Monofractal Dimension in Quantifying the Image of Neurons in the Plane: Analysis of Image Features of Multipolar Neurons from the Principal Olivary Nucleus in Humans with Age
Abstract
:1. Introduction
2. Materials and Methods
2.1. Image Acquisition
2.2. Post-Processing
2.3. Presentation and Quantification
2.4. Statistics
3. Results
3.1. Comparison of Monofractal and Computational Parameters
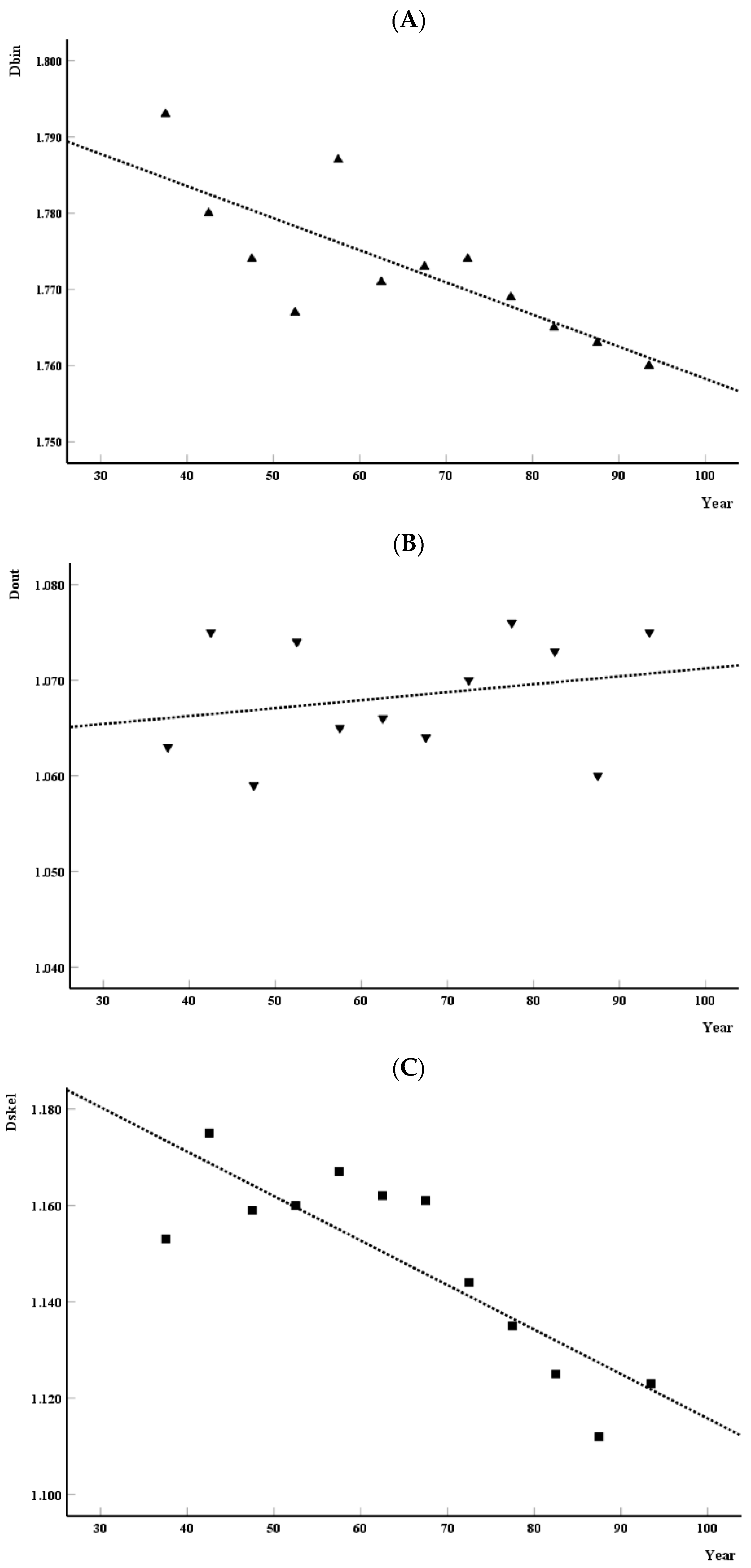
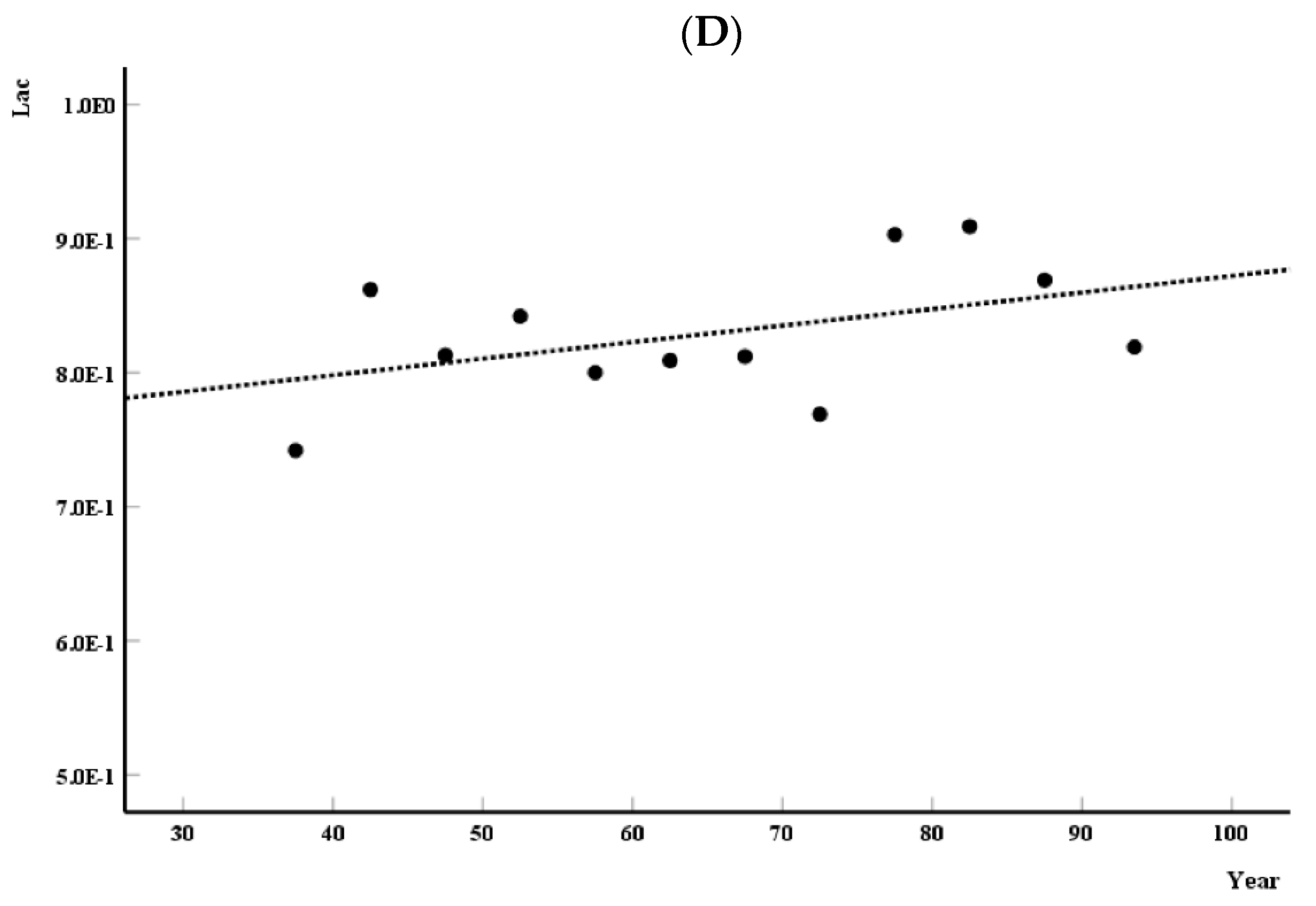
3.2. Analysis of the Distribution of Monofractal Parameters over the Years
3.3. Examining the Characteristics of the Image between Age Groups
4. Discussion
5. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Krstonošiċ, B. Morphological Analysis of Two Dimensional Projection of Neurons in the Human Neostriatum. Ph.D. Thesis, University of Novi Sad, Novi Sad, Serbia, 2013. [Google Scholar]
- Schierwagen, A.; Costa, L.F.; Alpar, A.; Gärtner, U.; Arendt, T. Neuromorphological phenotyping in transgenic mice: A multiscale fractal analysis, Volume II. In Mathematical Modeling of Biological Systems; Deutsch, A., de la Parra, R.B., De Boer, R.J., Diekmann, O., Jagers, P., Kisdi, E., Kretzschmar, M., Lansky, P., Meta, H., Eds.; Birkhäuser: Boston, MA, USA, 2008; pp. 185–192. [Google Scholar]
- Mason, A. Larkman, Correlations between morphology and electrophysiology of pyramidal neurons in slices of rat visual cortex. II. Electrophysiology. J. Neurosci. 1990, 10, 1415–1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herculano-Houzel, S. The glia/neuron ratio: How it varies uniformly across brain structures and species and what that means for brain physiology and evolution. Glia 2014, 62, 1377–1391. [Google Scholar] [CrossRef]
- Ausim Azizi, S. And the olive said to the cerebellum: Organization and functional significance of the olivo-cerebellar system. Neuroscientist 2007, 13, 616–625. [Google Scholar] [CrossRef] [PubMed]
- Bozhilova-Pastirova, A.; Ovtscharoff, W. The Inferior Olivary Complex; Advances in Anatomy, Embryology and Cell Biology; Springer: Berlin/Heidelberg, Germany, 2000; Volume 155, pp. 3–84. [Google Scholar] [CrossRef]
