A Plant Disease Classification Algorithm Based on Attention MobileNet V2
Abstract
:1. Introduction
2. Materials and Methods
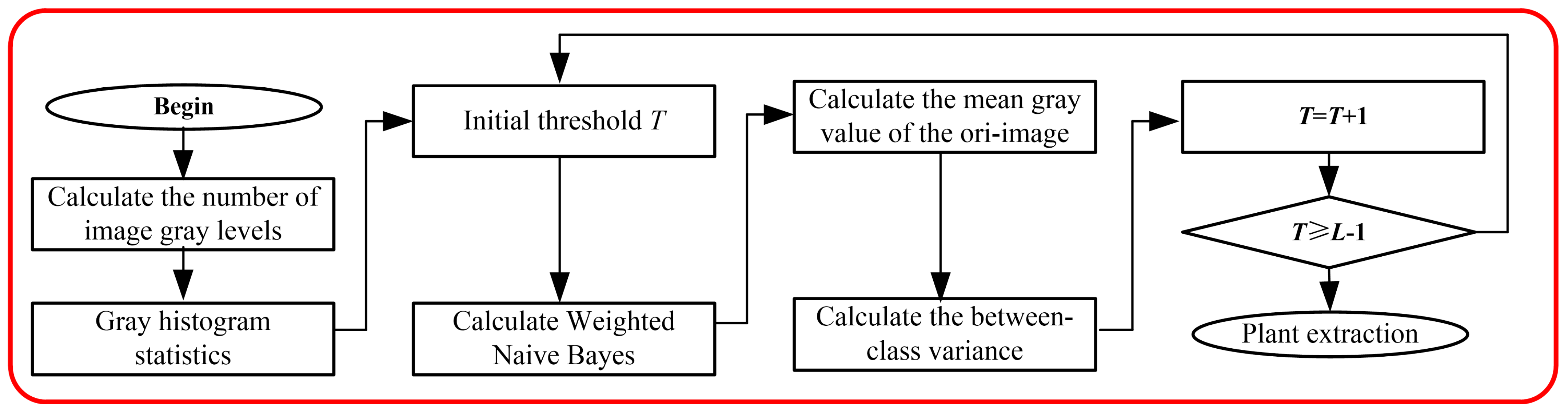
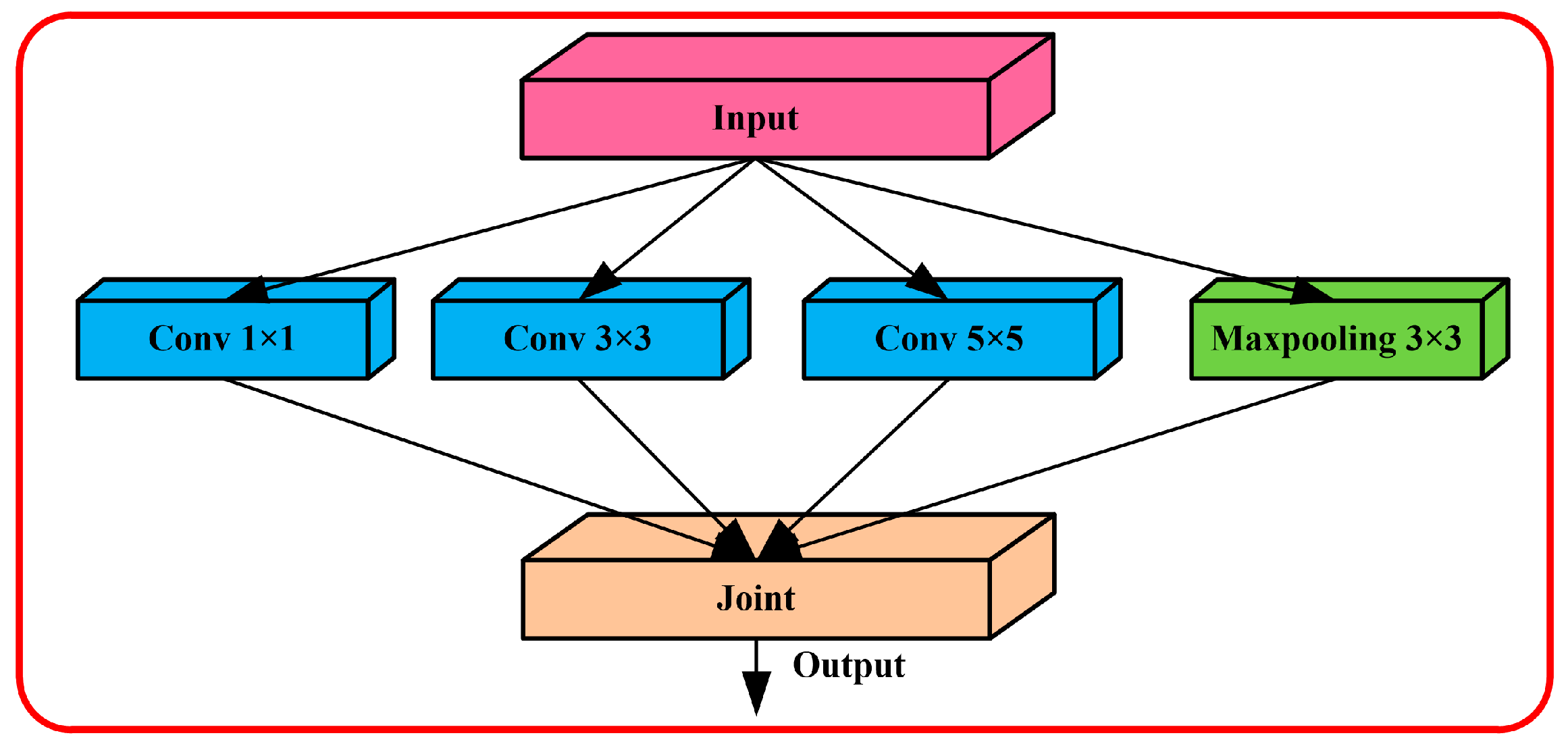
2.1. Multilevel Feature Extraction Algorithm
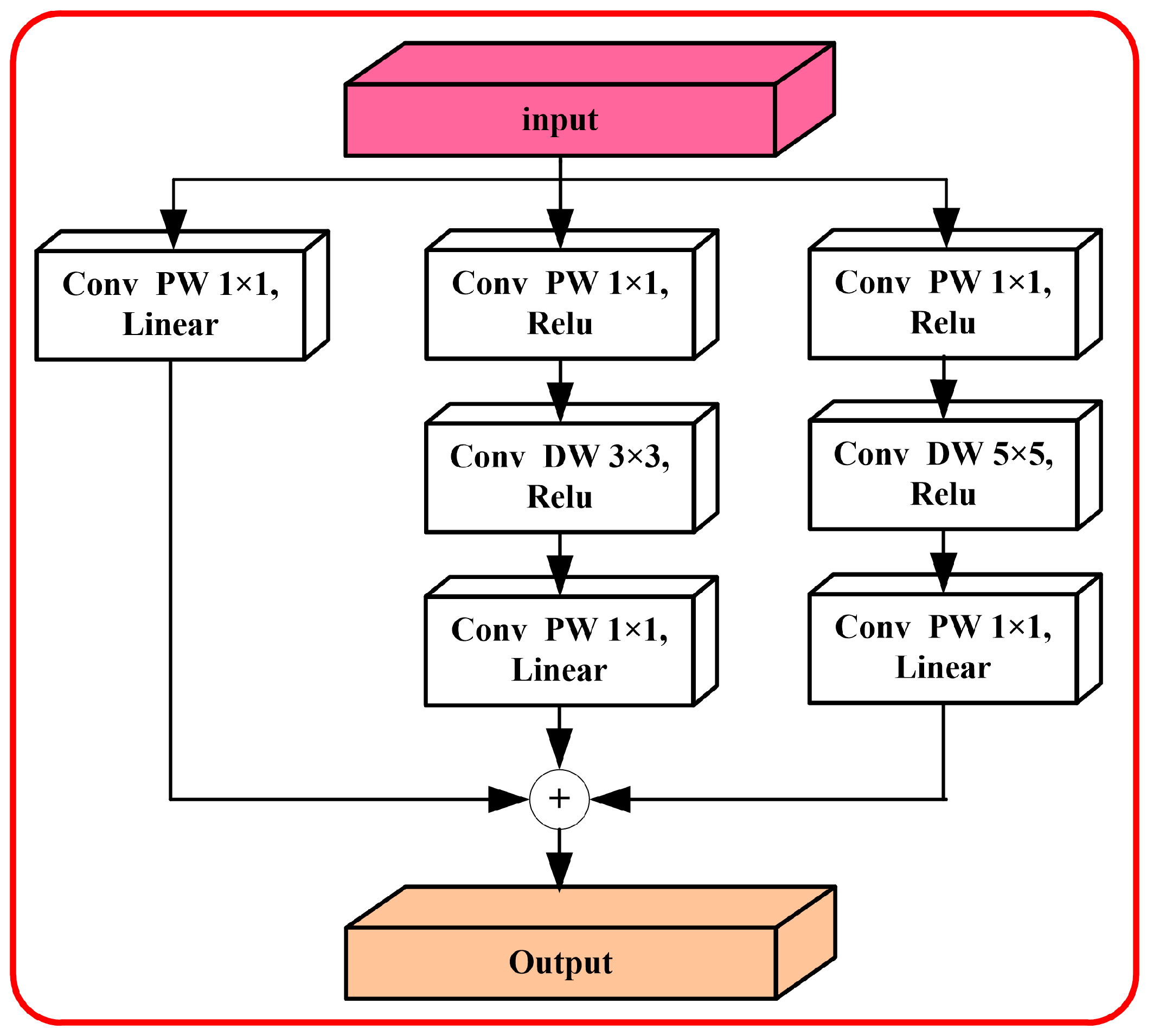
2.2. MobileNet Algorithm Based on Dual Attention
3. Experimental Results and Analysis
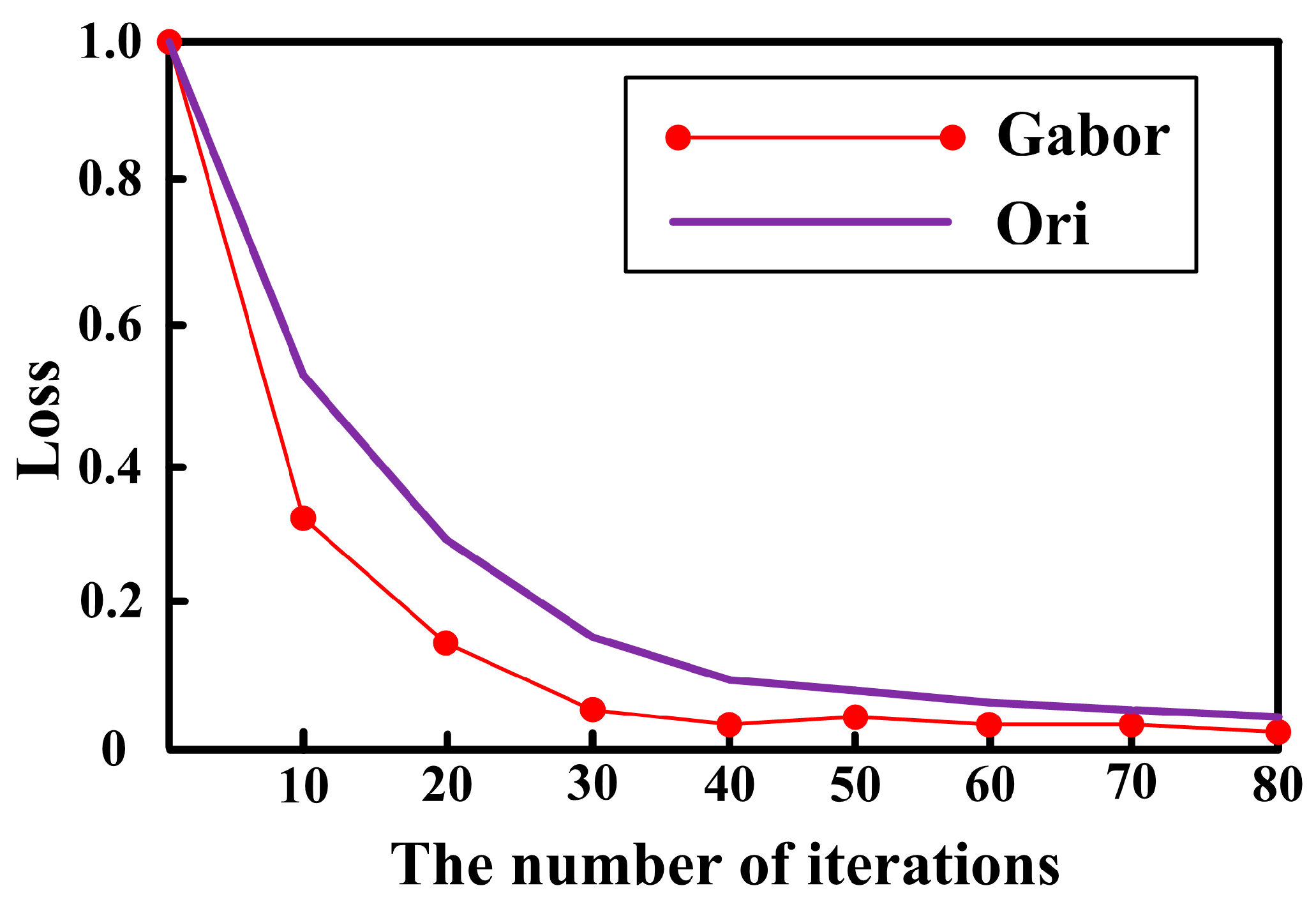
3.1. Feature Extraction Algorithm
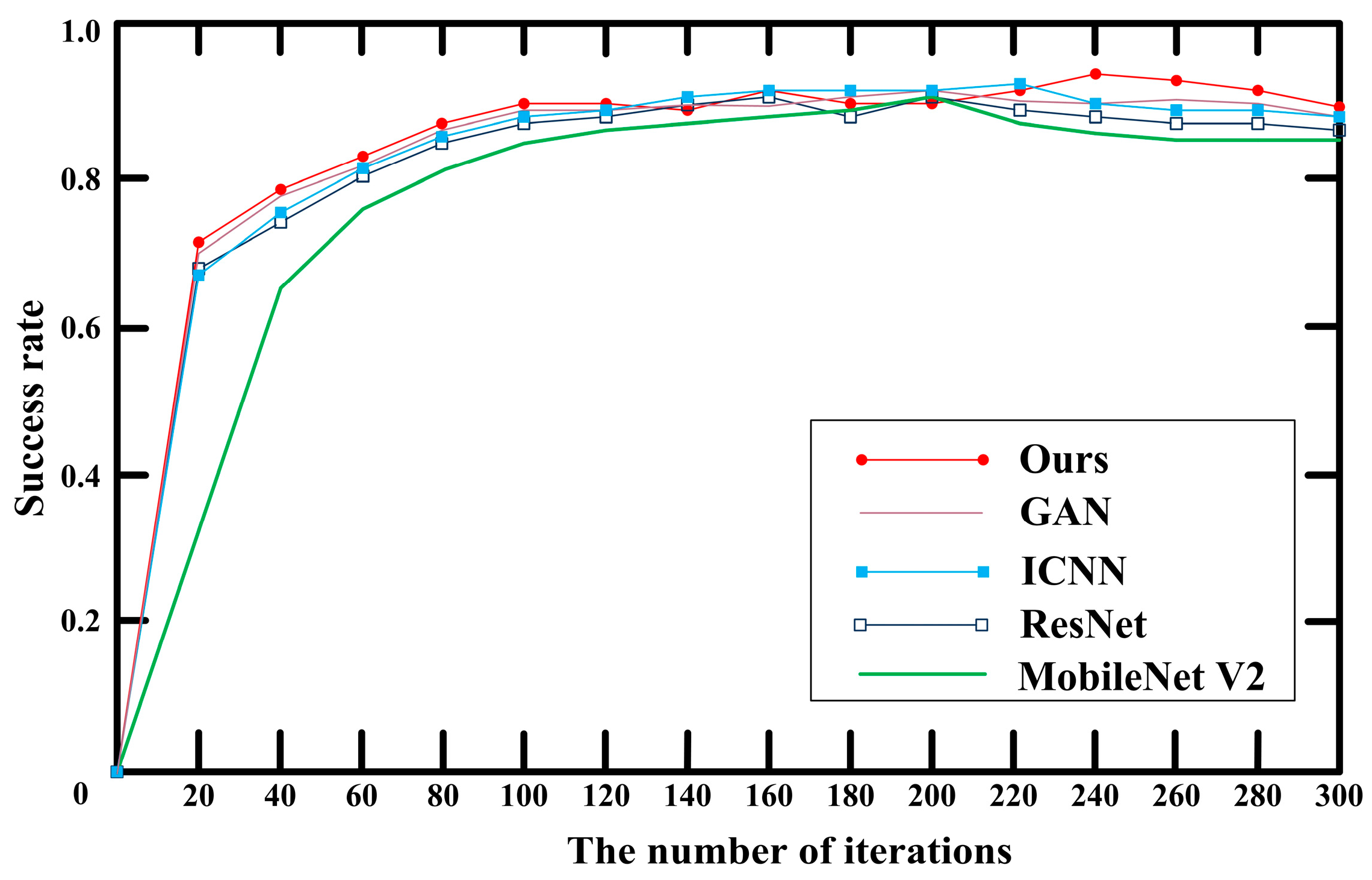
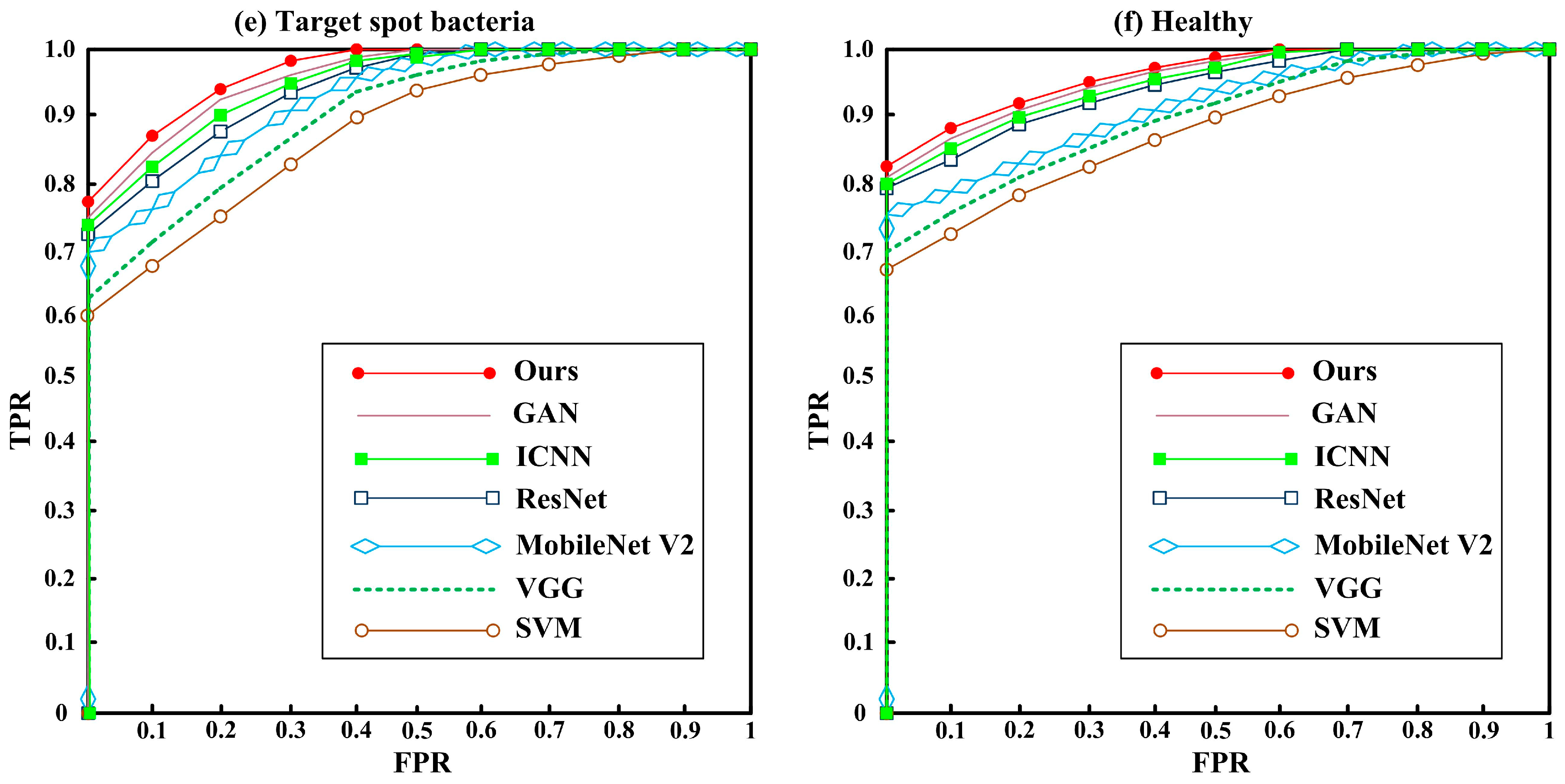
3.2. Comparison of Classification Algorithms
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Al-Hiary, H.; Bani-Ahmad, S.; Reyalat, M.; Braik, M.; Alrahamneh, Z. Fast and accurate detection and classification of plant diseases. Int. J. Comput. Appl. 2011, 17, 31–38. [Google Scholar] [CrossRef]
- Kulkarni, A.H.; Patil, A. Applying image processing technique to detect plant diseases. Int. J. Mod. Eng. Res. 2012, 2, 3661–3664. [Google Scholar]
- Arivazhagan, S.; Shebiah, R.N.; Ananthi, S.; Varthini, S.V. Detection of unhealthy region of plant leaves and classification of plant leaf diseases using texture features. Agric. Eng. Int. CIGR J. 2013, 15, 211–217. [Google Scholar]
- Hossain, E.; Hossain, M.F.; Rahaman, M.A. A color and texture based approach for the detection and classification of plant leaf disease using KNN classifier. In Proceedings of the 2019 International Conference on Electrical, Computer and Communication Engineering (ECCE), Cox’s Bazar, Bangladesh, 7–9 February 2019; pp. 1–6. [Google Scholar]
- Singh, V.; Misra, A.K. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef]
