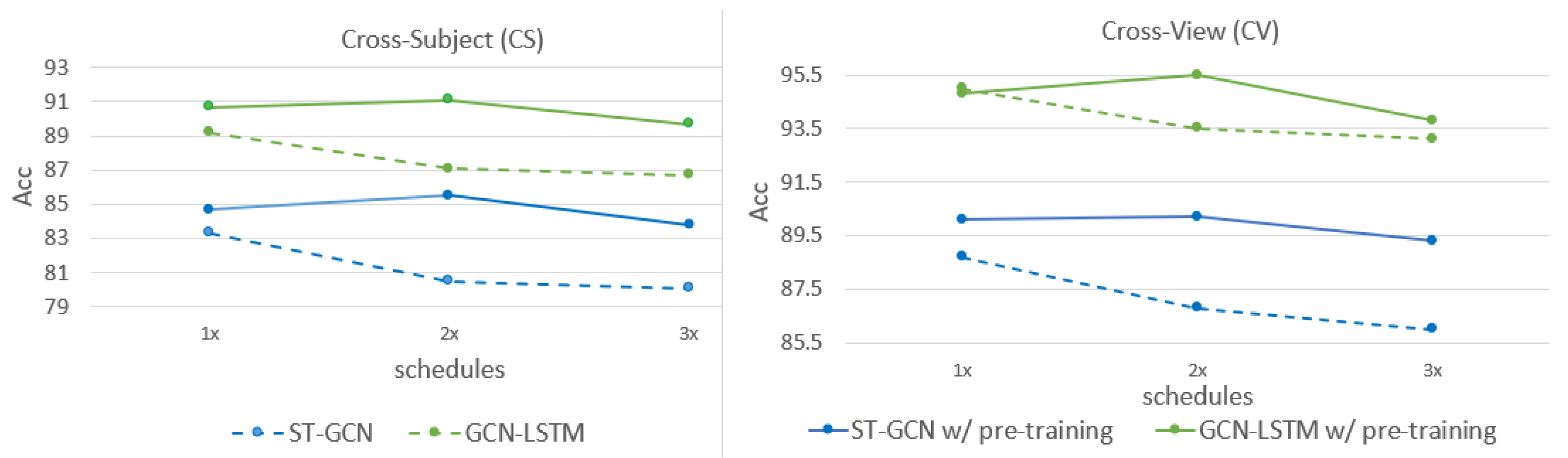
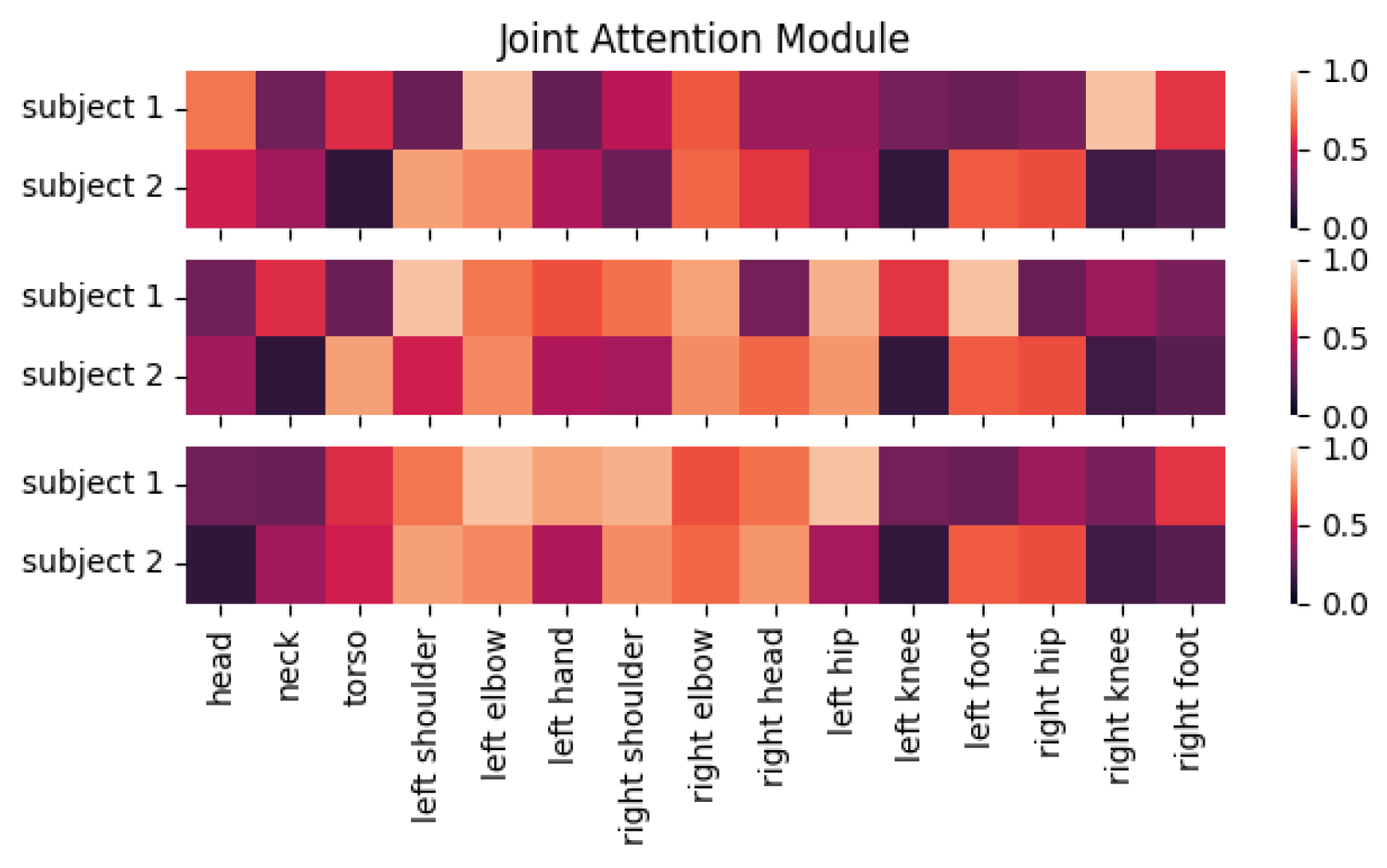
3.1. Preliminaries
Deep learning methods have been used to model and represent graph data. Traditional neural networks are limited to processing Euclidean data. By using representational learning, graph neural networks have generalized deep learning models to show good performance on structured graph data [
37]. Graph neural network (GNN) models have been shown to be a powerful family of networks that learn representations by aggregating features of entities and neighbors [
38], The extension of neural networks to data with graph structures is a new topic in deep learning research, and the application of convolutional and recurrent neural networks to graph is widely mentioned. Graph Convolutional Network (GCN), as an extension of convolutional neural networks to graph structures, is a general and effective framework for learning graph-structured data representations. In skeletal data action recognition, the human body is considered as a hinge system consisting of joints and bones and is represented as a skeletal graph. We use
to denote the skeleton data at
t frame, where
represents the set of all
N joints and
represents the joint-adjacency edges set. Define the neighboring set of
as
, where
is the shortest path distance from
to
. In addition, A predefined labelling function
sets the label
for each node
in the graph, which divides the neighboring set
into a fixed number of
K subsets. The graph convolution is calculated in the form of
where
denotes the feature representation of node
,
is the weight parameters assigned by the weight function according to the label of each node.
is the number of elements in the subset where the node is located, which is used to normalize the feature representation. The representation of
after the graph convolution is denoted as
. Using the adjacency matrix, the Equation (
1) can be expressed as:
where
is the adjacency matrix formed by the nodes labeled
in the spatial configuration and
is the degree matrix of the adjacency matrix.
