Abstract
The aim of this informative review was to investigate the application of radiomics in cancer imaging and to summarize the results of recent studies to support oncological imaging with particular attention to breast cancer, rectal cancer and primitive and secondary liver cancer. This review also aims to provide the main findings, challenges and limitations of the current methodologies. Clinical studies published in the last four years (2019–2022) were included in this review. Among the 19 studies analyzed, none assessed the differences between scanners and vendor-dependent characteristics, collected images of individuals at additional points in time, performed calibration statistics, represented a prospective study performed and registered in a study database, conducted a cost-effectiveness analysis, reported on the cost-effectiveness of the clinical application, or performed multivariable analysis with also non-radiomics features. Seven studies reached a high radiomic quality score (RQS), and seventeen earned additional points by using validation steps considering two datasets from two distinct institutes and open science and data domains (radiomics features calculated on a set of representative ROIs are open source). The potential of radiomics is increasingly establishing itself, even if there are still several aspects to be evaluated before the passage of radiomics into routine clinical practice. There are several challenges, including the need for standardization across all stages of the workflow and the potential for cross-site validation using real-world heterogeneous datasets. Moreover, multiple centers and prospective radiomics studies with more samples that add inter-scanner differences and vendor-dependent characteristics will be needed in the future, as well as the collecting of images of individuals at additional time points, the reporting of calibration statistics and the performing of prospective studies registered in a study database.
1. Introduction
Cancer presents an exclusive medical decision-making environment when considering its multiple forms during the disease course, the patient’s situation, available treatment options and treatment response. Technological developments in oncology imaging offer advantages in addressing the challenges associated with accurately detecting, characterizing and monitoring cancer, but conventional imaging assessment of cancer classically relies on visual assessments whose interpretations can be enhanced by innovative computational techniques. Radiomics promises major progress in the quantitative interpretation of images.
Radiomics is the analysis of medical images to obtain multiple quantitative data that cannot be identified by the human eye [1,2,3,4,5,6,7,8]. It provides insight into underlying pathophysiological phenomena not accessible to simple visual analysis.
Radiomics can be divided into two categories [7]: handcrafted radiomics and deep learning-based radiomics. The conventional radiomics workflow is typically based on extracting predesigned “features” (also referred to as handcrafted or engineered features) by a segmented region of interest (ROI). Nevertheless, recent advancements in deep learning have inspired trends toward deep learning-based radiomics (DLRs), which is also referred to as discovery radiomics.
In medicine, handcrafted radiomic models use data analytics to extract many features from medical images and is made up of several steps: (1) segmentation of the target lesion with manual segmentation by radiologists or with automatic and semi-automatic tools, (2) feature extraction to obtain multiple quantitative metrics and parameters from medical images, (3) feature selection with the aim of reducing the number of extracted features by avoiding correlated or redundant metrics, (4) analysis/classification by creating a predictive model using machine and deep learning approaches and (5) the validation of the results [9,10,11,12,13,14,15,16,17,18].
The extracted radiomics parameters can be morphological or statistical, and can be of first-order, second order and/or higher-order statistics. The morphological features characterize the target’s segmented lesion shape and its geometric features. Statistical features define the individual voxel values distribution, the associations between neighboring voxels allowing for extraction from medical image features linked to lesion heterogeneity and the quantification of successive voxels with equal intensities along certain directions. Higher order statistical metrics are acquired through the application of filters or mathematical transformations to the images [18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33].
Radiomics could be associated with clinical, pathological or genetic data to provide a model with predictive ability in order to offer a tailored precision medicine using these features as input of pattern recognition and artificial intelligence [34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59]. Currently, the main kind of artificial intelligence techniques that could be used are machine learning (ML) and deep learning (DL). MLs are widely used in medical imaging and have proven to be brilliant tools to assist general clinical cancer research [27,28] and could be used considering radiomics metrics as input data. However, some of the ML algorithms are not capable of using unstructured data. DL is the best technique for analyzing unstructured data built by multiple representation learning models on raw data [29,30,31,32,33]. The radiomics hypothesis is that different imaging features could be used in diagnosis, in prognosis predicting and therapeutic response in different cancer types. In fact, radiomics features provide data on the tumor microenvironment that can relate to histologic grade, prognosis, response to treatment and survival [22,23,24,25,26]. The automation brought by radiomics analysis and artificial intelligence models offers the opportunity to enhance the radiologists qualitative judgment, therefore improving tasks such as tumor detection, volumetry delineation, segmentation of lesions, linking intralesional imaging characteristics to genotypes and prediction of outcomes.
The aim of this review was to investigate the application of radiomics analysis in cancer imaging with the particular aim to summarize the results of recent studies to support oncological imaging, specifically in regards to breast cancer, rectal cancer and primitive and secondary liver cancer. Furthermore, we have proposed suggestions to increase reproducibility and robustness in radiomics applications.
2. Materials and Methods
2.1. Literature Search
This review resulted in a self-contained study without protocol and without a registration number.
To select the studies for this informative review, different electronic databases were considered, such as: “PubMed (US National Library of Medicine, http://www.ncbi.nlm.nih.gov/pubmed, accessed on 16 December 2022), Scopus (Elsevier, http://www.scopus.com/, accessed on 16 December 2022) and Web of Science (Thomson Reuters, http://apps.webofknowledge.com/ accessed on 16 December 2022)”.