- Kaur, R.; Singla, S. Classification of plant leaf diseases using gradient and texture feature. In Proceedings of the International Conference on Advances in Information Communication Technology & Computing, Thai Nguyen, Vietnam, 12–13 December 2016; pp. 1–7. [Google Scholar]
- Nanehkaran, Y.A.; Zhang, D.; Chen, J.; Tian, Y.; Al-Nabhan, N. Recognition of plant leaf diseases based on computer vision. J. Ambient. Intell. Humaniz. Comput. 2020, 11, 1–18. [Google Scholar] [CrossRef]
- Pujari, D.; Yakkundimath, R.; Byadgi, A.S. SVM and ANN based classification of plant diseases using feature reduction technique. IJIMAI 2016, 3, 6–14. [Google Scholar] [CrossRef]
- Brahimi, M.; Arsenovic, M.; Laraba, S.; Sladojevic, S.; Boukhalfa, K.; Moussaoui, A. Deep learning for plant diseases: Detection and saliency map visualization. In Human and Machine Learning; Springer: Cham, Switzerland, 2018; pp. 93–117. [Google Scholar]
- Mahmoud MA, B.; Guo, P.; Wang, K. Pseudoinverse learning autoencoder with DCGAN for plant diseases classification. Multimed. Tools Appl. 2020, 79, 26245–26263. [Google Scholar] [CrossRef]
- Sandesh Kumar, C.; Sharma, V.K.; Yadav, A.K.; Singh, A. Perception of plant diseases in color images through adaboost. In Innovations in Computational Intelligence and Computer Vision; Springer: Singapore, 2021; pp. 506–511. [Google Scholar]
- Hang, J.; Zhang, D.; Chen, P.; Zhang, J.; Wang, B. Classification of plant leaf diseases based on improved convolutional neural network. Sensors 2019, 19, 4161. [Google Scholar] [CrossRef]
- Atila, Ü.; Uçar, M.; Akyol, K.; Uçar, E. Plant leaf disease classification using EfficientNet deep learning model. Ecol. Inform. 2021, 61, 101182. [Google Scholar] [CrossRef]
