Machine Learning-Based Hyperspectral and RGB Discrimination of Three Polyphagous Fungi Species Grown on Culture Media
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Cultures
2.2. Hyperspectral Measurements
2.3. RGB Images
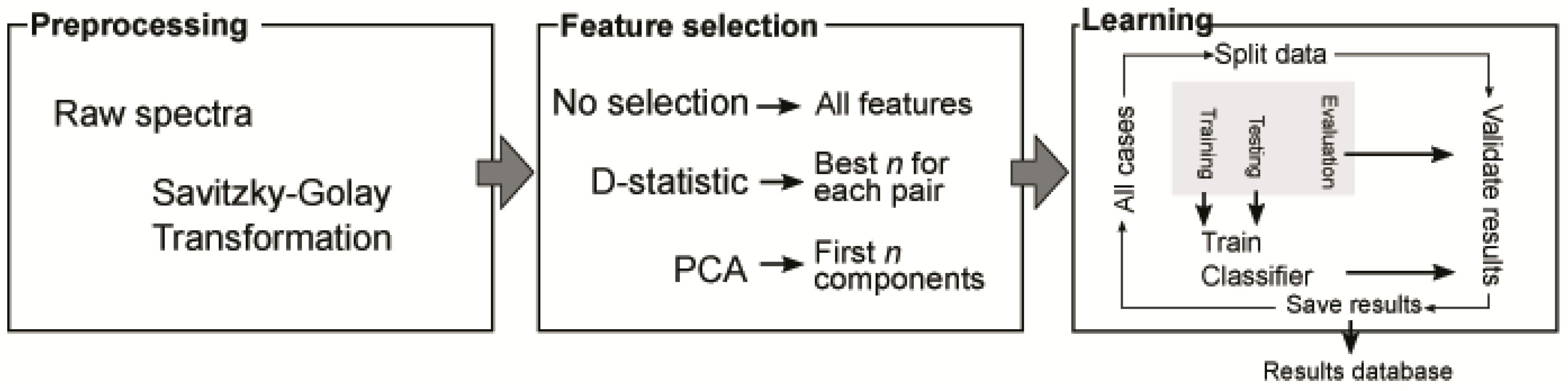
2.4. Data Processing and Models Created
2.4.1. Preprocessing and Feature Selection
2.4.2. Learning Process
3. Results and Discussion
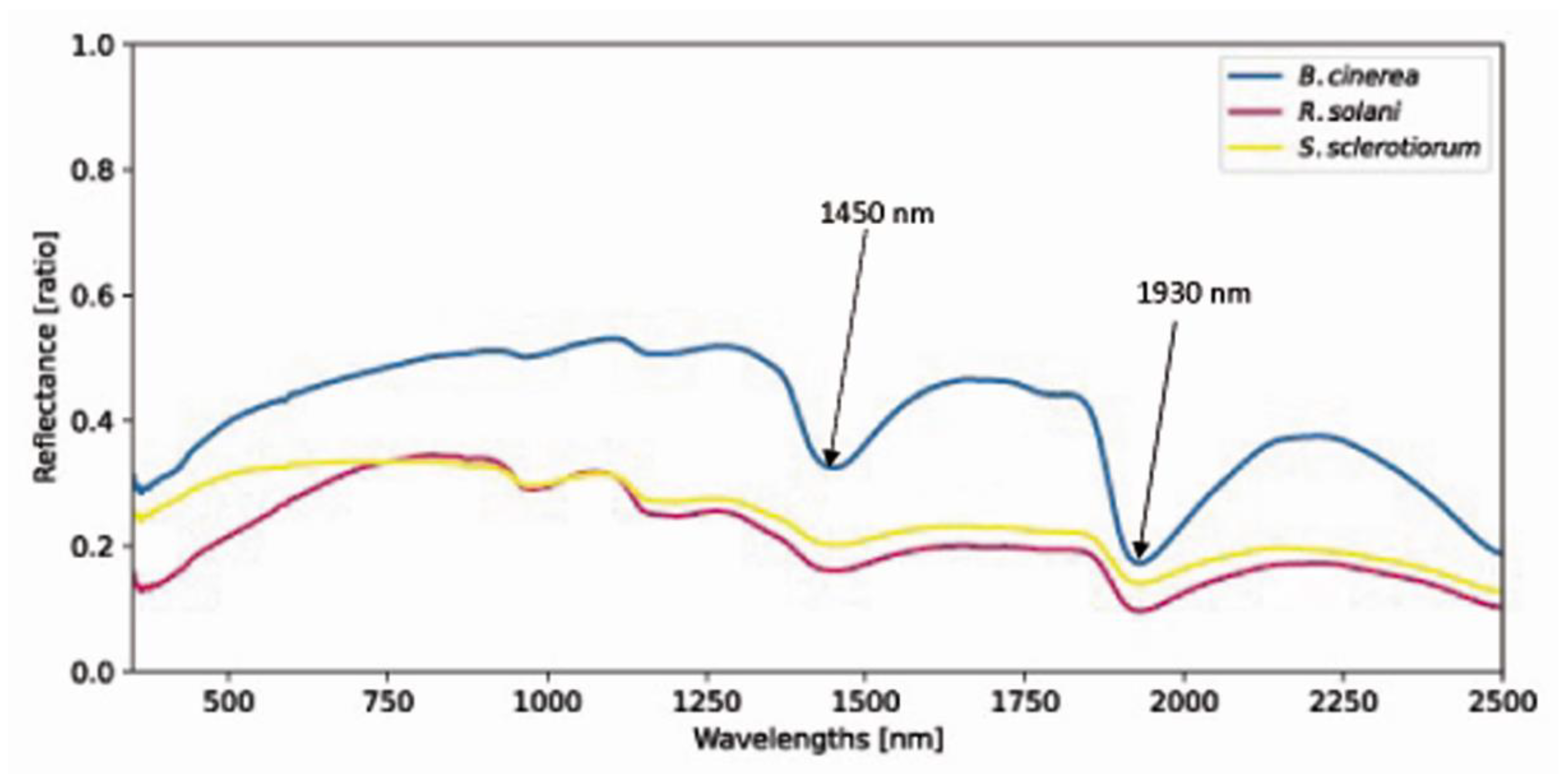
3.1. Hyperspectral Characteristics of Fungi
3.2. RGB Characteristics of Fungi
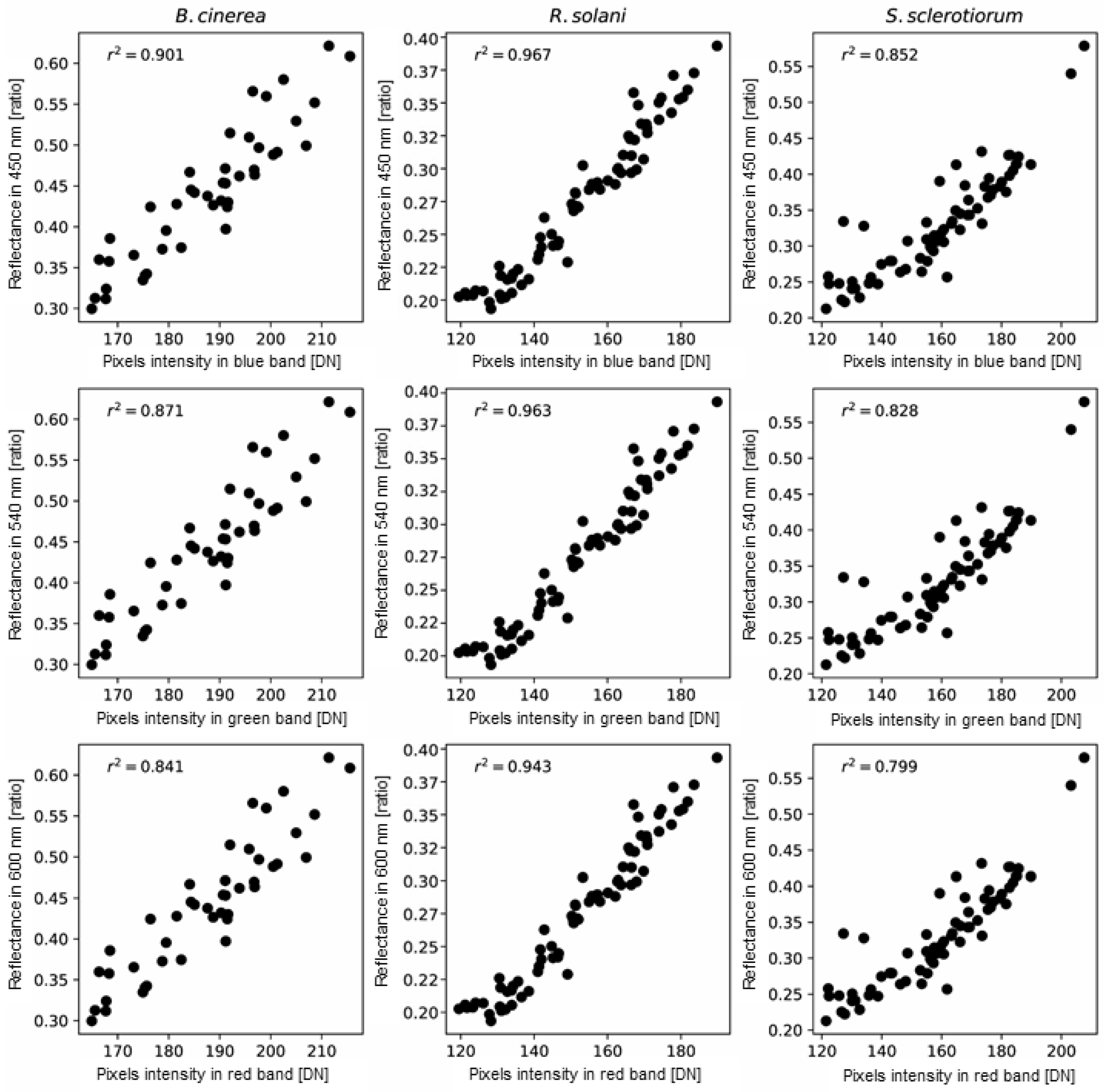
3.3. Relationship between Hyperspectral and RGB Characteristics
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karimi, K.; Arzanlou, M.; Pertot, I. Development of novel species-specific primers for the specific identification of Colletotrichum nymphaeae based on conventional PCR and LAMP techniques. Eur. J. Plant Pathol. 2020, 156, 463–475. [Google Scholar] [CrossRef]
- Baturo-Cieśniewska, A.; Groves, C.L.; Albrecht, K.A.; Grau, C.R.; Willis, D.K.; Smith, D.L. Molecular identification of Sclerotinia trifoliorum and Sclerotinia sclerotiorum isolates from the United States and Poland. Plant Dis. 2017, 101, 192–199. [Google Scholar] [CrossRef]
- Šišić, A.; Oberhänsli, T.; Baćanović-Šišić, J.; Hohmann, P.; Finckh, M.R. A novel real time PCR method for the detection and quantification of Didymella pinodella in symptomatic and asymptomatic plant hosts. J. Fungi 2022, 8, 41. [Google Scholar] [CrossRef] [PubMed]
- Pieczul, K.; Perek, A.; Kubiak, K. Detection of Tilletia caries, Tilletia laevis and Tilletia controversa wheat grain contamination using loop-mediated isothermal DNA amplification (LAMP). J. Microbiol. Methods 2018, 154, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Baysal-Gurel, F.; Ivey, M.L.L.; Dorrance, A.; Luster, D.; Frederick, R.; Czarnecki, J.; Miller, S.A. An immunofluorescence assay to detect urediniospores of Phakopsora pachyrhizi. Plant Dis. 2008, 92, 1387–1393. [Google Scholar] [CrossRef]
- Milner, H.; Ji, P.; Sabula, M.; Wu, T. Quantitative polymerase chain reaction (Q-PCR) and fluorescent in situ hybridization (FISH) detection of soilborne pathogen Sclerotium rolfsii. Appl. Soil Ecol. 2019, 136, 86–92. [Google Scholar] [CrossRef]
- Thornton, C.R.; Slaughter, D.C.; Davis, R.M. Detection of the sour-rot pathogen Geotrichum candidum in tomato fruit and juice by using a highly specific monoclonal antibody-based ELISA. Int. J. Food Microbiol. 2010, 143, 166–172. [Google Scholar] [CrossRef]
- Prigione, V.; Lingua, G.; Marchisio, V.F. Development and use of flow cytometry for detection of airborne fungi. Appl. Environ. Microbiol. 2004, 70, 1360–1365. [Google Scholar] [CrossRef][Green Version]