- Marić, D. Qualitative and Quantitative Analysis of Adult Human Dentate Nucleus Neurons Morphology. Ph.D. Thesis, University of Novi Sad, Novi Sad, Serbia, 2010. [Google Scholar]
- Radošević, D. A Morphological Analysis of the Neuronal and Glial Cells in the Human Principal Olivary Nucleus. Ph.D. Dissertation, University of Novi Sad, Novi Sad, Serbia, 2019. [Google Scholar]
- Stojić, D.; Radošević, D.; Rajković, N.; Marić, D.L.; Milošević, N.T. Classification by morphology of multipolar neurons of the human principal olivary nucleus. Neurosci. Res. 2021, 170, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An Epigenetic Biomarker of Aging for Lifespan and Healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ely, J.T.; Krone, C.A. Aging: Predictions of a New Perspective on Old Data. Exp. Biol. Med. 2002, 227, 939–942. [Google Scholar] [CrossRef] [PubMed]
- Sultan, F.; Glickstein, M. The cerebellum: Comparative and animal studies. Cerebellum 2007, 6, 168–176. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.; Wearne, S.L.; Rocher, A.B.; Macedo, A.; Morrison, J.H.; Hof, P.R. Age-related dendritic and spine changes in corticocortically projecting neurons in macaque monkeys. Cereb. Cortex 2003, 13, 950–961. [Google Scholar] [CrossRef] [PubMed]
- Renehan, W.E.; Jin, Z.; Zhang, X.; Schweitzer, L. Structure and function of gustatory neurons in the nucleus of the solitary tract. I. A classification of neurons based on morphological features. J. Comp. Neurol. 1994, 347, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Stojić, D.; Radošević, D.; Rajković, N.; Milošević, N.T. 2D Images of astrocytes in the human principal olivary nucleus: Monofractal analysis of the morphology. J. Biosci. Med. 2021, 9, 38–48. [Google Scholar] [CrossRef]
- Krstonošić, B.; Milošević, N.T.; Gudović, R.; Marić, D.L.; Ristanović, D. Neuronal images of the putamen in the adult human neostriatum: A revised classification supported by a qualitative and quantitative analysis. Anat. Sci. Int. 2012, 87, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Karperien, A.; Jelinek, H.F. ImageJ in Computational Fractal-Based Neuroscience: Pattern Extraction and Translation Research. In The Fractal Geometry of the Brain; Di Ieva, A., Ed.; Springer: New York, NY, USA, 2016; pp. 503–522. [Google Scholar]