In our work, we partition the neighboring sets of nodes based on the shortest path distance between them, which is similar to the relative distance used in convolutional neural networks. The partition function can be expressed as
. When the neighbouring set is defined as the shortest path less than 2, it is possible to give a matrix representation of each subset after partitioning in a simpler way, such as [
17]. In this paper, we give how to generate the adjacency matrix representation of the partitioned subsets under arbitrary distance settings. We define the polynomial of the adjacency matrix
as
, where
k is the number of highest terms. When
, we have
for the unit matrix, indicating that the neighboring nodes of each joint are only itself; when
, we have
indicating that the neighboring nodes of each joint are constructed according to the body structure
. It is important to note that in a single-subject action scene,
is represented by the adjacency matrix of the skeletal graph constructed by only one subject, but in our work, we focus on scenes with multi-subjects actions, so
represents the adjacency edges of multiple independent skeletal graphs constructed by multiple subjects in one frame. When the highest number of polynomials
, according to the definition of the distance partitioning strategy we have
Equation (
3) shows that we have
only if the shortest path distance between
and
is equal to
k, where
denote the row and column coordinates of the matrix, respectively. When
, we can compute the value of each position in the matrix
by traversing all nodes, but the time complexity of doing so is quadratic. Recall the meaning of multiplying adjacency matrices, i.e., each element in
represents the number of paths from
i to
j that pass through exactly
k edges. To this end, we can obtain
by
and
:
where
is a function whose output is zero or one:
The partitioning strategy based on distance can avoid dividing the same points into different subsets, while the partitioning strategy has a hierarchical structure that can learn finer-grained local features and learn the dependencies between nodes at longer distances as the network deepens, which is consistent with the idea and practice of convolutional neural networks.
3.2. Interaction Embedding Graph
Because graph convolution is used for spatial domain feature extraction, a predefined graph structure for the graph convolution is required, and the traditional graph structure cannot meet the requirements of accurate interaction recognition for modeling interaction information. Since only the coordinates of joints are used as known data in the skeletal data, the interaction between subjects can be understood as a feature representation of the implicit relationship between different subject joints. While graph convolution can model the relationship between joints within a single subject, it can also model interaction information. The problem is that multiple subjects form multiple independently connected graphs, but information cannot be propagated between them, which is why single-subject action recognition methods cannot achieve superior performance in interaction recognition.
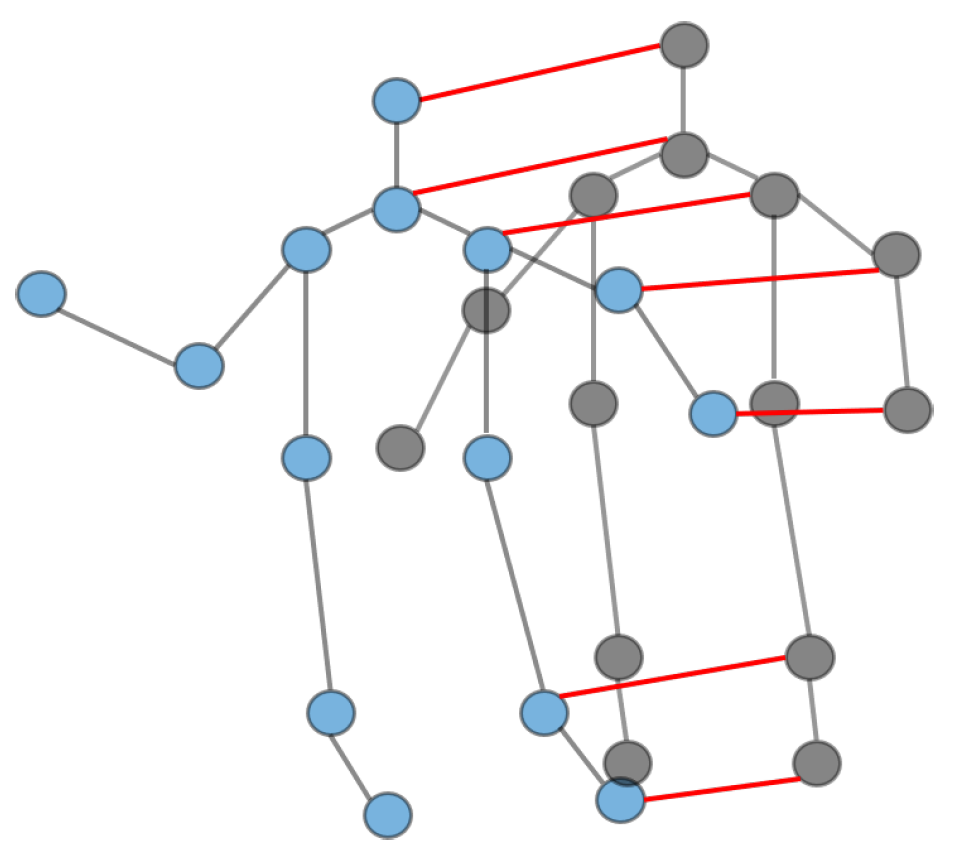
First we need to construct a graph representation of each subject, which is the same as the traditional skeleton graph representation, except that there are multiple subjects in the scene, and secondly, we introduce another kind of edge to model the interaction information. To distinguish between these two kinds of edges, we call the naturally connected edge within a subject a joint edge—which connects neighboring joints within the same subject—and call the other kind of edge that connects the same joints between different subjects a subject edge. As shown in
Figure 1, we use blue and black nodes to represent the joints of the different subjects, and the corresponding colored edges to represent the joint edges within the different subjects. In addition, we use subject edges to connect the skeleton graphs of different subject constructions. For brevity, we do not label all subject edges in the figure. Intuitively, the subject edge converts the independent skeletal graphs composed of multiple subjects into a unified connected graph, which enables modeling of interaction information and feature fusion between multiple subjects after graph convolution; Objectively, the addition of subject edges increases the number of neighbors of each joint, and as mentioned above, the increase of neighbors in the graph convolution means the increase of the receptive field, which means that each joint can not only learn the feature representation within the same subject, but also obtain useful information from other subjects.