- Pan, L.; Zhang, W.; Zhu, N.; Mao, S.; Tu, K. Early detection and classification of pathogenic fungal disease in post-harvest strawberry fruit by electronic nose and gas chromatography–mass spectrometry. Food Res. Int. 2014, 62, 162–168. [Google Scholar] [CrossRef]
- Hariharan, G.; Prasannath, K. Recent advances in molecular diagnostics of fungal plant pathogens: A mini review. Front. Cell. Infect. Microbiol. 2021, 10, 600234. [Google Scholar] [CrossRef]
- D’Hondt, L.; Höfte, M.; Van Bockstaele, E.; Leus, L. Applications of flow cytometry in plant pathology for genome size determination, detection and physiological status. Mol. Plant Pathol. 2011, 12, 815–828. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Ramasamy, R.P. Current and prospective methods for plant disease detection. Biosensors 2015, 5, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Gold, K.M.; Townsend, P.A.; Chlus, A.; Herrmann, I.; Couture, J.J.; Larson, E.R.; Gevens, A.J. Hyperspectral measurements enable pre-symptomatic detection and differentiation of contrasting physiological effects of late blight and early blight in potato. Remote Sens. 2020, 12, 286. [Google Scholar] [CrossRef]
- Sancho-Adamson, M.; Trillas, M.I.; Bort, J.; Fernandez-Gallego, J.A.; Romanyà, J. Use of RGB vegetation indexes in assessing early effects of Verticillium wilt of olive in asymptomatic plants in high and low fertility scenarios. Remote Sens. 2019, 11, 607. [Google Scholar] [CrossRef]
- Yao, Z.; Lei, Y.; He, D. Early visual detection of wheat stripe rust using visible/near-infrared hyperspectral imaging. Sensors 2019, 19, 952. [Google Scholar] [CrossRef]
- Yu, K.; Anderegg, J.; Mikaberidze, A.; Karisto, P.; Mascher, F.; McDonald, B.A.; Hund, A. Hyperspectral canopy sensing of wheat Septoria tritici blotch disease. Front. Plant Sci. 2018, 9, 1195. [Google Scholar] [CrossRef]
- Wijekoon, C.P.; Goodwin, P.H.; Hsiang, T. Quantifying fungal infection of plant leaves by digital image analysis using Scion Image software. J. Microbiol. Methods 2008, 74, 94–101. [Google Scholar] [CrossRef]
- Corkidi, G.; Balderas-Ruíz, K.A.; Taboada, B.; Serrano-Carreón, L.; Galindo, E. Assessing mango anthracnose using a new three-dimensional image-analysis technique to quantify lesions on fruit. Plant Pathol. 2006, 55, 250–257. [Google Scholar] [CrossRef]
- Thomas, S.; Kuska, M.T.; Bohnenkamp, D.; Brugger, A.; Alisaac, E.; Wahabzada, M.; Mahlein, A.K. Benefits of hyperspectral imaging for plant disease detection and plant protection: A technical perspective. J. Plant Dis. Prot. 2018, 125, 5–20. [Google Scholar] [CrossRef]
