0.04) | 1.3 (0.7) | 2.8 (0.8) | - | - | - | 0.81 (0.06) | 2.4 (1.1) | 5.5 (1.5) | 0.81 (0.05) | 2.3 (1.2) | 5.6 (1.6) | 0.75 (0.05) | 2.1 (0.9) | 6.2 (1.2) | 0.70 (0.05) | 2.0 (0.9) | 6.6 (1.2) |
| M21 | 0.81 (0.05) | 2.4 (1.1) | 5.2 (1.5) | 0.81 (0.06) | 2.4 (1.1) | 5.5 (1.5) | - | - | - | 0.81 (0.06) | 2.2 (1.2) | 4.3 (1.4) | 0.74 (0.06) | 2.4 (1.2) | 5.5 (1.5) | 0.70 (0.06) | 2.1 (1.0) | 5.7 (1.4) |
| M22 | 0.81 (0.05) | 2.2 (1.2) | 5.3 (1.5) | 0.81 (0.05) | 2.3 (1.2) | 5.6 (1.6) | 0.81 (0.06) | 2.2 (1.2) | 4.3 (1.4) | - | - | - | 0.70 (0.05) | 2.6 (1.2) | 6.1 (1.3) | 0.66 (0.05) | 2.3 (1.1) | 6.3 (1.3) |
| M31 | 0.76 (0.05) | 2.1 (0.9) | 5.8 (1.3) | 0.75 (0.05) | 2.1 (0.9) | 6.2 (1.2) | 0.74 (0.06) | 2.4 (1.2) | 5.5 (1.5) | 0.70 (0.05) | 2.6 (1.2) | 6.1 (1.3) | - | - | - | 0.80 (0.07) | 1.9 (1.0) | 3.7 (1.1) |
| M32 | 0.72(0.05) | 1.8 (0.9) | 6.2 (1.3) | 0.70 (0.05) | 2.0 (0.9) | 6.6 (1.2) | 0.70 (0.06) | 2.1 (1.0) | 5.7 (1.4) | 0.66 (0.05) | 2.3 (1.1) | 6.3 (1.3) | 0.80 (0.07) | 1.9 (1.0) | 3.7 (1.1) | - | - | - |
References
- Plathow, C.; Fink, C.; Ley, S.; Puderbach, M.; Eichinger, M.; Zuna, I.; Schmähl, A.; Kauczor, H.U. Measurement of tumor diameter-dependent mobility of lung tumors by dynamic MRI. Radiother. Oncol. 2004, 73, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Shirato, H.; Suzuki, K.; Sharp, G.C.; Fujita, K.; Onimaru, R.; Fujino, M.; Kato, N.; Osaka, Y.; Kinoshita, R.; Taguchi, H.; et al. Speed and amplitude of lung tumor motion precisely detected in fourdimensional setup and in real-time tumor-tracking radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2006, 64, 1229–1236. [Google Scholar] [CrossRef]
- Shirato, H.; Seppenwoolde, Y.; Kitamura, K.; Onimura, R.; Shimizu, S. Intrafractional tumor motion: Lung and liver. Semin. Radiat. Oncol. 2004, 14, 10–18. [Google Scholar] [CrossRef]
- Plathow, C.; Klopp, M.; Fink, C.; Sandner, A.; Hof, H.; Puderbach, M.; Herth, F.; Schmähl, A.; Kauczor, H.U. Quantitative analysis of lung and tumour mobility: Comparison of two time-resolved MRI sequences. Br. J. Radiol. 2005, 78, 836–840. [Google Scholar] [CrossRef]
- Fallone, B.G.; Murray, B.; Rathee, S.; Stanescu, T.; Steciw, S.; Vidakovic, S.; Blosser, E.; Tymofichuk, D. First MR images obtained during megavoltage photon irradiation from a prototype integrated linac-MR system. Med. Phys. 2009, 36, 2084–2088. [Google Scholar] [CrossRef] [PubMed]
- Fallone, B.G. The rotating biplanarlinac magnetic resonance imaging system. Semin. Radiat. Oncol. 2014, 24, 200–202. [Google Scholar] [CrossRef]
- Mutic, S.; Dempsey, J.F. The ViewRay system: Magnetic resonance-guided and controlled radiotherapy. Semin. Radiat. Oncol. 2014, 24, 196–199. [Google Scholar] [CrossRef] [PubMed]
- Raaymakers, B.W.; Lagendijk, J.J.W.; Overweg, J.; Kok, J.G.M.; Raaijmakers, A.J.E.; Kerkhof, E.M.; van der Put, R.W.; Meijsing, I.; Crijins, S.P.M.; Benedosso, F.; et al. Integrating a 1.5 T MRI scanner with a 6 MV accelerator: Proof of concept. Phys. Med. Biol. 2009, 54, N229–N237. [Google Scholar] [CrossRef]