- Sardogan, M.; Tuncer, A.; Ozen, Y. Plant leaf disease detection and classification based on CNN with LVQ algorithm. In Proceedings of the 2018 3rd International Conference on Computer Science and Engineering (UBMK), Sarajevo, Bosnia and Herzegovina, 20–23 September 2018; pp. 382–385. [Google Scholar]
- Deepa, N.R.; Nagarajan, N. Kuan noise filter with Hough transformation based reweighted linear program boost classification for plant leaf disease detection. J. Ambient. Intell. Humaniz. Comput. 2021, 12, 5979–5992. [Google Scholar] [CrossRef]
- Altan, G. Performance evaluation of capsule networks for classification of plant leaf diseases. Int. J. Appl. Math. Electron. Comput. 2020, 8, 57–63. [Google Scholar] [CrossRef]
- Pal, A.; Kumar, V. AgriDet: Plant leaf disease severity classification using agriculture detection framework. Eng. Appl. Artif. Intell. 2023, 119, 105754. [Google Scholar] [CrossRef]
- Liang, Q.; Xiang, S.; Hu, Y.; Coppola, G.; Zhang, D.; Sun, W. PD2SE-Net: Computer-assisted plant disease diagnosis and severity estimation network. Comput. Electron. Agric. 2019, 157, 518–529. [Google Scholar] [CrossRef]
- Yu, H.; Liu, J.; Chen, C.; Heidari, A.A.; Zhang, Q.; Chen, H.; Mafarja, M.; Turabieh, H. Corn leaf diseases diagnosis based on K-means clustering and deep learning. IEEE Access 2021, 9, 143824–143835. [Google Scholar] [CrossRef]
- Padol, P.B.; Yadav, A.A. SVM classifier based grape leaf disease detection. In Proceedings of the 2016 Conference on Advances in Signal Processing (CASP), Pune, India, 9–11 June 2016; pp. 175–179. [Google Scholar]
- Rani, F.P.; Kumar, S.N.; Fred, A.L.; Dyson, C.; Suresh, V.; Jeba, P.S. K-means clustering and SVM for plant leaf disease detection and classification. In Proceedings of the 2019 International Conference on Recent Advances in Energy-Efficient Computing and Communication (ICRAECC), Nagercoil, India, 7–20 March 2019; pp. 1–4. [Google Scholar]