- Padmavathi, K.; Thangadurai, K. Implementation of RGB and grayscale images in plant leaves disease detection—Comparative study. Indian J. Sci. Technol. 2016, 9, 1–6. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Abdullah, N.E.; Rahim, A.A.; Hashim, H.; Kamal, M.M. Classification of rubber tree leaf diseases using multilayer perceptron neural network. In Proceedings of the 2007 5th Student Conference on Research and Evelopment, Selangor, Malaysia, 11–12 December 2007; IEEE: Manhattan, NY, USA, 2007; pp. 1–6. [Google Scholar]
- Yu, H.; Cheng, X.; Chen, C.; Heidari, A.A.; Liu, J.; Cai, Z.; Chen, H. Apple leaf disease recognition method with improved residual network. Multimed Tools Appl. 2022, 81, 7759–7782. [Google Scholar] [CrossRef]
- Price, T.V.; Gross, R.; Wey, J.H.; Osborne, C.F. A comparison of visual and digital image-processing methods in quantifying the severity of coffee leaf rust (Hemileia vastatrix). Aust. J. Exp. Agric. 1993, 33, 97–101. [Google Scholar] [CrossRef]
- Tucker, C.C.; Chakraborty, S. Quantitative assessment of lesion characteristics and disease severity using digital image processing. J. Phytopathol. 1997, 145, 273–278. [Google Scholar] [CrossRef]
- Martin, D.P.; Rybicki, E.P. Microcomputer-based quantification of maize streak virus symptoms in zea mays. Phytopathology 1998, 88, 422–427. [Google Scholar] [CrossRef]
- Weizheng, S.; Yachun, W.; Zhanliang, C.; Hongda, W. Grading method of leaf spot disease based on image processing. In Proceedings of the 2008 International Conference on Computer Science and Software Engineering, Wuhan, China, 12–14 December 2008; IEEE: Manhattan, NY, USA, 2008; pp. 491–494. [Google Scholar]
- Camargo, A.; Smith, J.S. Image pattern classification for the identification of disease causing agents in plants. Comput. Electron. Agric. 2009, 66, 121–125. [Google Scholar] [CrossRef]
- Herrero-Latorre, C.; Barciela-García, J.; García-Martín, S.; Peña-Crecente, R.M. Detection and quantification of adulterations in aged wine using RGB digital images combined with multivariate chemometric techniques. Food Chem. X 2019, 3, 100046. [Google Scholar] [CrossRef]
- Schleder, G.R.; Padilha, A.C.M.; Acosta, C.M.; Costa, M.; Fazzio, A. From DFT to machine learning: Recent approaches to materials science–a review. J. Phys. Mater. 2019, 2, 032001. [Google Scholar] [CrossRef]
- van Dijk, A.D.J.; Kootstra, G.; Kruijer, W.; de Ridder, D. Machine learning in plant science and plant breeding. Iscience 2021, 24, 101890. [Google Scholar] [CrossRef]