- Yun, J.; Wachowicz, K.; Mackenzie, M.; Rathee, S.; Robinson, D.; Fallone, B.G. First demonstration of intrafractional tumor-tracked irradiation using 2D phantom MR images on a prototype linac-MR. Med. Phys. 2013, 40, 051718. [Google Scholar] [CrossRef]
- Tacke, M.B.; Nill, S.; Krauss, A.; Oelfke, U. Real-time tumor tracking: Automatic compensation of target motion using the Siemens 160 MLC. Med. Phys. 2010, 37, 753–761. [Google Scholar] [CrossRef]
- Cho, B.; Poulsen, P.R.; Sloutsky, A.; Sawant, A.; Keall, P.J. First demonstration of combined kV/MV image-guided real-time dynamic multileaf-collimator target tracking. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 859–867. [Google Scholar] [CrossRef] [PubMed]
- Sawant, A.; Venkat, R.; Srivastava, V.; Carlson, D.; Povzner, S.; Cattell, H.; Keall, P. Management of three-dimensional intrafraction motion through real-time DMLC tracking. Med. Phys. 2008, 35, 2050–2061. [Google Scholar] [CrossRef] [PubMed]
- Keall, P.J.; Mageras, G.S.; Balter, J.M.; Emery, R.S.; Forster, K.M.; Jiang, S.B.; Kapatoes, J.M.; Low, D.A.; Murphy, M.J.; Murray, B.R.; et al. The management of respiratory motion in radiation oncology report of AAPM Task Group 76. Med. Phys. 2006, 33, 3874–3900. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.; Park, J.C.; Gach, H.M.; Chun, J.; Mutic, S. Technical note: Realtime 3D MRI in the presence of motion for MRI-guided radiotherapy: 3D dynamic keyhole imaging with super-resolution. Med. Phys. 2019, 46, 4631–4638. [Google Scholar] [CrossRef] [PubMed]
- Bjerre, T.; Crijns, S.; af Rosenschöld, P.M.; Aznar, M.; Specht, L.; Larsen, R.; Keall, P. Three-dimensional MRI-linac intra-fraction guidance using multiple orthogonal cine-MRI planes. Phys. Med. Biol. 2013, 58, 4943–4950. [Google Scholar] [CrossRef]
- Yun, J.; Yip, E.; Gabos, Z.; Wachowicz, K.; Rathee, S.; Fallone, B.G. Neural-network based autocontouring algorithm for intrafractional lung-tumor tracking using Linac-MR. Med. Phys. 2015, 42, 2296–2310. [Google Scholar] [CrossRef]
- Bourque, A.E.; Bedwani, S.; Filion, É.; Carrier, J.F. A particle filter based autocontouring algorithm for lung tumor tracking using dynamic magnetic resonance imaging. Med. Phys. 2016, 43, 5161. [Google Scholar] [CrossRef]
- Friedrich, F.; Hörner-Rieber, J.; Renkamp, C.K.; Klüter, S.; Bachert, P.; Ladd, M.E.; Knowles, B.R. Stability of conventional and machine learning-based tumor auto-segmentation techniques using undersampled dynamic radial bSSFP acquisitions on a 0.35 T hybrid MR-linac system. Med. Phys. 2021, 48, 587–596. [Google Scholar] [CrossRef]
- Fast, M.F.; Eiben, B.; Menten, M.J.; Wetscherek, A.; Hawkes, D.J.; McClelland, J.R.; Oelfke, U. Tumor auto-contouring on 2d cine MRI for locally advanced lung cancer: A comparative study. Radiother. Oncol. 2017, 125, 485–491. [Google Scholar] [CrossRef]
- Menten, M.J.; Fast, M.F.; Wetscherek, A.; Rank, C.M.; Kachelrieß, M.; Collins, D.J.; Nill, S.; Oelfke, U. The impact of 2D cine MR imaging parameters on automated tumor and organ localization for MR-guided real-time adaptive radiotherapy. Phys. Med. Biol. 2018, 63, 235005. [Google Scholar] [CrossRef]