Because joint edges and subject edges are discriminatively constructed for modeling spatial structure features and interaction features, we use two different adjacency matrices for separate representations. We also use
to denote the adjacency matrix composed of joint edges, where
M and
N are the number of subjects and the number of joints per subject, respectively. In general, the joints of each target labeled by the dataset are the same, so
is a diagonal matrix consisting of the joint adjacency matrix of each subject. In addition, we use the adjacency matrix
to store our predefined subject edges. Another reason we use two matrices for separate representations is that we use two different matrices of learnable parameters in the network to model joint and subject edges separately, one for learning spatial information and the other for interaction information. However, pre-defined subject edges are not sufficient, as they are less helpful for modeling relationships between joints at longer distances. Inspired by the edge-learnable weight matrix in graph convolution, we propose the subject edge-learnable matrix
, which has the same dimension size as
. Any graph convolution operation performed with a predefined subject edge
is accompanied by a
to learn the relational representation between distant joints. With the addition of predefined target edges and dynamic subject edges, the graph convolution operation also needs to be changed accordingly to complete the representation of the interaction information features, and the improved graph convolution can be expressed as:
Equation (
6) adds a new term to the Equation (
2), which completes the modeling of the interaction information.
is similar to
as a hyperparameter controlling the neighbouring set of each node defined by the subject edge.
is the degree matrix of
, which serves to perform normalization of the features by eliminating the influence of the number of neighboring joints on the output, which can be calculated by
.
is a matrix of learnable parameters formed by superimposing the weight vectors of multiple output channels, where
C and
represent the input and output feature dimensions, respectively. Similar to the definition of
, we define
as a polynomial of the adjacency matrix
and
.
The definition of is the same as its definition in . In the neighborhood defined by the joint edges, we use a distance partitioning strategy to partition the neighboring joints into subsets, and use the learnable weights in each subset to model the influence of the joints in each subset on the central joint to update the features so that it aggregates structural information from the other joints. We also partition the neighboring joints defined by the subject edges into subsets according to the distance partitioning strategy, and model the influence of the joints in each subset on the centroid using the learnable weights , but in this case the graph convolution operation models the interaction information between the subjects. While it is possible to simply increase to make distant joints also neighbors of the central node, there are several drawbacks to such an approach: (1) increased receptive fields increase the model memory and inference time cost; (2) excessively large can result in loops that make the model represent redundant joint dependencies. Therefore, we introduce a data-driven dynamic subject edge. In the implementation, we represent it as a learnable matrix with the same dimensions as , accompanied by at each occurrence of and denoted as . Note that is shared in each graph convolution operation and is initialized to an all-zero matrix.
3.3. Joint Attention Module
Most graph convolution methods use graph representations to represent skeletal data and model the relationships between joints using learnable weights in the networks. However, there is no network or operation that can be used to enhance key joint features throughout the process. We thus introduce an attention mechanism. Unlike previous work [
39] that introduced attention weights in graph convolutional networks, our JAM has three distinct advantages: (1) the module is designed independently of the backbone network, so it can be used by other backbone networks; (2) the independently designed JAM utilizes both temporal and spatial domain information, and these spatio-temporal features can help it learn more robust attention weights; (3) with the feature map of the whole skeletal sequence as input, the module can learn the importance of each joint in a global view.