- Bock, C.H.; Barbedo, J.G.; Del Ponte, E.M.; Bohnenkamp, D.; Mahlein, A.K. From visual estimates to fully automated sensor-based measurements of plant disease severity: Status and challenges for improving accuracy. Phytopathol. Res. 2020, 2, 9. [Google Scholar] [CrossRef]
- Singh, A.; Ganapathysubramanian, B.; Singh, A.K.; Sarkar, S. Machine learning for high-throughput stress phenotyping in plants. Trends Plant Sci. 2016, 21, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Alizadeh, M.; Jones, A.M.P.; Torkamaneh, D. Machine learning: Its challenges and opportunities in plant system biology. Appl. Microbiol. Biotechnol. 2022, 106, 3507–3530. [Google Scholar] [CrossRef] [PubMed]
- Grinblat, G.L.; Uzal, L.C.; Larese, M.G.; Granitto, P.M. Deep learning for plant identification using vein morphological patterns. Comput. Electron. Agric. 2016, 127, 418–424. [Google Scholar] [CrossRef]
- Mishra, B.; Kumar, N.; Mukhtar, M.S. Systems biology and machine learning in plant–pathogen interactions. Mol. Plant-Microbe Interact. 2019, 32, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Aboelghar, M.; Wahab, H.A. Spectral footprint of Botrytis cinerea, a novel way for fungal characterization. Adv. Biosci. Biotechnol. 2013, 4, 374–382. [Google Scholar] [CrossRef][Green Version]
- Aboelghar, M.; Moustafa, M.S.; Ali, A.M.; Wahab, H.A. Hyperspectral analysis of Botrytis cinerea infected lettuce. EPH-Int. J. Agric. Environ. Res. 2019, 5, 26–42. [Google Scholar]
- Reynolds, G.J.; Windels, C.E.; MacRae, I.V.; Laguette, S. Hyperspectral remote sensing for detection of Rhizoctonia crown and root rot in sugarbeet. Phytopathology 2009, 99, 108. [Google Scholar]
- Cao, F.; Liu, F.; Guo, H.; Kong, W.; Zhang, C.; He, Y. Fast detection of Sclerotinia sclerotiorum on oilseed rape leaves using low-altitude remote sensing technology. Sensors 2018, 18, 4464. [Google Scholar] [CrossRef]
- Cameron, M.; Kumar, L. The Depths of Cast Shadow. Remote Sens. 2019, 11, 1806. [Google Scholar] [CrossRef]
- Piekarczyk, J.; Ratajkiewicz, H.; Jasiewicz, J.; Sosnowska, D.; Wójtowicz, A. An application of reflectance spectroscopy to differentiate of entomopathogenic fungi species. J. Photochem. Photobiol. B Biol. 2019, 190, 32–41. [Google Scholar] [CrossRef]
- Savitzky, A.; Golay, M.J.E. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; ACM: New York, NY, USA, 2016; pp. 785–794. [Google Scholar]
- Nawar, S.; Mouazen, A.M. Comparison between random forests, artificial neural networks and gradient boosted machines methods of on-line Vis-NIR spectroscopy measurements of soil total nitrogen and total carbon. Sensors 2017, 17, 2428. [Google Scholar] [CrossRef] [PubMed]