- Cerviño, L.I.; Du, J.; Jiang, S.B. MRI-guided tumor tracking in lung cancer radiotherapy. Phys. Med. Biol. 2011, 56, 3773–3785. [Google Scholar] [CrossRef]
- Rueckert, D.; Sonoda, L.I.; Hayes, C.; Hill, D.L.; Leach, M.O.; Hawkes, D.J. Nonrigid registration using free-form deformations: Application to breast MR images. IEEE Trans. Med. Imaging 1999, 18, 712–721. [Google Scholar] [CrossRef] [PubMed]
- Thirion, J.P. Image matching as a diffusion process: An analogy with Maxwell’s demons. Med. Image Anal. 1998, 2, 243–260. [Google Scholar] [CrossRef] [PubMed]
- Vercauteren, T.; Pennec, X.; Perchant, A.; Ayache, N. Diffeomorphic demons: Efficient non-parametric image registration. Neuroimage 2009, 45, S61–S72. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18. Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar] [CrossRef]
- Wong, J.; Fong, A.; McVicar, N.; Smith, S.; Giambattista, J.; Wells, D.; Kolbeck, C.; Giambattista, J.; Gondara, L.; Alexander, A. Comparing deep learning-based auto-segmentation of organs at risk and clinical target volumes to expert inter-observer variability in radiotherapy planning. Radiother. Oncol. 2020, 144, 152–158. [Google Scholar] [CrossRef]
- Urago, Y.; Okamoto, H.; Kaneda, T.; Murakami, N.; Kashihara, T.; Takemori, M.; Nakayama, H.; Iijima, K.; Chiba, T.; Kuwahara, J. Evaluation of auto-segmentation accuracy of cloud-based artificial intelligence and atlas-based models. Radiat. Oncol. 2021, 16, 175. [Google Scholar] [CrossRef]
- Molière, S.; Hamzaoui, D.; Granger, B.; Montagne, S.; Allera, A.; Ezziane, M.; Luzurier, A.; Quint, R.; Kalai, M.; Ayache, N.; et al. Reference standard for the evaluation of automatic segmentation algorithms: Quantification of inter observer variability of manual delineation of prostate contour on MRI. Diagn. Interv. Imaging 2024, 105, 65–73. [Google Scholar] [CrossRef]
- Cunha, F.F.; Blüml, V.; Zopf, L.M.; Walter, A.; Wagner, M.; Weninger, W.J.; Thomaz, L.A.; Tavora, L.M.N.; da Silva Cruz, L.A.; Faria, S.M.M. Lossy Image Compression in a Preclinical Multimodal Imaging Study. J. Digit. Imaging 2023, 36, 1826–1850. [Google Scholar] [CrossRef]
- Lim, V.T.; Gacasan, A.C.; Tuan, J.K.L.; Tan, T.W.K.; Li, Y.; Nei, W.L.; Looi, W.S.; Lin, X.; Tan, H.Q.; Chua, E.C.P.; et al. Evaluation of inter- and intra-observer variations in prostate gland delineation using CT-alone versus CT/TPUS. Rep. Pract. Oncol. Radiother. 2022, 27, 97–103. [Google Scholar] [CrossRef]
- Palacios, M.A.; Gerganov, G.; Cobussen, P.; Tetar, S.U.; Finazzi, T.; Slotman, B.J.; Senan, S.; Haasbeek, C.J.A.; Kawrakow, I. Accuracy of deformable image registration-based intra-fraction motion management in Magnetic Resonance-guided radiotherapy. Phys. Imaging Radiat. Oncol. 2023, 26, 100437. [Google Scholar] [CrossRef]