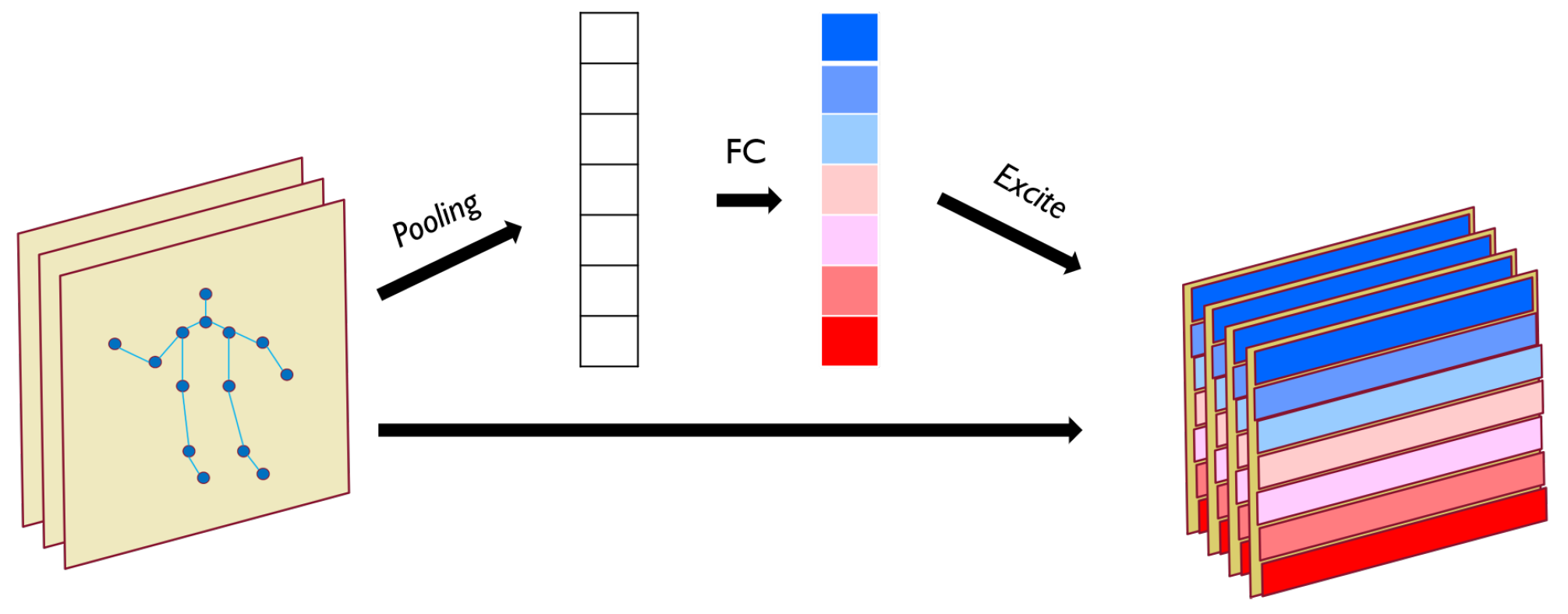
The JAM learns from the spatio-temporal information feature map output from the previous layer and arranges different attention weights for different joints. As shown in
Figure 2, for a given input feature map with dimensions
, the entire feature map first undergoes a pooling operation, which generates a joint description feature map by aggregating the feature maps in the temporal dimension. The role of joint description feature map is to generate embeddings corresponding to the global distribution of each joint feature, allowing the global information from the network to be used by later network layers. Following the feature aggregation is the excitation, which takes the global distribution embedding as input to generate modulation weights for each joint. These weights are then applied to the original input feature map to produce the output of the entire JAM.