- Golden, C.E.; Rothrock, M.J., Jr.; Mishra, A. Comparison between random forest and gradient boosting machine methods for predicting Listeria spp. prevalence in the environment of pastured poultry farms. Food Res. Int. 2019, 122, 47–55. [Google Scholar] [CrossRef] [PubMed]
- dos Santos, U.J.; de Melo Dematte, J.A.; Menezes, R.S.C.; Dotto, A.C.; Guimarães, C.C.B.; Alves, B.J.R.; Primoa, D.C.; Sampaio, E.V.D.S.B. Predicting carbon and nitrogen by visible near-infrared (Vis-NIR) and mid-infrared (MIR) spectroscopy in soils of Northeast Brazil. Geoderma Reg. 2020, 23, e00333. [Google Scholar] [CrossRef]
- Ivanov, A.; Riccardi, G. Kolmogorov-Smirnov test for feature selection in emotion recognition from speech. In Proceedings of the 2012 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Kyoto, Japan, 25–30 March 2012; IEEE: Manhattan, NY, USA, 2012; pp. 5125–5128. [Google Scholar]
- Japkowicz, N.; Shah, M. Evaluating Learning Algorithms: A Classification Perspective; Cambridge University Press: Cambridge, UK, 2011. [Google Scholar]
- Fabris, F.; Freitas, A.A. Analysing the Overfit of the Auto-sklearn Automated Machine Learning Tool; ACM: Nicosia, Cyprus, 2019. [Google Scholar]
- Tsamardinos, I.; Greasidou, E.; Borboudakis, G. Bootstrapping the out-of-sample predictions for efficient and accurate cross-validation. Mach. Learn. 2018, 107, 1895–1922. [Google Scholar] [CrossRef]
- Pituch, K.A.; Stapleton, L.M.; Kang, J.Y. A comparison of single sample and bootstrap methods to assess mediation in cluster randomized trials. Multivar. Behav. Res. 2006, 41, 367–400. [Google Scholar] [CrossRef]
- Conrad, A.O.; Li, W.; Lee, D.Y.; Wang, G.L.; Rodriguez-Saona, L.; Bonello, P. Machine learning-based presymptomatic detection of rice sheath blight using spectral profiles. Plant Phenomics 2020, 2020, 8954085. [Google Scholar] [CrossRef]
- Kong, W.; Zhang, C.; Huang, W.; Liu, F.; He, Y. Application of hyperspectral imaging to detect Sclerotinia sclerotiorum on oilseed rape stems. Sensors 2018, 18, 123. [Google Scholar] [CrossRef]
- Cambaza, E.; Koseki, S.; Kawamura, S. Why RGB imaging should be used to analyze Fusarium graminearum growth and estimate deoxynivalenol contamination. Methods Protoc. 2019, 2, 25. [Google Scholar] [CrossRef]
- Ponti, M.; Chaves, A.A.; Jorge, F.R.; Costa, G.B.; Colturato, A.; Branco, K.R. Precision agriculture: Using low-cost systems to acquire low-altitude images. IEEE Comput. Graph. Appl. 2016, 36, 14–20. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, X.; You, Z.; Zhang, L. Leaf image based cucumber disease recognition using sparse representation classification. Comput. Electron. Agric. 2017, 134, 135–141. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, S.; Zhang, C.; Wang, X.; Shi, Y. Cucumber leaf disease identification with global pooling dilated convolutional neural network. Comput. Electron. Agric. 2019, 162, 422–430. [Google Scholar] [CrossRef]