Articles published in the last four years were analyzed since this time window (January 2019–December 2022) and are consistent with the most recent developments and trends in the use of radiomics in oncology. For the paper search, the following keywords were combined: radiomics AND/OR rectal cancer/tumor AND/OR breast cancer AND/OR liver cancer/tumor/metastasis. Exclusion criteria was: (1) articles of radiomics in other fields different from cancer imaging; (2) type of article as commentary, updated article, editorial letter, review article meta-analysis or case report; (3) articles without sufficient information for consideration or if the paper did not provide the number of cases analyzed, the partitioning of the dataset, the segmentation method, the radiometric features extracted, the statistical model to evaluate performance or the analysis of findings in a quantitative form. Moreover, papers that were not written in the English language were excluded. PRISMA checklist [34] was used. The research was conducted according to PICOS model (population; interventions; comparator group; outcomes; study design—Table 1).

Table 1.
Inclusion and exclusion criteria adopted to select studies according to the PICOS model.
2.2. Data Extraction and Quality Analysis
Papers were selected by two investigators with over fifteen years of experience in radiomics analysis in cancer imaging (V.G. and R.F.) according to a specific procedure represented in the Figure 1. The two investigators performed data extraction and then recorded the outcome, field of application, number of cases analyzed, partitioning of the dataset, segmentation method, radiometric features extracted, feature selection approach and statistical model used to evaluate the performance of the extracted features and the paper results.
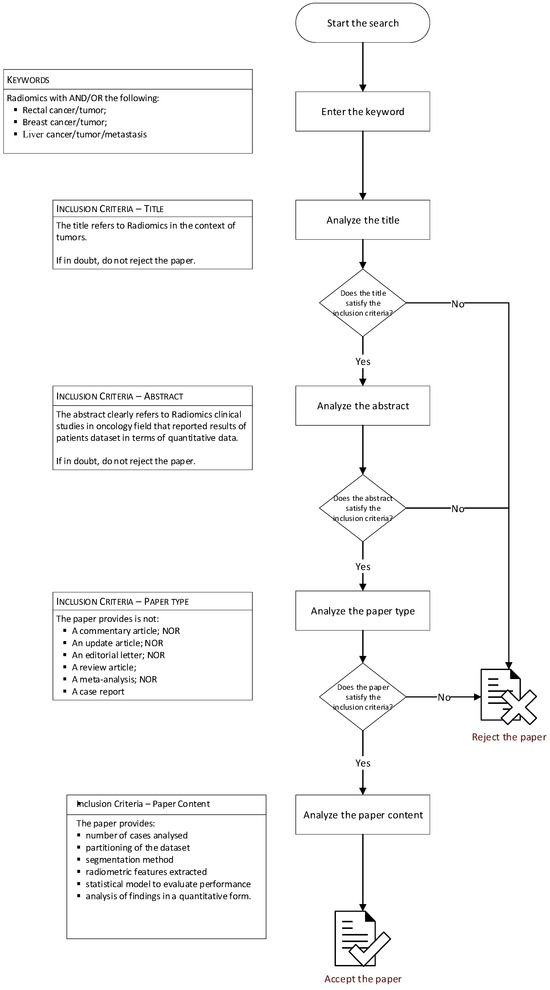
Figure 1.
Flowchart of research methods.
The methodological quality of each radiomics study was performed using the radiomic quality score (RQS) [35] by two different readers in consensus and by a third operator to solve disagreements between the two readers. The RQS includes 16 items that explore crucial steps of a radiomics pipeline: (1) image protocol quality; (2) multiple segmentations; (3) phantom study; (4) imaging at multiple time points; (5) feature reduction or adjustment for multiple testing; (6) multivariable analysis with non-radiomics features; (7) biologic correlates; (8) cut-off analyses; (9) discrimination statistics; (10) calibration statistics; (11) prospective studies registered in a trial database; (12) validation; (13) comparison to gold standard; (14) potential clinical applications; (15) cost-effectiveness analysis; (16) open science and data.
Each of these items has a different weight and can contribute positively or negatively in terms of points attributed, with −8 being the minimum and 36 being the maximum score that can be reached. The absolute score is then converted to a percentage value (with 36 = 100%). Figure 2 illustrates the values that can be attributed to the 16 items to obtain the RQS.
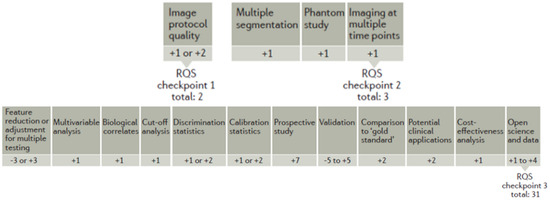
Figure 2.
Radiomic quality score illustration.
3. Results
Figure 3 shows a schematic representation of the included and excluded manuscripts. There were 591 articles analyzed. Of these articles, 142 were rejected because they did not consider the use of radiomics in the field of clinical oncology. Another 158 studies were excluded because they were commentary articles, update articles, editorial letters, review articles, meta-analyses or clinical cases. A further 102 studies were excluded due to insufficient data in methodology or results.