- Yip, E.; Yun, J.; Gabos, Z.; Baker, S.; Yee, D.; Wachowicz, K.; Rathee, S.; Fallone, B.G. Evaluating performance of a user-trained MR lung tumor autocontouring algorithm in the context of intra- and interobserver variations. Med. Phys. 2018, 45, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Eccles, C.L.; Patel, R.; Simeonov, A.K.; Lockwood, G.; Haider, M.; Dawson, L.A. Comparison of liver tumor motion with and without abdominal compression using cine-magnetic resonance imaging. Int. J. Radiat. Oncol. Biol. Phys. 2011, 79, 602–608. [Google Scholar] [CrossRef]
- Tong, X.; Chen, X.; Li, J.; Xu, Q.; Lin, M.H.; Chen, L.; Price, R.A.; Ma, C.M. Intrafractional prostate motion during external beam radiotherapy monitored by a real-time target localization system. J. Appl. Clin. Med. Phys. 2015, 16, 5013. [Google Scholar] [CrossRef]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the quantitative imaging network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Han, G.; Wachowicz, K.; Usmani, N.; Yee, D.; Wong, J.; Elangovan, A.; Yun, J.; Fallone, B.G. Patient-specific hyperparameter optimization of a deep learning-based tumor autocontouring algorithm on 2D liver, prostate, and lung cine MR images: A pilot study. Algorithms 2025, 18, 233. [Google Scholar] [CrossRef]
- Hansen, N.; Ostermeier, A. Completely Derandomized Self-Adaptation in Evolution Strategies. Evol. Comput. 2001, 9, 159–195. [Google Scholar] [CrossRef]
- Yun, J.; Yip, E.; Gabos, Z.; Usmani, N.; Yee, D.; Wachowicz, K.; Fallone, B.G. An AI-based tumor autocontouring algorithm for non-invasive intra-fractional tumor-tracked radiotherapy (nifteRT) on linac-MR. Med. Phys. 2020, 47, e576. [Google Scholar]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for evaluating 3D medical image segmentation: Analysis, selection, and tool. BMC Med. Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Huttenlocher, D.P.; Klanderman, G.A.; Rucklidge, W.J. Comparing images using the Hausdorff distance. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 850–863. [Google Scholar] [CrossRef]
- Louie, A.V.; Rodrigues, G.; Olsthoorn, J.; Palma, D.; Yu, E.; Yaremko, B.; Ahmad, B.; Aivas, I.; Gaede, S. Inter-observer and intraobserver reliability for lung cancer target volume delineation in the 4DCT era. Radiother. Oncol. 2010, 95, 166–171. [Google Scholar] [CrossRef]
- Hamilton, C.S.; Denham, J.W.; Joseph, D.J.; Lamb, D.S.; Spry, N.A.; Gray, A.J.; Atkinson, C.H.; Wynne, C.J.; Abdelaal, A.; Bydder, P.V.; et al. Treatment and planning decisions in non-small cell carcinoma of the lung: An Australasian patterns of practice study. Clin. Oncol. (R. Coll. Radiol.) 1992, 4, 141–147. [Google Scholar] [CrossRef]
- Senan, S.; de Koste, J.; Samson, M.; Tankink, H.; Jansen, P.; Nowak, P.J.; Krol, A.D.; Schmitz, P.; Lagerwaard, F.J. Evaluation of a target contouring protocol for 3D conformal radiotherapy in non-small cell lung cancer. Radiother. Oncol. 1999, 53, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Covert, E.C.; Fitzpatrick, K.; Mikell, J.; Kaza, R.K.; Millet, J.D.; Barkmeier, D.; Gemmete, J.; Christensen, J.; Schipper, M.J.; Dewaraja, Y.K. Intra- and inter-operator variability in MRI-based manual segmentation of HCC lesions and its impact on dosimetry. EJNMMI Phys. 2022, 9, 90. [Google Scholar] [CrossRef]