- Kumar, R.; Baloch, G.; Pankaj, A.B.B.; Abdul, B.B.; Bhatti, J. Fungal blast disease detection in rice seed using machine learning. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 248–258. [Google Scholar] [CrossRef]
- Fernández, C.I.; Leblon, B.; Wang, J.; Haddadi, A.; Wang, K. Detecting infected cucumber plants with close-range multispectral imagery. Remote Sens. 2021, 13, 2948. [Google Scholar] [CrossRef]
- Pozza, E.A.; de Carvalho Alves, M.; Sanches, L. Using computer vision to identify seed-borne fungi and other targets associated with common bean seeds based on red–green–blue spectral data. Trop. Plant Pathol. 2022, 47, 168–185. [Google Scholar] [CrossRef]
- Anthonys, G.; Wickramarachchi, N. An image recognition system for crop disease identification of paddy fields in Sri Lanka. In Proceedings of the 2009 International Conference on Industrial and Information Systems (ICIIS), Peradeniya, Sri Lanka, 28–31 December 2009; IEEE: Manhattan, NY, USA, 2009; pp. 403–407. [Google Scholar]
- Pavicic, M.; Overmyer, K.; Rehman, A.; Jones, P.; Jacobson, D.; Himanen, K. Image-based methods to score fungal pathogen symptom progression and severity in excised Arabidopsis leaves. Plants 2021, 10, 158. [Google Scholar] [CrossRef]
- Sun, Y.; Gu, X.; Wang, Z.; Huang, Y.; Wei, Y.; Zhang, M.; Tu, K. Leiqing Pan Growth simulation and discrimination of Botrytis cinerea, Rhizopus stolonifer and Colletotrichum acutatum using hyperspectral reflectance imaging. PLoS ONE 2015, 10, e0143400. [Google Scholar]
- Zhou, X.G.; Zhang, D.; Lin, F. UAV remote sensing: An innovative tool for detection and management of rice diseases. In Diagnostics of Plant Diseases; Kurouski, D., Ed.; IntechOpen: London, UK, 2021; p. 95535. [Google Scholar]
- Rego, C.H.Q.; França-Silva, F.; Gomes-Junior, F.G.; Moraes, M.H.D.D.; Medeiros, A.D.D.; Silva, C.B.D. Using multispectral imaging for detecting seed-borne fungi in cowpea. Agriculture 2020, 10, 361. [Google Scholar] [CrossRef]
- Fahrentrapp, J.; Ria, F.; Geilhausen, M.; Panassiti, B. Detection of gray mold leaf infections prior to visual symptom appearance using a five-band multispectral sensor. Front. Plant Sci. 2019, 10, 628. [Google Scholar] [CrossRef]
- Du, Q.; Raksuntorn, N.; Cai, S.; Moorhead, R.J. Color display for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1858–1866. [Google Scholar] [CrossRef]