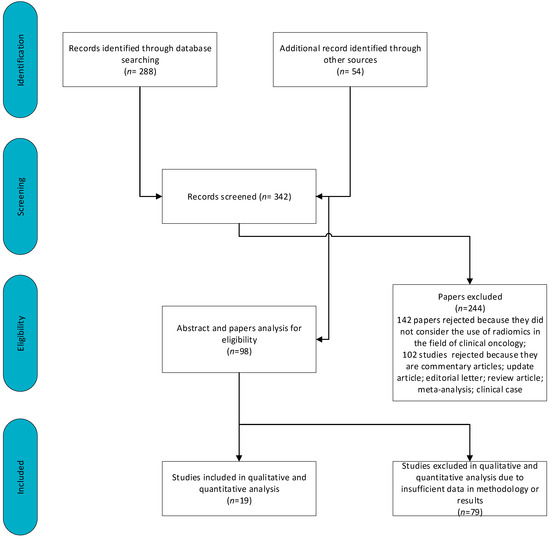
Figure 3.
Schematic representation of included and excluded papers.
Therefore, 19 manuscripts are included in this review. Table 2 reports the data collected by radiologists for these articles.

Table 2.
Radiomics quality score (RQS) assessment for all included articles.
The studies included used features calculated by several imaging modalities including computed tomography (CT), positron emission tomography/CT (PET/CT), magnetic resonance imaging (MRI) and contrast-enhanced mammography (CEM) with different objective such as differential diagnosis, prognosis prediction, therapy assessment, etc. [9,11,13,22,23,36,37,38,39,40,46,47,48,49,50,53,54,58,59].
Table 1 reported the RQS assessment for each included study. The median RQS score was 15, which translates to 41.67% of the ideal score of 36. The lowest score was 8, which translates to 22.22% of the ideal quality score. Compared to the ideal score, the RQS of the following studies were the lowest in imaging at multiple time points: phantom study on all scanners, calibration statistics and cost-effectiveness analysis (0%), followed by multivariable analysis with non-radiomics features, prospective study registered in a trial database and validation. Seven studies with high score of seventeen (47.22% of the ideal quality score) [9,11,13,36,37,40,54]) earned additional points by using validation steps considering two datasets from two distinct institutes and open science and data domains (radiomics features calculated on a set of representative ROIs are open source).
3.1. Radiomics Studies in Rectal Cancer
Radiomics analysis in rectal cancer was used to assess and predict chemo-radiation therapy in locally advanced rectal cancer (LARC) patients using an MRI. It was also used in the prognosis prediction.
The five included studies in rectal cancer were retrospective studies (Table 3). The lowest score was 13, which is 36.11% of the ideal quality score. The highest score was 15, which is 41.67% of the ideal quality score. None of the included studies detected inter-scanner differences and vendor-dependent features, collected images of individuals at additional time points, reported calibration statistics, performed prospective studies registered in a trial database, performed a cost-effectiveness analysis report on the cost-effectiveness of the clinical application or made the code and data publicly available.

Table 3.
Radiomics Clinical Studies in rectal cancer.
Xue et al. [46] demonstrated that the integrated model based on T2 weighted imaging and apparent diffusion coefficient maps had the potential for preoperative immunoscore expectations in rectal cancer. They found a model based on T2-weighted imaging and apparent diffusion coefficient images in the prognosis prediction and in the individualized immunotherapy, guiding the integrated model showed in the validation cohort an AUC of 0.768.
Chiloiro et al. [47] demonstrated that radiomics analysis achieved a good performance in identifying complete responders in rectal cancer and demonstrated that the diagnostic performance of radiomics improves when combined with standard clinical evaluation. Three models were produced: a radiomics model, a multidisciplinary tumor board model and a combined model that predicted with AUCs of 0.82, 0.73 and 0.84—the complete pathological response.
Cusumano et al. [48] investigated a MR radiomics model to detect complete pathologic response in LARC showing good performances both using 1.5 T and 3 T scanners. The predictive model AUC applied to the whole data set was 0.72, while values of 0.70 and 0.83 were obtained when the patient subgroups obtained with 1.5 T and 3 T MRI scanners were considered. Chiloiro et al. [49] supported a possible role of delta radiomics in predicting following occurrences of distant metastasis in patients with LARC. A logistic regression proved to be the best performing one with a testing set that balanced accuracy, sensitivity and specificity of 78.5%, 71.4% and 85.7%, respectively, to predict distant metastasis.
Chen et al. [50] reported that pre chemo radiation therapy MRI, post chemo radiation therapy MRI and delta radiomics-based models could predict tumor responses in LARC. The GLRLM-GLN calculated before therapy was able to classify pathological complete response groups with an accuracy at 88.5% on the training set and of 57.1% on the test set. When combined with 3D diameter, the accuracy increased on training data to 92.3%. The best predictors for a good response were the pre-global minimum combined with the clinical N stage in the multivariate analysis that obtained an accuracy of 100% on training and test sets.
The findings of these studies suggest that radiomics has the potential to provide valuable information in evaluating therapy and predicting prognosis in rectal cancer. However, there are still challenges in terms of standardization of imaging protocols, feature extraction and validation of radiomics models. Further research is needed to validate the clinical utility of radiomics and to establish its role in routine clinical practice.
3.2. Radiomics Studies in Breast Cancer
The usefulness of radiomics in distinguishing malignant from benign breast lesions as well as in predicting histopathological type, estimating tumor grade and assisting the staging procedure was explored in this manuscript. Therefore, the application of radiomics strategies as prediction tools for treatment response will be explored alongside the risk of recurrence.