- Trivedi, V.K.; Shukla, P.K.; Pandey, A. Automatic segmentation of plant leaves disease using min-max hue histogram and k-mean clustering. Multimed. Tools Appl. 2022, 81, 20201–20228. [Google Scholar] [CrossRef]
- Faithpraise, F.; Birch, P.; Young, R.; Obu, J.; Faithpraise, B.; Chatwin, C. Automatic plant pest detection and recognition using k-means clustering algorithm and correspondence filters. Int. J. Adv. Biotechnol. Res. 2013, 4, 189–199. [Google Scholar]
- Tamilselvi, P.; Kumar, K.A. Unsupervised machine learning for clustering the infected leaves based on the leaf-colours. In Proceedings of the 2017 Third International Conference on Science Technology Engineering & Management (ICONSTEM), Chennai, India, 23–24 March 2017; pp. 106–110. [Google Scholar]
- Hasan, R.I.; Yusuf, S.M.; Mohd Rahim, M.S.; Alzubaidi, L. Automatic clustering and classification of coffee leaf diseases based on an extended kernel density estimation approach. Plants 2023, 12, 1603. [Google Scholar] [CrossRef]
- Yadhav, S.Y.; Senthilkumar, T.; Jayanthy, S.; Kovilpillai, J.J.A. Plant disease detection and classification using cnn model with optimized activation function. In Proceedings of the 2020 International Conference on Electronics and Sustainable Communication Systems (ICESC), Coimbatore, India, 2–4 July 2020; pp. 564–569. [Google Scholar]
- Bhimavarapu, U. Prediction and classification of rice leaves using the improved PSO clustering and improved CNN. Multimed. Tools Appl. 2023, 82, 21701–21714. [Google Scholar] [CrossRef]