- Magnusson, M.; Sigurdsson, J.; Armansson, S.E.; Ulfarsson, M.O.; Deborah, H.; Sveinsson, J.R. Creating RGB images from hyperspectral images using a color matching function. In Proceedings of the IGARSS 2020-2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; IEEE: Manhattan, NY, USA, 2020; pp. 2045–2048. [Google Scholar]
- Cao, X.; Tong, X.; Dai, Q.; Lin, S. High resolution multispectral video capture with a hybrid camera system. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; IEEE: Manhattan, NY, USA, 2011; pp. 297–304. [Google Scholar]
- Kawakami, R.; Matsushita, Y.; Wright, J.; Ben-Ezra, M.; Tai, Y.W.; Ikeuchi, K. High-resolution hyperspectral imaging via matrix factorization. In Proceedings of the CVPR 2011, Colorado Springs, CO, USA, 20–25 June 2011; IEEE: Manhattan, NY, USA, 2011; pp. 2329–2336. [Google Scholar]
- Arad, B.; Ben-Shahar, O. Sparse recovery of hyperspectral signal from natural RGB images. In European Conference on Computer Vision; Leibe, B., Matas, J., Sebe, N., Welling, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 19–34. [Google Scholar]
- Shi, Z.; Chen, C.; Xiong, Z.; Liu, D.; Wu, F.H. Advanced cnn-based hyperspectral recovery from RGB images. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition Workshops, Salt Lake City, UT, USA, 18–22 June 2018; IEEE: Manhattan, NY, USA, 2018; pp. 939–947. [Google Scholar]
- Barbedo, J.G.A. Digital image processing techniques for detecting, quantifying and classifying plant diseases. SpringerPlus 2013, 2, 660. [Google Scholar] [CrossRef] [PubMed]
- Bardsley, S.J.; Ngugi, H.K. Reliability and accuracy of visual methods to quantify severity of foliar bacterial spot symptoms on peach and nectarine. Plant Pathol. 2013, 62, 460–474. [Google Scholar] [CrossRef]
- Abd-El-Haliem, A. An unbiased method for the quantitation of disease phenotypes using a custom-built macro plugin for the program ImageJ. In Plant Fungal Pathogens. Methods in Molecular Biology (Methods and Protocols), Bolton, M., Thomma, B., Eds.; Humana Press: New York, NY, USA, 2012; pp. 635–644. [Google Scholar]
- Hunt, E.R., Jr.; Daughtry, C.S.T.; Eitel, J.U.; Long, D.S. Remote sensing leaf chlorophyll content using a visible band index. Agron. J. 2011, 103, 1090–1099. [Google Scholar] [CrossRef]

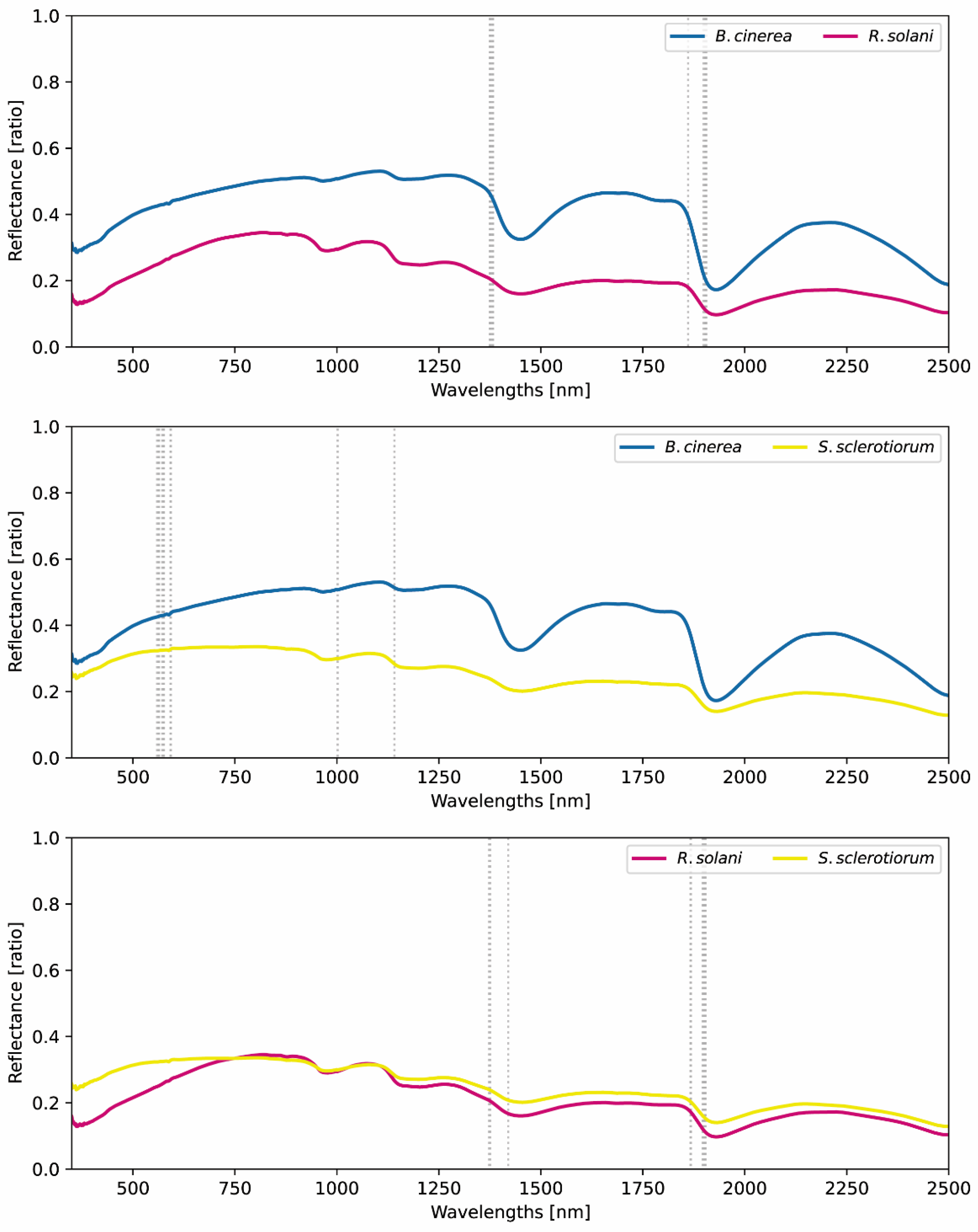
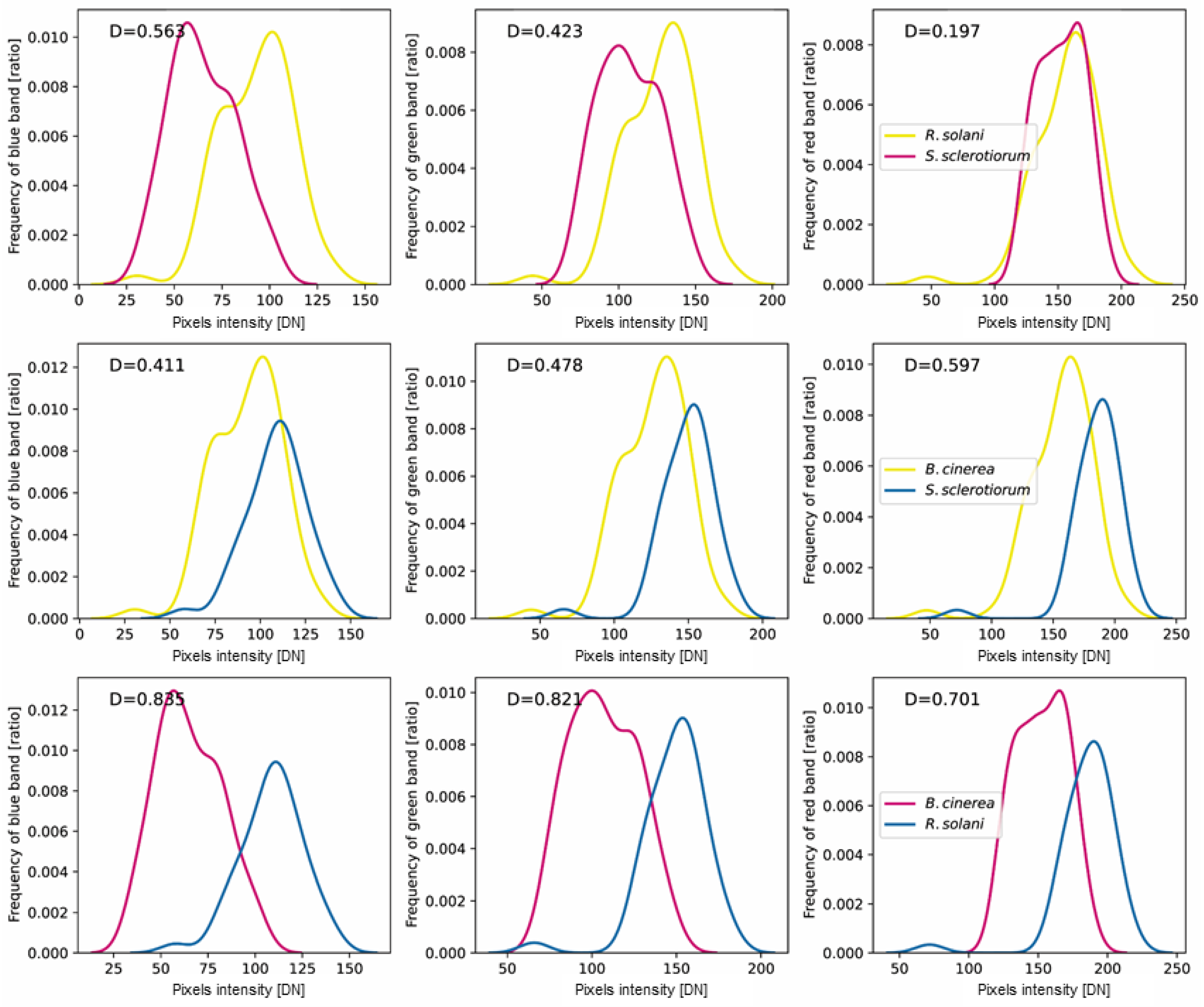
| B. cinerea-R. solani | B. cinerea-S. sclerotiorum | R. solani-S. sclerotiorum | |||
|---|---|---|---|---|---|
| Band | D-Statistic | Band | D-Statistic | Band | D-Statistic |
| 1904 | 0.791 | 1904 | 0.818 | 572 | 0.525 |
| 1906 | 0.789 | 1376 | 0.815 | 575 | 0.511 |
| 1382 | 0.789 | 1901 | 0.800 | 563 | 0.496 |
| 1899 | 0.779 | 1420 | 0.799 | 576 | 0.475 |