The five included studies in breast cancer were conducted retrospectively (Table 4). The lowest score was 11, which is 30.56% of the ideal quality score, and the highest score was 17, which is 47.22% of the ideal quality score obtained published as open source extracted radiomics features [54]. None of the included studies detected inter-scanner differences and vendor-dependent features, collected images of individuals at additional time points, reported calibration statistics, performed prospective study registered in a trial database or performed an analysis report on the cost-effectiveness of the clinical application.

Table 4.
Radiomics Clinical Studies in breast cancer.
Fusco et al. [23] evaluated the possibility of using radiomics metrics by CEM and dynamic contrast enhanced MRI in the benign and malignant breast lesion discrimination through different classifiers performing balancing and feature selection procedures. The best performance was obtained considering 18 robust characteristics and a linear discriminant analysis with a precision of 0.84 and an AUC of 0.88.
Tsuchiya et al. [53] assessed the MRI-based radiomics model to differentiate phyllodes breast tumors from fibroadenomas, investigating several machine models. A support vector machine reached the best AUC of 0.96, and the combined model, which was constructed using both radiomics features and radiological features, had a significantly improved performance in the validation set (AUC of 0.97).
Petrillo et al. [54] used the CEM and the radiomics analysis in the classification of suspicious breast lesions and performed both univariate analysis and multivariate analysis to investigate the better approach and the higher accuracy in the classification of malignant and benign lesions. At univariate analysis, the best accuracy in the differentiation of benign and malignant breast lesions was obtained using the original_gldm_DependenceNonUniformity with an accuracy of 89%, while in the classification of the hormone receptor presence, a lower level of accuracy was found (81.65%). For multivariate analysis using features extracted from cranio-caudal images, the maximum test accuracy in the malignant and benign lesion differentiation was 96% with logistic regression. For features extracted from mediolateral oblique images, the best test accuracy was 92% and was always in the classification of breast lesions obtained using a classification tree algorithm.
Feng et al. [58] demonstrated that a radiomics feature set combining three DCE-MRI parametric maps and ADC maps yielded an area under the ROC curve of 0.839 within the training set and 0.795 within the independent validation set in breast cancer KI-67 determination.
Wang et al. [59] constructed a radiomics score significantly associated with disease-free survival (DFS) for locally advanced breast cancer (LABC) patients in training cohorts, validation cohorts and external validation cohorts (p < 0.001, p = 0.014 and p = 0.041, respectively). The radiomics-based nomogram showed better predictive performance of DFS compared with the TNM model. They demonstrated that radiomics scores could effectively predict the prognosis of LABC after neoadjuvant chemotherapy and radiotherapy.
3.3. Radiomics Studies in Liver Primitive and Secondary Cancer
The main potential applications of radiomic models in liver primitive and secondary carcinoma are to predict histology, predict response to treatment, predict genetic signature, predict recurrence and predict survival.
The nine included studies in liver primitive and secondary cancer were retrospective studies (Table 5). The lowest score was 8, which is 22% of the ideal quality score. The highest score was 17, which is 47.22% of the ideal quality score. None of the included studies detected inter-scanner differences and vendor-dependent features, collected images of individuals at additional time points, reported calibration statistics, performed prospective study registered in a trial database or performed an analysis report on the cost-effectiveness of the clinical application. However, the studies [9,11,13,36,37,40] earned additional points by using multivariable analysis with non-radiomics features or validation steps considering two datasets from two distinct institutes or open science and data domain published extracted radiomics features.

Table 5.
Radiomics Clinical Studies in Liver primitive and secondary cancer.
Granata et al. [9] demonstrated that radiomics and machine learning analysis, based on the Gd-EOB-DTPA-enhanced magnetic resonance imaging (EOB-MRI) study, allow the identification of several biomarkers for detection of the different growth patterns in colorectal cancer liver metastases. Study [11] reported that using univariate analysis was not possible to accurately discriminate the RAS mutation status. Instead, considering a multivariate analysis and classification approaches, a k-nearest neighbors (KNN) exclusively with texture parameters as predictors achieved the best results (an accuracy of 87.5% with 91.7% of sensitivity and 83.3% of specificity on external validation cohort).
In another study, Granata et al. [13] confirmed that radiomics data can be used to detect several features that may have an impact on the treatment choice for patients with liver metastases, obtaining a more tailored approach. The radiomics metric Wavelet_HHL_glcm_Imc2 alone showed the best accuracy in discriminating between expansive and infiltrative tumor growth equal to 79%. Wavelet_LLL_firstorder_Mean showed the best accuracy in budding tumor detection equal to 86%, Original_firstorder_RobustMeanAbsoluteDeviation showed the best accuracy in identifying mucinous tumor types equal to 88% and Wavelet_HLH_glcm_Idmn showed the best accuracy in identifying tumor recurrence equal to 85%. The best linear regression model was achieved in the recurrence detection, combining linearly 16 radiomics metrics (accuracy of 97%). However, the best results were reached in the tumor front growth detection, combining seven radiomics features with an accuracy of 97%, a sensitivity of 90% and specificity of 100%. In addition, Granata et al. [36] investigated radiomics and ML approaches in the mucinous colorectal liver metastases evaluation by MRI, demonstrating that radiomics metrics could permit the characterization of the lesion subtype with a more tailored therapeutic approach. They showed that the best performance was obtained by T2-weighted combining linearly radiomics features using a linear regression (accuracy of 94%). Moreover, Granata et al. [37] demonstrated that the best performance in the discrimination of tumor budding was obtained by a KNN considering four radiomics predictors by T2-weighted MRI, yielding an accuracy of 93%, a sensitivity of 81% and a specificity of 97%. In all studies [9,13,36,37] the authors used multiple segmentations from different radiologists, an external validation dataset, detected and discussed biologic correlates and published as open data extracted radiomics features.