- Hatuwal, B.K.; Shakya, A.; Joshi, B. Plant leaf disease recognition using random Forest, KNN, SVM and CNN. Polibits 2020, 62, 13–19. [Google Scholar]
- Pareek, P.K.; Ramya, I.M.; Jagadeesh, B.N.; LeenaShruthi, H.M. Clustering based segmentation with 1D-CNN model for grape fruit disease detection. In Proceedings of the 2023 IEEE International Conference on Integrated Circuits and Communication Systems (ICICACS), Raichur, India, 24–25 February 2023; pp. 1–7. [Google Scholar]
- Mukti, I.Z.; Biswas, D. Transfer learning based plant diseases detection using ResNet50. In Proceedings of the 2019 4th International Conference on Electrical Information and Communication Technology (EICT), Khulna, Bangladesh, 20–22 December 2019; pp. 1–6. [Google Scholar]
- Li, M.; Cheng, S.; Cui, J.; Li, C.; Li, Z.; Zhou, C.; Lv, C. High-performance plant pest and disease detection based on model ensemble with inception module and cluster algorithm. Plants 2023, 12, 200. [Google Scholar] [CrossRef]
- Muammer, T.; Hanbay, D. Plant disease and pest detection using deep learning-based features. Turk. J. Electr. Eng. Comput. Sci. 2019, 27, 1636–1651. [Google Scholar]
- Ramesh, S.; Hebbar, R.; Niveditha, M.; Pooja, R.; Shashank, N.; Vinod, P.V. Plant disease detection using machine learning. In Proceedings of the 2018 International Conference on Design Innovations for 3Cs Compute Communicate Control (ICDI3C), Bangalore, India, 25–28 April 2018; pp. 41–45. [Google Scholar]
- Hoang, N.D. Detection of surface crack in building structures using image processing technique with an improved Otsu method for image thresholding. Adv. Civ. Eng. 2018, 2018, 3924120. [Google Scholar] [CrossRef]