- Klüter, S. Technical design and concept of a 0.35 T MR-Linac. Clin. Transl. Radiat. Oncol. 2019, 18, 98–101. [Google Scholar] [CrossRef]
- Chun, J.; Zhang, H.; Gach, H.M.; Olberg, S.; Mazur, T.; Green, O.; Kim, T.; Kim, H.; Kim, J.S.; Mutic, S.; et al. MRI super-resolution reconstruction for MRI-guided adaptive radiotherapy using cascaded deep learning: In the presence of limited training data and unknown translation model. Med. Phys. 2019, 46, 4148–4164. [Google Scholar] [CrossRef]
- Grover, J.; Liu, P.; Dong, B.; Shan, S.; Whelan, B.; Keall, P.; Waddington, D.E.J. Super-resolution neural networks improve the spatiotemporal resolution of adaptive MRI-guided radiation therapy. Commun. Med. 2024, 4, 64. [Google Scholar] [CrossRef]

| Site | Patient | Gender | Age | Tumor Area (cm2) | Overall Stage | TNM Stage | Primary Cancer |
|---|---|---|---|---|---|---|---|
| Liver | 1 | F | 65 | 36.2 | III | TXNXM1 | Rectal adenocarcinoma |
| 2 | M | 56 | 0.9 | II | N/A | HCC | |
| 3 | M | 70 | 24.2 | IV | pT4pN2MX | Sigmoid colon adenocarcinoma | |
| 4 | M | 57 | 2.8 | I | N/A | HCC | |
| 5 | M | 64 | 2.0 | II | N/A | HCC | |
| 6 | M | 63 | 3.7 | IVB | T2N1M1 | Nasopharyngeal carcinoma | |
| 7 | M | 65 | 3.1 | IVA | T3N2M1 | Colorectal carcinoma | |
| 8 | M | 59 | 2.4 | IV | T3N0M1 | Rectal adenocarcinoma | |
| 9 | M | 68 | 1.5 | IIB | TXNXM1 | Rectal adenocarcinoma | |
| 10 | F | 82 | 6.0 | IV | T3N2M1 | Colorectal cancer | |
| Prostate | 11 | M | 69 | 25.0 | IIA | T1c | Prostatic adenocarcinoma |
| 12 | M | 69 | 24.4 | IIA | T1c | Prostatic adenocarcinoma | |
| 13 | M | 69 | 8.4 | IIIC | T3aN0M0 | Prostatic adenocarcinoma | |
| 14 | M | 69 | 8.4 | IIIC | T3aN0M0 | Prostatic adenocarcinoma | |
| 15 | M | 73 | 14.8 | IIB | N/A | Prostatic adenocarcinoma | |
| 16 | M | 73 | 12.8 | IIB | N/A | Prostatic adenocarcinoma | |
| 17 | M | 69 | 33.1 | IIA | T1c | Prostatic adenocarcinoma | |
| 18 | M | 75 | 26.4 | IIB | T1c | Prostatic adenocarcinoma | |
| 19 | M | 68 | 15.0 | IIB | T1c | Prostatic adenocarcinoma | |
| 20 | M | 77 | 18.9 | IIIA | T1c | Prostatic adenocarcinoma | |
| Lung | 21 | M | 81 | 1.3 | I | T1N0M0 | NSCLC |
| 22 | M | 79 | 3.8 | I | pT2pN1pMX | NSCLC | |
| 23 | F | 73 | 6.4 | II | T2N0M0 | NSCLC | |
| 24 | M | 72 | 4.8 | IVA | T1NXM1a | NSCLC | |
| 25 | F | 78 | 7.4 | I | T1N0M0 | Lung cancer unspecified | |
| 26 | M | 65 | 1.4 | IA | T1N0M0 | NSCLC | |
| 27 | M | 65 | 5.1 | I | cT1cN0M0 | NSCLC | |
| 28 | M | 70 | 1.7 | IB | N0M0 | SCLC | |
| 29 | M | 75 | 3.0 | IIA | T2bN0M0 | NSCLC | |
| 30 | M | 65 | 3.8 | IA | T1bN0M0 | NSCLC | |
| Mean (SD) | 10.3 (10.4) |
| Same Expert, Same Session (SESS) | Same Expert, Different Session (SEDS) | Different Experts (DE) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Median | SD | Worst | Mean | Median | SD | Worst | Mean | Median | SD | Worst | |||
| Liver | Automatic vs. Manual | DC | 0.90 | 0.89 | 0.05 | 0.87 | 0.79 | 0.79 | 0.04 | 0.73 | 0.70 | 0.68 | 0.04 | 0.60 |