- Milošević, N.T. The Morphology of Brain Neurons: Box Counting Method in Quantitative Analysis of 2D Image. In The Fractal Geometry of the Brain; Di Ieva, A., Ed.; Springer: New York, NY, USA, 2016; pp. 109–126. [Google Scholar]
- Kesić, S.; Spasić, S.Z. Application of Higuchi’s fractal dimension from basic to clinical neurophysiology: A review. Comput. Methods Programs Biomed. 2016, 133, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Jelinek, H.F.; Elston, G.N.; Zietsch, B. Fractal analysis: Pitfalls and revelations in neuroscience. In Fractals in Biology and Medicine IV; Losa, G.A., Merlini, D., Nonnenmacher, T.F., Weibel, E.R., Eds.; Birkhäuser: Basel, Switzerland, 2005; pp. 85–94. [Google Scholar]
- Fernandez, E.; Jelinek, H.F. Use of fractal theory in neuroscience: Methods, advantages, and potential problems. Methods 2001, 24, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Ristanović, D.; Milošević, N.T.; Štulić, V. Application of modified Sholl analysis to neuronal dendritic arborization of the cat spinal cord. J. Neurosci. Methods 2006, 158, 212–218. [Google Scholar]
- Karperien, A. FracLac for ImageJ. Version 2.0. 2007. Available online: https://imagej.nih.gov/ij/plugins/fraclac/fraclac-manual.pdf (accessed on 10 May 2022).
- Riffenburgh, R.H. Statistics in Medicine; Academic Press: London, UK, 1999. [Google Scholar]
- Vranes, V.; Milošević, N.T. Monofractal analysis of digital images of the human dentate nucleus: Correlation, classification and morphometric analysis. In Proceeding of Arithmetic Methods in Mathematical Physics and Biology; Banaszak, G., Milewski, J., Waliszewski, P., Eds.; University of Poznan: Bedlewo, Poland, 2018; p. 14.10. [Google Scholar]
- Favela, L.H.; Coey, C.A.; Griff, E.R.; Richardson, M.J. Fractal analysis reveals subclasses of neurons and suggests an explanation of their spontaneous activity. Neurosci. Lett. 2016, 626, 54–58. [Google Scholar] [CrossRef] [PubMed]