- Vembandasamy, K.; Sasipriya, R.; Deepa, E. Heart diseases detection using Naive Bayes algorithm. Int. J. Innov. Sci. Eng. Technol. 2015, 2, 441–444. [Google Scholar]
- Yuan, Y.; Wang, L.N.; Zhong, G.; Gao, W.; Jiao, W.; Dong, J.; Shen, B.; Xia, D.; Xiang, W. Adaptive Gabor convolutional networks. Pattern Recognit. 2022, 124, 108495. [Google Scholar] [CrossRef]
- Srinivasu, P.N.; SivaSai, J.G.; Ijaz, M.F.; Bhoi, A.K.; Kim, W.; Kang, J.J. Classification of skin disease using deep learning neural networks with MobileNet V2 and LSTM. Sensors 2021, 21, 2852. [Google Scholar] [CrossRef]
- Dang, L.; Pang, P.; Lee, J. Depth-wise separable convolution neural network with residual connection for hyperspectral image classification. Remote Sens. 2020, 12, 3408. [Google Scholar] [CrossRef]
- Fu, H.; Song, G.; Wang, Y. Improved YOLOv4 marine target detection combined with CBAM. Symmetry 2021, 13, 623. [Google Scholar] [CrossRef]
- Qiu, S.; Jin, Y.; Feng, S.; Zhou, T.; Li, Y. Dwarfism computer-aided diagnosis algorithm based on multimodal pyradiomics. Inf. Fusion 2022, 80, 137–145. [Google Scholar] [CrossRef]
- Selvanambi, R.; Natarajan, J.; Karuppiah, M.; Islam, S.H.; Hassan, M.M.; Fortino, G. Lung cancer prediction using higher-order recurrent neural network based on glowworm swarm optimization. Neural Comput. Appl. 2020, 32, 4373–4386. [Google Scholar] [CrossRef]
- Kaya, Y.; Gürsoy, E. A novel multi-head CNN design to identify plant diseases using the fusion of RGB images. Ecol. Inform. 2023, 75, 101998. [Google Scholar] [CrossRef]
- Lamba, S.; Saini, P.; Kaur, J.; Kukreja, V. Optimized classification model for plant diseases using generative adversarial networks. Innov. Syst. Softw. Eng. 2023, 19, 103–115. [Google Scholar] [CrossRef]