| CD (mm) | 1.4 | 1.4 | 0.8 | 1.8 | 2.3 | 2.4 | 0.9 | 2.7 | 3.3 | 3.2 | 1.0 | 4.0 | ||
| HD (mm) | 3.0 | 2.8 | 0.9 | 3.7 | 5.3 | 5.6 | 1.1 | 5.7 | 7.7 | 7.9 | 1.3 | 9.3 | ||
| Manual vs. Manual | DC | - | - | - | - | 0.78 | 0.79 | 0.05 | 0.72 | 0.69 | 0.67 | 0.05 | 0.60 | |
| CD (mm) | - | - | - | - | 2.7 | 2.6 | 1.1 | 2.9 | 3.5 | 3.5 | 1.2 | 4.1 | ||
| HD (mm) | - | - | - | - | 5.7 | 5.8 | 1.4 | 6.1 | 7.8 | 7.9 | 1.6 | 9.3 | ||
| Prostate | Automatic vs. Manual | DC | 0.95 | 0.95 | 0.01 | 0.94 | 0.93 | 0.93 | 0.01 | 0.92 | 0.87 | 0.88 | 0.02 | 0.82 |
| CD (mm) | 1.2 | 1.1 | 0.6 | 2.0 | 1.7 | 1.7 | 0.7 | 2.6 | 2.8 | 2.8 | 0.7 | 3.4 | ||
| HD (mm) | 3.6 | 3.3 | 0.9 | 4.9 | 5.1 | 5.3 | 0.9 | 6.3 | 7.9 | 7.6 | 1.0 | 10.5 | ||
| Manual vs. Manual | DC | - | - | - | - | 0.92 | 0.92 | 0.02 | 0.91 | 0.87 | 0.87 | 0.02 | 0.83 | |
| CD (mm) | - | - | - | - | 1.7 | 1.4 | 0.8 | 2.5 | 2.8 | 2.9 | 0.8 | 3.4 | ||
| HD (mm) | - | - | - | - | 5.3 | 5.1 | 1.1 | 6.9 | 7.9 | 7.5 | 1.2 | 10.6 | ||
| Lung | Automatic vs. Manual | DC | 0.89 | 0.89 | 0.05 | 0.87 | 0.85 | 0.84 | 0.05 | 0.81 | 0.76 | 0.76 | 0.04 | 0.68 |
| CD (mm) | 1.3 | 1.3 | 0.7 | 1.6 | 1.5 | 1.6 | 0.8 | 2.0 | 2.0 | 2.0 | 0.9 | 2.4 | ||
| HD (mm) | 2.6 | 2.5 | 0.9 | 3.1 | 3.2 | 3.4 | 1.0 | 4.1 | 5.4 | 5.5 | 1.1 | 6.4 | ||
| Manual vs. Manual | DC | - | - | - | - | 0.84 | 0.81 | 0.06 | 0.80 | 0.75 | 0.75 | 0.05 | 0.66 | |
| CD (mm) | - | - | - | - | 1.8 | 1.9 | 1.0 | 2.2 | 2.2 | 2.3 | 1.1 | 2.6 | ||
| HD (mm) | - | - | - | - | 3.6 | 3.7 | 1.1 | 4.3 | 5.8 | 5.8 | 1.4 | 6.6 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, G.; Elangovan, A.; Wong, J.; Waheed, A.; Wachowicz, K.; Usmani, N.; Gabos, Z.; Yun, J.; Fallone, B.G. Quantifying Intra- and Inter-Observer Variabilities in Manual Contours for Radiotherapy: Evaluation of an MR Tumor Autocontouring Algorithm for Liver, Prostate, and Lung Cancer Patients. Algorithms 2025, 18, 290. https://doi.org/10.3390/a18050290
Han G, Elangovan A, Wong J, Waheed A, Wachowicz K, Usmani N, Gabos Z, Yun J, Fallone BG. Quantifying Intra- and Inter-Observer Variabilities in Manual Contours for Radiotherapy: Evaluation of an MR Tumor Autocontouring Algorithm for Liver, Prostate, and Lung Cancer Patients. Algorithms. 2025; 18(5):290. https://doi.org/10.3390/a18050290
Chicago/Turabian StyleHan, Gawon, Arun Elangovan, Jordan Wong, Asmara Waheed, Keith Wachowicz, Nawaid Usmani, Zsolt Gabos, Jihyun Yun, and B. Gino Fallone. 2025. "Quantifying Intra- and Inter-Observer Variabilities in Manual Contours for Radiotherapy: Evaluation of an MR Tumor Autocontouring Algorithm for Liver, Prostate, and Lung Cancer Patients" Algorithms 18, no. 5: 290. https://doi.org/10.3390/a18050290
APA StyleHan, G., Elangovan, A., Wong, J., Waheed, A., Wachowicz, K., Usmani, N., Gabos, Z., Yun, J., & Fallone, B. G. (2025). Quantifying Intra- and Inter-Observer Variabilities in Manual Contours for Radiotherapy: Evaluation of an MR Tumor Autocontouring Algorithm for Liver, Prostate, and Lung Cancer Patients. Algorithms, 18(5), 290. https://doi.org/10.3390/a18050290