Yang et al. [22] established a predictive integrated model for early recurrence of hepato-cellular carcinoma (HCC) after ablation, and the model presented good predictive performance. Multivariate analyses suggested that the rad-score including four radiomics features, number of lesions, integrity of the capsule, pathological type and alpha-fetoprotein were independent influencing factors of HCC recurrence. The AUC of predicting early recurrence at 1, 2 and 3 years in the validation group was 0.72 (95% CI: 0.58–0.84), 0.61 (95% CI: 0.45–0.78) and 0.64 (95% CI: 0.40–0.87).
Gao et al. [38] investigated the ability of radiomics and deep features by MRI in the identification of a predictive model for the early recurrence of HCC after surgery. The authors integrated radiomics and deep features into a combined model and demonstrated improved performance in the detection of patients at high risk of early recurrence (area under curve (AUC) 0.840, accuracy 77.7%).
De Robertis et al. [39] demonstrated that CT texture analysis of pancreatic adenocarcinoma could identify features able to predict the liver metastases likelihood. This study included 220 patients. Tumor size, arterial HU_MAX, arterial GLZLM_SZHGE and portal GLCM_CORRELATION were significant predictors of the likelihood of liver metastases, with odds ratios of 1.1, 0.9, 1 and 1.49, respectively.
Shi et al. [40] reported that RAS and BRAF mutated tumors show discriminatory CT radiomic features which, if combined with semantic features, could help in the detection of tumors harboring RAS and BRAF mutations in patients with colorectal liver metastasis in order to increase patient stratification and customized treatments. The combined score of radiomic and semantic features could discriminate between wild-type and mutant patients with an AUC = 0.95 in primary cohorts and =0.79 in validation cohorts.
Radiomics studies have demonstrated its predictive value such as the grade of hepatocellular carcinoma, the ability to predict recurrence or the differential diagnosis of other primary or secondary liver tumors and the correlation with genetic mutations. However, once again the added value of radiomics in modifying therapeutic choices, and therefore as a decision-making model, is not clear.
4. Discussion
This manuscript summarizes recent radiomics studies in cancer imaging and discuss the challenges and limitations of the methodologies employed.
Radiomics looks to be very promising, as it has been applied in oncology to improve diagnosis and prognosis with the intention of helping the clinician, and is increasingly oriented towards precision medicine. The foundation on which radiomics is built is that imaging data indirectly carry significant information about tumor biology, behavior and pathophysiology, and can provide information that would otherwise not be apparent to purely visual radiological interpretation.
Radiomics presupposes an alternative non-invasive tool for characterizing tumors, which has experienced growing interest with the advent of more powerful and more sophisticated computer machine learning algorithms. However, the incorporation of radiomics into cancer clinical decision support systems still needs in-depth analysis of its relationship with tumor biology.
Moreover, there were many differences in the methods used in image segmentation, feature extraction and prediction model construction. Furthermore, some important aspects have not always been considered by the authors such as the importance of the external validation of the set in the evaluation of the intra and inter-observer variability and in the balancing of the data set. In fact, the critical problems in radiomics use are the insufficient standardization and generalization of radiomics results, data quality control, repeatability, reproducibility, database matching and model overfitting issues [55,57].
A key attention is to determine the availability of sufficient data to support the development of a radiomics signature. As a rule, for binary classification studies, one should aim to obtain at least 10–15 samples for each feature that is provided in the final radiomics signature [55,57].
In medicine, two different approaches can be applied to cancer imaging: radiomic features extracted from the target lesion that can be used as inputs for machine learning algorithms, or an entire medical image or a series of images to train a deep learning model to directly perform tumor detection, characterization and monitoring [60,61,62,63,64,65,66,67,68,69].
This review reported that radiomics metrics can extract biological and path-physiological evidence from target lesions from images, and the equivalent quantitative features can offer a precise non-invasive biomarker for oncological diagnosis, prognosis and outcome monitoring. Artificial intelligence has been used in conjunction with radiomics features to solve difficult problems that would have been intractable using traditional statistical approaches. In addition, artificial intelligence-based methods have made important advancements in the field of radiological oncological medical imaging [70,71].
However, great variability has been observed in methodologies for radiomics extraction, reduction and classification models. This aspect influences the reproducibility and generalizability of the results and a large level of variability can be observed in the scientific reports of the different authors. Many authors have not partitioned the data set. In general, the dataset should be divided into training datasets (70% of samples), test datasets (20% of samples) and validation datasets (10% of samples) [71]. Furthermore, the samples should also include external datasets to better validate the results of the radiomics and artificial intelligence procedures [72,73,74,75].
In addition, another major problem is the repeatability and replicability of artificial intelligence and radiomics techniques [76]. This problem is mainly linked to the variability of the image acquisition equipment and of the protocol itself, as well as to the variability of the reconstruction and pre-processing techniques of the images used, for example, to optimize the signal-to-noise ratio. Furthermore, image segmentation or feature extraction methods are not absolute and are problematic to apply to be completely standardized, which means that the implementation of several tasks simultaneously with deep learning and machine learning methods is still restricted [76,77,78,79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101].