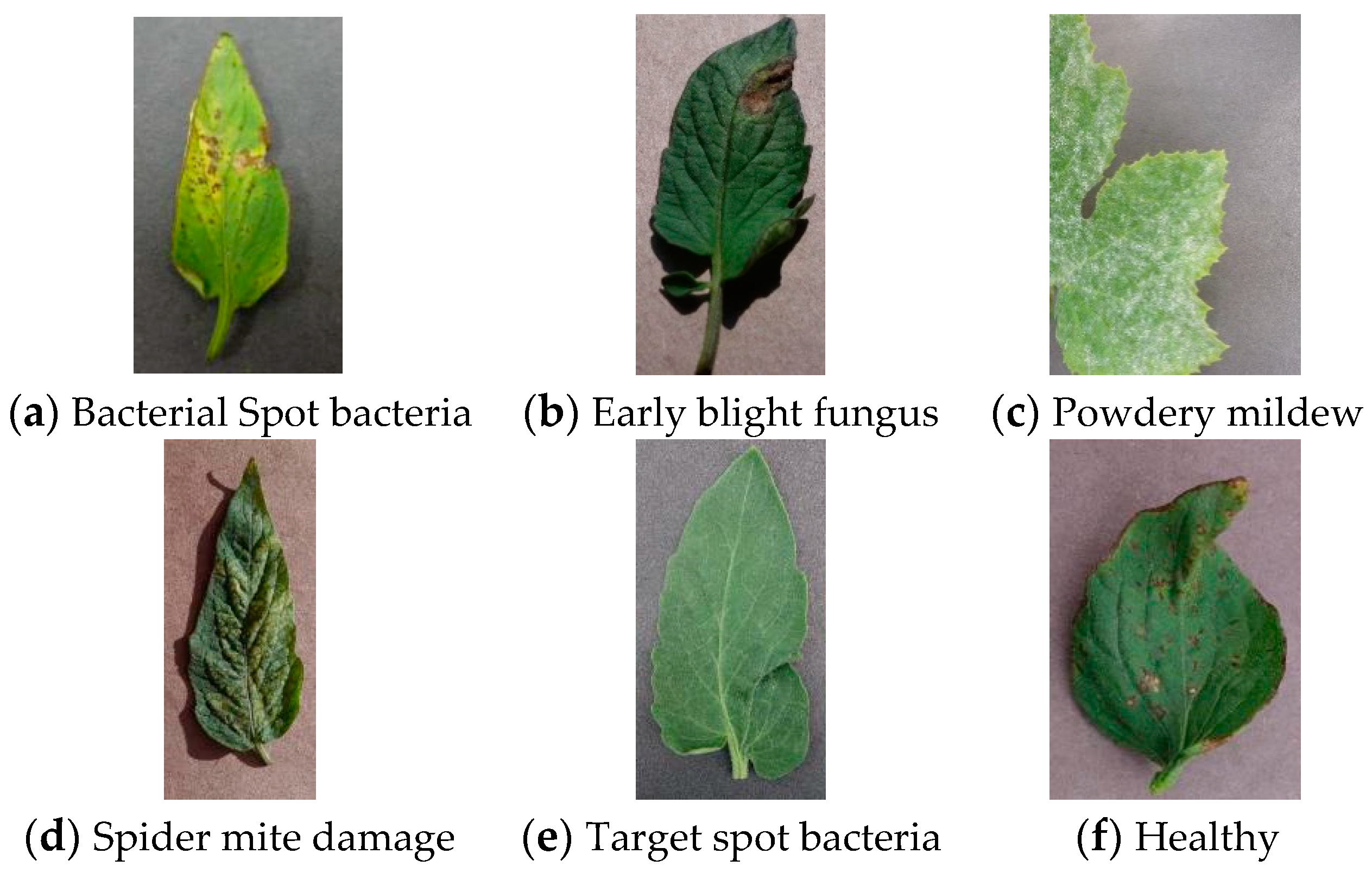


| Apple | Healthy | Sreawberry | Healthy | ||
| Scab | General | Scorch | General | ||
| Serious | Serious | ||||
| Cedar Rust | General | Tomato | Bacterial Spot Bacteria | General | |
| Serious | Serious | ||||
| Cherry | Healthy | Early Blight Fungus | General | ||
| Powdery Mildew | General | Serious | |||
| Serious | Late Blight Water Mold | General | |||
| Corn | Healthy | Serious | |||
| Cercospora Zeaemaydis Techon and Daniels | General | Leaf Mold Fungus | General | ||
| Serious | Serious | ||||
| Puccinia Polvsora | General | Target Spot Bacteria | General | ||
| Serious | Serious | ||||
| Corn Curvularia Leaf Spot Fungus | General | Septoria Leaf Spot Fungus | General | ||
| Serious | Serious | ||||
| Maize dwarf mosaic virus | Spider Mite Damage | General | |||
| Grape | Healthy | Serious | |||
| Black Rot Fungus | General | YLCV Virus | General | ||
| Serious | Serious | ||||
| Black Measles Fungus | General | Tomv | |||
| Serious | Pepper | Healthy | |||
| Leaf Blight Fungus | General | Scab | General | ||
| Serious | Serious | ||||
| Citrus | Healthy | Potato | Healthy | ||
| Greening June | General | Early Blight Fungus | General | ||
| Serious | Serious | ||||
| Peach | Healthy | Late Blight Fungus | General | ||
| Bacterial Spot | General | Serious | |||
| Serious | Pepper | Scab | General | ||
| Pepper | Healthy | Serious | |||
| (1) | ||||
| Algorithm | AOM | AVM | AUM | CM |
| T | 0.71 | 0.41 | 0.34 | 0.65 |
| OSTU [34] | 0.76 | 0.35 | 0.33 | 0.70 |
| GSO [41] | 0.82 | 0.34 | 0.31 | 0.72 |
| Ours | 0.85 | 0.31 | 0.29 | 0.75 |
| (2) | ||||
| Algorithm | AOM | AVM | AUM | CM |
| T | 0.74 | 0.33 | 0.35 | 0.69 |