| 1903 | 0.776 | 1899 | 0.798 | 564 | 0.458 |
| 1376 | 0.775 | 1373 | 0.798 | 593 | 0.437 |
| 1861 | 0.773 | 1897 | 0.797 | 559 | 0.433 |
| 1378 | 0.772 | 1868 | 0.794 | 1002 | 0.430 |
| 1375 | 0.770 | 1903 | 0.790 | 592 | 0.412 |
| 1383 | 0.770 | 1867 | 0.788 | 1141 | 0.394 |
| Methods | Classification Assesment | B. cinerea | R. solani | S. sclerotiorum | Mean |
|---|---|---|---|---|---|
| All features | Wrong a | 9 | 1 | 8 | 6 |
| Vague b | 11 | 13 | 39 | 21 | |
| Good c | 160 | 270 | 237 | 222.333 | |
| Rec d | 0.915 | 0.967 | 0.918 | 0.933 | |
| Acc e | 0.889 | 0.951 | 0.835 | 0.891 | |
| Selected fetures | Wrong | 8 | 6 | 18 | 10.667 |
| Vague | 15 | 29 | 36 | 26.667 | |
| Good | 157 | 249 | 230 | 212 | |
| Rec | 0.904 | 0.922 | 0.874 | 0.900 | |
| Acc | 0.872 | 0.877 | 0.810 | 0.853 | |
| PCA | Wrong | 6 | 5 | 4 | 5 |
| Vague | 11 | 13 | 11 | 11,667 | |
| Good | 163 | 266 | 269 | 232.667 | |
| Rec | 0.940 | 0.962 | 0.963 | 0.955 | |
| Acc | 0.910 | 0.940 | 0.950 | 0.933 |
| Classification Assesment | B. cinerea | R. solani | S. sclerotiorum |
|---|---|---|---|
| Wrong a | 6 | 1 | 5 |
| Vague b | 16 | 14 | 12 |
| Good c | 23 | 57 | 54 |
| Rec d | 0.717 | 0.898 | 0.835 |
| Acc e | 0.511 | 0.792 | 0.761 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piekarczyk, J.; Wójtowicz, A.; Wójtowicz, M.; Jasiewicz, J.; Sadowska, K.; Łukaszewska-Skrzypniak, N.; Świerczyńska, I.; Pieczul, K. Machine Learning-Based Hyperspectral and RGB Discrimination of Three Polyphagous Fungi Species Grown on Culture Media. Agronomy 2022, 12, 1965. https://doi.org/10.3390/agronomy12081965
Piekarczyk J, Wójtowicz A, Wójtowicz M, Jasiewicz J, Sadowska K, Łukaszewska-Skrzypniak N, Świerczyńska I, Pieczul K. Machine Learning-Based Hyperspectral and RGB Discrimination of Three Polyphagous Fungi Species Grown on Culture Media. Agronomy. 2022; 12(8):1965. https://doi.org/10.3390/agronomy12081965
Chicago/Turabian StylePiekarczyk, Jan, Andrzej Wójtowicz, Marek Wójtowicz, Jarosław Jasiewicz, Katarzyna Sadowska, Natalia Łukaszewska-Skrzypniak, Ilona Świerczyńska, and Katarzyna Pieczul. 2022. "Machine Learning-Based Hyperspectral and RGB Discrimination of Three Polyphagous Fungi Species Grown on Culture Media" Agronomy 12, no. 8: 1965. https://doi.org/10.3390/agronomy12081965
APA StylePiekarczyk, J., Wójtowicz, A., Wójtowicz, M., Jasiewicz, J., Sadowska, K., Łukaszewska-Skrzypniak, N., Świerczyńska, I., & Pieczul, K. (2022). Machine Learning-Based Hyperspectral and RGB Discrimination of Three Polyphagous Fungi Species Grown on Culture Media. Agronomy, 12(8), 1965. https://doi.org/10.3390/agronomy12081965