To resolve this and to construct reproducible and standardized radiomics features extraction and building models, the following aspects should be considered: (1) to obtain reproducible radiomics features, image biomarker standardization initiative (IBSI) [77], a radiomics standardization initiative of the international community, should be considered and a common tool for feature extraction should be used to avoid lack of robustness and reproducibility in the steps of definition, implementation and pre-processing of images of radiomics features. (2) To build robust and generalizable models, the data can be enlarged by shearing, rotating or inverting original images.
Also, it is important to check if the data is balanced. When the proportions between classes are unequal in a classification problem, the data can be severely biased and a larger sample size may be required for the developed model to be generalizable. Data balancing techniques that include creating artificial data with algorithms such as the synthetic minority oversampling technique may be considered [40,78,79,80]. However, only a few papers have considered balancing techniques to reduce the unbalanced dataset problem [23].
In addition to the technical, regulatory and ethical requirements, problems need to be evaluated. As any big data project requires access to huge data sets, the collection of critical issues such as patient privacy and informed permission need to be addressed. This is critical not only by a medico-legal point of view, but also by a “human” point of view, because, like Coppola et al. [81] stressed, we must not superintend the meaning of the irreplaceable doctor–patient bond. Connecting doctors and patients directly will always be an important phase of healthcare services that artificial intelligence can never replace [2,20,68,102,103,104,105,106,107,108,109].
Among the studies analyzed, none assessed the differences between scanners and vendor-dependent characteristics, collected images of individuals at additional points in time, performed calibration statistics, represented a prospective study performed registered in a study database, conducted an analysis report on the cost-effectiveness of the clinical application or reported multivariable analysis with also non radiomics features. Seven studies reached a high score of 17 [9,11,13,36,37,40,54]) and earned additional points by using validation step considering two datasets from two distinct institutes and open science and data domains (radiomics features calculated on a set of representative ROIs are open source).
Our study has some limitations that should be taken into consideration. First, all studies included in this meta-analysis were retrospective in study design, which was subject to selection bias, underscoring the need for prospective validation. Second, in the absence of direct comparisons between radiomics models and other scoring or non-radiomics models, it is difficult to draw a conclusion that radiomics models are superior to other non-radiomics models. However, radiomics studies have had marked heterogeneity in their workflow. In the future, it will be necessary to establish and promote an imaging data acquisition protocol, standardize the research workflow, and conduct prospective multicenter quality control studies. Furthermore, combining radiomics with multiomics could lead to a breakthrough in the individualized medical treatment of tumors.
5. Conclusions
In conclusion, this research topic has involved many works which have made full use of radiomics in cancer imaging, but even in this case many aspects regarding the methodologies used should be considered. The potential of radiomics is becoming increasingly established, although there are still several aspects to be evaluated before the transition to routine clinical practice.
There are several challenges to address, including the need for standardization at all stages of workflow and the potential for cross-site validation using heterogeneous real-world datasets. Furthermore, multiple centers and prospective radiomics studies with more samples that add inter-scanner differences and vendor-dependent characteristics will be needed in the future, along with collected images of individuals at additional time points, reported calibration statistics and performed prospective studies registered in a study database.
Author Contributions
R.F. and V.G. wrote the initial manuscript and revised it. All authors defined the methodology, revised the manuscript for intellectual content and approved the final manuscript as submitted. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Italian Ministry of Health Ricerca Corrente funds.
Data Availability Statement
Data are available at link https://zenodo.org/records/10477650 (accessed on 13 November 2023).
Acknowledgments
The authors are grateful to Alessandra Trocino, librarian at the National Cancer Institute of Naples, Italy.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.T.H.M.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- He, X.; Li, K.; Wei, R.; Zuo, M.; Yao, W.; Zheng, Z.; He, X.; Fu, Y.; Li, C.; An, C.; et al. A multitask deep learning radiomics model for predicting the macrotrabecular-massive subtype and prognosis of hepatocellular carcinoma after hepatic arterial infusion chemotherapy. Radiol. Med. 2023, 128, 1508–1520. [Google Scholar] [CrossRef]