Specifically, the JAM is a computational unit that maps the input
to the feature map
. We define
, where
is the feature representation corresponding to the joint
i. The signal in each output feature map is obtained by a locality operation, so that each cell of the transformed output feature map cannot make use of information from the context outside that region. Therefore, we use a pooling operation to convert the input features associated with the global temporal domain into a joint description feature map, which is achieved by performing a global average pooling. Formally, the joint description feature map
is generated by collapsing the temporal dimension
T. For example, the description feature
of the
i joint is computed as follows
To take advantage of the information aggregated in the pooling operation, we follow the activation operation with the aim of obtaining joint attention weights. To achieve this target, the activation operation must satisfy two requirements: (1) it must be flexible enough to learn nonlinear interactions between joints, and (2) it needs to learn non-reciprocal relationships, since we want to allow the existence of multiple key joints, rather than emphasizing only one of them. Therefore, we select to use a gated network with a
activation function.
where
and
are the
and
activation functions, respectively,
,
. To limit the size and complexity of the JAM, we implement the gate mechanism through a bottleneck structure that consists of two fully connected strata and two nonlinear activation functions. The number of channels of the middle-state feature map in the bottleneck structure is determined by the scaling factor
a, and then the feature dimension is reduced to 1 by the
activation function and the fully-connected layer. The final output of JAM is the modulated feature map.
where the dimension of
is the same as
. The JAM can be integrated into our backbone network. In addition, the flexibility of the block means that it can be applied to other networks.
3.4. Pre-Training and Contrastive Loss
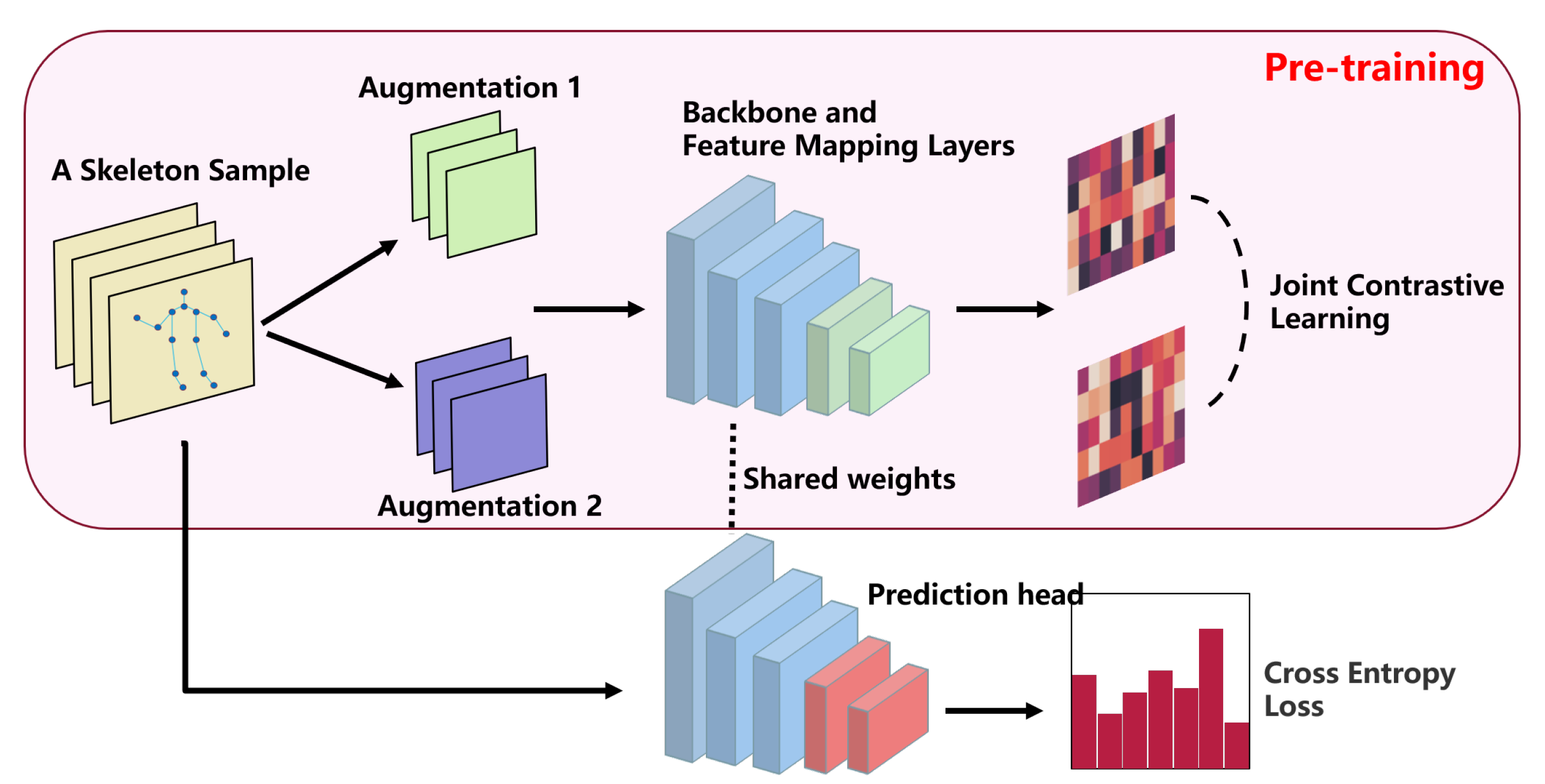
A well-designed pretext task for pre-training aims to learn network weights that are universally applicable and useful to multiple downstream tasks. In terms of architecture, speed of inference is a major consideration in some tasks where the network used is a lightweight network. In contrast, the success of pre-training relies on networks with an excess of parameters. In terms of data, a sufficiently large amount of data is one of the keys to successful pre-training, so a dataset like NTU60 or NTU120 is needed. Finally, in terms of loss design, a contrast loss function needs to be designed according to the pretext task. In
Figure 3, we conclude the pre-training framework we explore in this paper, and call the pre-training framework JointContrast. Specifically, given a skeletal sample
, we first generate two new samples
and
aligned in the same world coordinate system by common data augmentation, including random flips, rotations, translations, and scaling. We then feed the samples
and
to the shared backbone network to extract spatio-temporal feature representations
and
. Finally, a contrast loss applied to these two spatio-temporal features is defined: we minimize the feature distance between corresponding joints and maximize the feature distance between non-corresponding joints.