- Rowland, C.; Harland, B.; Smith, J.H.; Moslehi, S.; Dalrymple-Alford, J.; Taylor, R.P. Investigating Fractal Analysis as a Diagnostic Tool That Probes the Connectivity of Hippocampal Neurons. Front. Physiol. 2022, 23, 932598. [Google Scholar] [CrossRef] [PubMed]
- Di Ieva, A.; Grizzi, F.; Jelinek, H.F.; Pellionisz, A.J.; Losa, G.A. Fractals in the neurosciences, part I: General principles and basic neurosciences. Neuroscientist 2014, 20, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Losa, G.A.; Di Ieva, A.; Grizzi, F.; De Vico, G. On the fractal nature of nervous cell system. Front. Neuroanat. 2011, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Cornforth, D.; Jelinek, H.F. Monofractal and multifractal analysis for interpretation of function-structure relationships in finite size biological material. In Classification and Application of Fractals; Mitchell, E.W., Murray, E.R., Eds.; Nova Science Publishers: New York, NY, USA, 2011; pp. 1–22. [Google Scholar]
- Puškaš, N.; Zaletel, I.; Stefanović, B.D.; Ristanović, D. Fractal dimension of apical dendritic arborization differs in the superficial and the deep pyramidal neurons of the rat cerebral neocortex. Neurosci. Lett. 2015, 589, 88–91. [Google Scholar] [CrossRef] [PubMed]

| Size | Shape | Complexity | ||||||
|---|---|---|---|---|---|---|---|---|
| N | As (pix2) | Dbin | N | Pnf (pix) | Dout | N | Nm | Dskel |
| 5 | 15408 | 1.737 | 1 | 1184 | 1.094 | 2 | 6.5 | 1.137 |
| 13 | 16046 | 1.747 | 6 | 1487 | 1.068 | 8 | 7.6 | 1.113 |
| 12 | 16732 | 1.763 | 18 | 1677 | 1.060 | 12 | 8.5 | 1.113 |
| 13 | 17361 | 1.765 | 21 | 1891 | 1.062 | 25 | 9.5 | 1.137 |
| 14 | 18120 | 1.762 | 27 | 2120 | 1.070 | 20 | 10.6 | 1.149 |
| 14 | 18885 | 1.767 | 22 | 2279 | 1.075 | 21 | 11.5 | 1.149 |
| 15 | 19535 | 1.778 | 17 | 2494 | 1.074 | 23 | 12.4 | 1.155 |
| 14 | 20314 | 1.772 | 9 | 2705 | 1.068 | 13 | 13.4 | 1.166 |
| 14 | 20968 | 1.787 | 8 | 2905 | 1.070 | 8 | 14.4 | 1.173 |
| 11 | 21705 | 1.787 | 5 | 3093 | 1.058 | 3 | 15.3 | 1.154 |