- Limkin, E.J.; Sun, R.; Dercle, L.; Zacharaki, E.I.; Robert, C.; Reuzé, S.; Schernberg, A.; Paragios, N.; Deutsch, E.; Ferté, C. Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann. Oncol. 2017, 28, 1191–1206. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-J.; Wu, F.-Z.; Yang, S.-C.; Tang, E.-K.; Liang, C.-H. Radiomics in Early Lung Cancer Diagnosis: From Diagnosis to Clinical Decision Support and Education. Diagnostics 2022, 12, 1064. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Simone, C.B., II; Krishnan, S.; Lin, S.H.; Yang, J.; Hahn, S.M. The Rise of Radiomics and Implications for Oncologic Management. J. Natl. Cancer Inst. 2017, 1, 109. [Google Scholar] [CrossRef] [PubMed]
- Committeri, U.; Fusco, R.; Di Bernardo, E.; Abbate, V.; Salzano, G.; Maglitto, F.; Orabona, G.D.; Piombino, P.; Bonavolontà, P.; Arena, A.; et al. Radiomics Metrics Combined with Clinical Data in the Surgical Management of Early-Stage (cT1–T2 N0) Tongue Squamous Cell Carcinomas: A Preliminary Study. Biology 2022, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Afshar, P.; Mohammadi, A.; Plataniotis, K.N.; Oikonomou, A.; Benali, H. From Handcrafted to Deep-Learning-Based Cancer Radiomics: Challenges and Opportunities. IEEE Signal Process. Mag. 2019, 36, 132–160. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Lin, G.; Pandey, S.; Yeh, C.-H.; Wang, J.-J.; Lin, C.-Y.; Ho, T.-Y.; Ko, S.-F.; Ng, S.-H. Fully automated segmentation and radiomics feature extraction of hypopharyngeal cancer on MRI using deep learning. Eur. Radiol. 2023, 33, 6548–6556. [Google Scholar] [CrossRef] [PubMed]
- Granata, V.; Fusco, R.; De Muzio, F.; Cutolo, C.; Raso, M.M.; Gabelloni, M.; Avallone, A.; Ottaiano, A.; Tatangelo, F.; Brunese, M.C.; et al. Radiomics and Machine Learning Analysis Based on Magnetic Resonance Imaging in the Assessment of Colorectal Liver Metastases Growth Pattern. Diagnostics 2022, 12, 1115. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Barretta, M.L.; Picone, C.; Avallone, A.; Belli, A.; Patrone, R.; Ferrante, M.; Cozzi, D.; Grassi, R.; et al. Radiomics in hepatic metastasis by colorectal cancer. Infect. Agents Cancer 2021, 16, 39. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Avallone, A.; De Stefano, A.; Ottaiano, A.; Sbordone, C.; Brunese, L.; Izzo, F.; Petrillo, A. Radiomics-Derived Data by Contrast Enhanced Magnetic Resonance in RAS Mutations Detection in Colorectal Liver Metastases. Cancers 2021, 13, 453. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Catalano, O.; Avallone, A.; Palaia, R.; Botti, G.; Tatangelo, F.; Granata, F.; Cascella, M.; Izzo, F.; et al. Diagnostic accuracy of magnetic resonance, computed tomography and contrast enhanced ultrasound in radiological multimodality assessment of peribiliary liver metastases. PLoS ONE 2017, 12, e0179951. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Setola, S.V.; De Muzio, F.; Aversana, F.D.; Cutolo, C.; Faggioni, L.; Miele, V.; Izzo, F.; Petrillo, A. CT-Based Radiomics Analysis to Predict Histopathological Outcomes Following Liver Resection in Colorectal Liver Metastases. Cancers 2022, 14, 1648. [Google Scholar] [CrossRef] [PubMed]
- Alongi, P.; Rovera, G.; Stracuzzi, F.; Popescu, C.E.; Minutoli, F.; Arnone, G.; Baldari, S.; Deandreis, D.; Caobelli, F. Artificial Intelligence in Breast Cancer: A Systematic Review on PET Imaging Clinical Applications. Curr. Med. Imaging 2023, 19, 832–843. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Yang, J.; Shen, N.; Xu, Q.; Zhao, Q. Artificial intelligence in lung cancer diagnosis and prognosis: Current application and future perspective. Semin. Cancer Biol. 2023, 89, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Weng, Y.; Wang, B.; Li, Z. Natural Language Processing Applications for Computer-Aided Diagnosis in Oncology. Diagnostics 2023, 13, 286. [Google Scholar] [CrossRef]
- Derevianko, A.; Pizzoli, S.F.M.; Pesapane, F.; Rotili, A.; Monzani, D.; Grasso, R.; Cassano, E.; Pravettoni, G. The Use of Artificial Intelligence (AI) in the Radiology Field: What Is the State of Doctor–Patient Communication in Cancer Diagnosis? Cancers 2023, 15, 470. [Google Scholar] [CrossRef]
- Wang, L.; Chen, X.; Zhang, L.; Li, L.; Huang, Y.B.; Sun, Y.; Yuan, X. Artificial intelligence in clinical decision support systems for oncology. Int. J. Med. Sci. 2023, 20, 79–86. [Google Scholar] [CrossRef]
- Wei, J.; Jiang, H.; Gu, D.; Niu, M.; Fu, F.; Han, Y.; Song, B.; Tian, J. Radiomics in liver diseases: Current progress and future opportunities. Liver Int. 2020, 40, 2050–2063. [Google Scholar] [CrossRef] [PubMed]
- Hajianfar, G.; Haddadi Avval, A.; Hosseini, S.A.; Nazari, M.; Oveisi, M.; Shiri, I.; Zaidi, H. Time-to-event overall survival prediction in glioblastoma multiforme patients using magnetic resonance imaging radiomics. Radiol. Med. 2023, 128, 1521–1534. [Google Scholar] [CrossRef]