InfoNCE, proposed in [
40], is used by most of the unsupervised representational learning methods applied to scene understanding. By treating contrast learning as a dictionary query process, InfoNCE views contrast learning as a classification problem of classifying positive sample pairs into the same class and negative sample pairs into different classes, with a concrete implementation using
loss. Our proposed contrast loss function based on corresponding joints is an improvement on the original InfoNCE.
where
is the set of all positive joint pairs. In this form, we only consider joints that have at least one corresponding joint and do not use additional non-corresponding joints as negative. For a positive joint pair
, the joint feature
will be used as the query and
will be used as the positive key. In addition, we use all joint features
where
and
as the set of negative keys. We summarize the above pre-training process as Algorithm 1.
| Algorithm 1 Joint contrast learning algorithm. |
Require: Backbone NN Require: Skeleton dataset Require: Channels for feature maps D Ensure: Pre-training parameters of the backbone network for Each skeletal sample in the dataset do Generate two views from , and Sampling two transformations Compute the features of the skeletal data by and Compute the loss and update the parameters of the network NN by back propagation using the contrast loss function on the corresponding joints end for
|
3.5. Backbone and Fine-Tuning
In this paper, we use the Graph Convolutional embedding LSTM (GC-LSTM) network as the backbone, which is originally designed in [
39] that achieved significant improvement over prior methods. Specifically, we utilize one LSTM layer as joint encoder to model joint representation, and then three GC-LSTM blocks with proposed JAM model discriminative spatialtemporal features. Finally, temporal average pooling is the implementation of average pooling in the temporal domain, and we use the global feature of all joints to predict the class of human action. The graph convolution operation embedded in GC-LSTM enables memory cells to process graph-structured data. By combining our proposed interaction information embedding graph representation, it is capable of representing the joint dependencies between different subjects.
In the previous section we use the correspondence between joints and complete unsupervised pre-training using contrast learning to obtain the pre-trained network parameters. The effects are (1) to make the features corresponding to each joint more dispersed in the feature space and to highlight the unique role played by each joint in the action; (2) to close the feature distance between samples of the same category, making the model more robust. After unsupervised pre-training of the model using data from the source dataset, we initialize the network using the pre-trained parameters and then fine-tune it on the target training set. Unsupervised pre-training work requires a large amount of data, even if that data is unlabeled. We chose NTU120 as the pre-training dataset, which is the largest dataset related to skeletal data with a rich sample of action classes and inter-class diversity.