| OSTU [34] | 0.81 | 0.31 | 0.32 | 0.73 |
| GSO [41] | 0.85 | 0.27 | 0.29 | 0.76 |
| Ours | 0.87 | 0.26 | 0.27 | 0.78 |
| (3) | ||||
| Algorithm | AOM | AVM | AUM | CM |
| T | 0.75 | 0.31 | 0.33 | 0.70 |
| OSTU [34] | 0.78 | 0.27 | 0.31 | 0.73 |
| GSO [41] | 0.84 | 0.24 | 0.28 | 0.77 |
| Ours | 0.88 | 0.23 | 0.24 | 0.80 |
| (4) | ||||
| Algorithm | AOM | AVM | AUM | CM |
| T | 0.74 | 0.34 | 0.27 | 0.71 |
| OSTU [34] | 0.81 | 0.35 | 0.25 | 0.74 |
| GSO [41] | 0.86 | 0.24 | 0.22 | 0.8 |
| Ours | 0.91 | 0.21 | 0.19 | 0.84 |
| (5) | ||||
| Algorithm | AOM | AVM | AUM | CM |
| T | 0.79 | 0.31 | 0.25 | 0.74 |
| OSTU [34] | 0.87 | 0.28 | 0.23 | 0.79 |
| GSO [41] | 0.91 | 0.23 | 0.19 | 0.83 |
| Ours | 0.93 | 0.18 | 0.17 | 0.86 |
| (6) | ||||
| Algorithm | AOM | AVM | AUM | CM |
| T | 0.86 | 0.26 | 0.23 | 0.79 |
| OSTU [34] | 0.89 | 0.23 | 0.22 | 0.81 |
| GSO [41] | 0.92 | 0.17 | 0.18 | 0.86 |
| Ours | 0.95 | 0.15 | 0.16 | 0.88 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Qiu, S.; Ye, H.; Liao, X. A Plant Disease Classification Algorithm Based on Attention MobileNet V2. Algorithms 2023, 16, 442. https://doi.org/10.3390/a16090442
Wang H, Qiu S, Ye H, Liao X. A Plant Disease Classification Algorithm Based on Attention MobileNet V2. Algorithms. 2023; 16(9):442. https://doi.org/10.3390/a16090442
Chicago/Turabian StyleWang, Huan, Shi Qiu, Huping Ye, and Xiaohan Liao. 2023. "A Plant Disease Classification Algorithm Based on Attention MobileNet V2" Algorithms 16, no. 9: 442. https://doi.org/10.3390/a16090442
APA StyleWang, H., Qiu, S., Ye, H., & Liao, X. (2023). A Plant Disease Classification Algorithm Based on Attention MobileNet V2. Algorithms, 16(9), 442. https://doi.org/10.3390/a16090442