| 7 | 22300 | 1.794 | 3 | 3298 | 1.064 | 3 | 17.3 | 1.218 |
| 7 | 22984 | 1.803 | 2 | 3621 | 1.088 | 1 | 18.2 | 1.250 |
| R | 0.604 | 0.173 | 0.423 | |||||
| p | <0.001 | 0.041 | <0.001 | |||||
| Parameters | CV (%) | m3 | m4 | p |
|---|---|---|---|---|
| Dbin | 1.41 | 0.429 | 0.797 | 0.062 |
| Dout | 1.50 | −0.097 | −0.432 | 0.482 |
| Dskel | 3.75 | −0.199 | 0.014 | 0.737 |
| Λ | 15.88 | −0.162 | 0.471 | 0.027 |
| Dbin | Dout | Dskel | Λ | |
|---|---|---|---|---|
| Group 1 (46 cells) | ||||
| Mean | 1.775 | 1.068 | 1.162 | 0.82 |
| SD | 0.026 | 0.016 | 0.040 | 0.10 |
| SE | 0.004 | 0.002 | 0.006 | 0.01 |
| Group 2 (44 cells) | ||||
| Mean | 1.774 | 1.067 | 1.157 | 0.79 |
| SD | 0.025 | 0.017 | 0.040 | 0.13 |
| SE | 0.004 | 0.002 | 0.006 | 0.02 |
| Group 3 (49 cells) | ||||
| Mean | 1.766 | 1.070 | 1.126 | 0.88 |
| SD | 0.023 | 0.016 | 0.041 | 0.15 |
| SE | 0.003 | 0.002 | 0.006 | 0.02 |
| Difference | / | / | *** 1–3; ** 2–3 | * 2–3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milošević, N. Monofractal Dimension in Quantifying the Image of Neurons in the Plane: Analysis of Image Features of Multipolar Neurons from the Principal Olivary Nucleus in Humans with Age. Fractal Fract. 2022, 6, 408. https://doi.org/10.3390/fractalfract6080408
Milošević N. Monofractal Dimension in Quantifying the Image of Neurons in the Plane: Analysis of Image Features of Multipolar Neurons from the Principal Olivary Nucleus in Humans with Age. Fractal and Fractional. 2022; 6(8):408. https://doi.org/10.3390/fractalfract6080408
Chicago/Turabian StyleMilošević, Nebojša. 2022. "Monofractal Dimension in Quantifying the Image of Neurons in the Plane: Analysis of Image Features of Multipolar Neurons from the Principal Olivary Nucleus in Humans with Age" Fractal and Fractional 6, no. 8: 408. https://doi.org/10.3390/fractalfract6080408
APA StyleMilošević, N. (2022). Monofractal Dimension in Quantifying the Image of Neurons in the Plane: Analysis of Image Features of Multipolar Neurons from the Principal Olivary Nucleus in Humans with Age. Fractal and Fractional, 6(8), 408. https://doi.org/10.3390/fractalfract6080408