- Saini, A.; Breen, I.; Pershad, Y.; Naidu, S.; Knuttinen, M.G.; Alzubaidi, S.; Sheth, R.; Albadawi, H.; Kuo, M.; Oklu, R. Radiogenomics and Radiomics in Liver Cancers. Diagnostics 2018, 9, 4. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yuan, C.; Zhang, Y.; Li, K.; Wang, Z. Predicting hepatocellular carcinoma early recurrence after ablation based on magnetic resonance imaging radiomics nomogram. Medicine 2022, 101, e32584. [Google Scholar] [CrossRef]
- Fusco, R.; Di Bernardo, E.; Piccirillo, A.; Rubulotta, M.R.; Petrosino, T.; Barretta, M.L.; Raso, M.M.; Vallone, P.; Raiano, C.; Di Giacomo, R.; et al. Radiomic and Artificial Intelligence Analysis with Textural Metrics Extracted by Contrast-Enhanced Mammography and Dynamic Contrast Magnetic Resonance Imaging to Detect Breast Malignant Lesions. Curr. Oncol. 2022, 29, 1947–1966. [Google Scholar] [CrossRef] [PubMed]
- Alahmari, S.S.; Cherezov, D.; Goldgof, D.B.; Hall, L.O.; Gillies, R.J.; Schabath, M.B. Delta Radiomics Improves Pulmonary Nodule Malignancy Prediction in Lung Cancer Screening. IEEE Access 2018, 6, 77796–77806. [Google Scholar] [CrossRef] [PubMed]
- Mei, D.; Luo, Y.; Wang, Y.; Gong, J. CT texture analysis of lung adenocarcinoma: Can Radiomic features be surrogate biomarkers for EGFR mutation statuses. Cancer Imaging 2018, 18, 52. [Google Scholar] [CrossRef] [PubMed]
- Granata, V.; Fusco, R.; Maio, F.; Avallone, A.; Nasti, G.; Palaia, R.; Albino, V.; Grassi, R.; Izzo, F.; Petrillo, A. Qualitative assessment of EOB-GD-DTPA and Gd-BT-DO3A MR contrast studies in HCC patients and colorectal liver metastases. Infect. Agents Cancer 2019, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-G.; Jun, S.; Cho, Y.-W.; Lee, H.; Kim, G.B.; Seo, J.B.; Kim, N. Deep learning in medical imaging: General overview. Korean J. Radiol. 2017, 18, 570–584. [Google Scholar] [CrossRef]
- Kourou, K.; Exarchos, T.P.; Exarchos, K.P.; Karamouzis, M.V.; Fotiadis, D.I. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 2015, 13, 8–17. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Alongi, P.; Stefano, A.; Comelli, A.; Laudicella, R.; Scalisi, S.; Arnone, G.; Barone, S.; Spada, M.; Purpura, P.; Bartolotta, T.V.; et al. Radiomics analysis of 18F-Choline PET/CT in the prediction of disease outcome in high-risk prostate cancer: An explorative study on machine learning feature classification in 94 patients. Eur. Radiol. 2021, 31, 4595–4605. [Google Scholar] [CrossRef]
- Vernuccio, F.; Arnone, F.; Cannella, R.; Verro, B.; Comelli, A.; Agnello, F.; Stefano, A.; Gargano, R.; Rodolico, V.; Salvaggio, G.; et al. Diagnostic performance of qualitative and radiomics approach to parotid gland tumors: Which is the added benefit of texture analysis? Br. J. Radiol. 2021, 94, 20210340. [Google Scholar] [CrossRef] [PubMed]
- Fusco, R.; Sansone, M.; Granata, V.; Grimm, R.; Pace, U.; Delrio, P.; Tatangelo, F.; Botti, G.; Avallone, A.; Pecori, B.; et al. Diffusion and perfusion MR parameters to assess preoperative short-course radiotherapy response in locally advanced rectal cancer: A comparative explorative study among Standardized Index of Shape by DCE-MRI, intravoxel incoherent motion- and diffusion kurtosis imaging-derived parameters. Abdom. Imaging 2018, 44, 3683–3700. [Google Scholar] [CrossRef]
- Shafiq-Ul-Hassan, M.; Zhang, G.G.; Latifi, K.; Ullah, G.; Hunt, D.C.; Balagurunathan, Y.; Abdalah, M.A.; Schabath, M.B.; Goldgof, D.G.; Mackin, D.; et al. Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels. Med. Phys. 2017, 44, 1050–1062. [Google Scholar] [CrossRef] [PubMed]
- Available online: http://prisma-statement.org/prismastatement/checklist.aspx (accessed on 16 December 2022).
- Moore, C.; Murphy, A. Radiomics Quality Score. Reference Article. Available online: https://www.radiomics.world/rqs (accessed on 23 June 2023).
- Granata, V.; Fusco, R.; De Muzio, F.; Cutolo, C.; Setola, S.V.; Dell’aversana, F.; Grassi, F.; Belli, A.; Silvestro, L.; Ottaiano, A.; et al. Radiomics and machine learning analysis based on magnetic resonance imaging in the assessment of liver mucinous colorectal metastases. Radiol. Med. 2022, 127, 763–772. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; De Muzio, F.; Cutolo, C.; Setola, S.V.; Grassi, R.; Grassi, F.; Ottaiano, A.; Nasti, G.; Tatangelo, F.; et al. Radiomics textural features by MR imaging to assess clinical outcomes following liver resection in colorectal liver metastases. Radiol. Med. 2022, 127, 461–470. [Google Scholar] [CrossRef]
- Gao, W.; Wang, W.; Song, D.; Yang, C.; Zhu, K.; Zeng, M.; Rao, S.-X.; Wang, M. A predictive model integrating deep and radiomics features based on gadobenate dimeglumine-enhanced MRI for postoperative early recurrence of hepatocellular carcinoma. Radiol. Med. 2022, 127, 259–271. [Google Scholar] [CrossRef]
- De Robertis, R.; Geraci, L.; Tomaiuolo, L.; Bortoli, L.; Beleù, A.; Malleo, G.; D’onofrio, M. Liver metastases in pancreatic ductal adenocarcinoma: A predictive model based on CT texture analysis. Radiol. Med. 2022, 127, 1079–1084. [Google Scholar] [CrossRef]
- Shi, R.; Chen, W.; Yang, B.; Qu, J.; Cheng, Y.; Zhu, Z.; Gao, Y.; Wang, Q.; Liu, Y.; Li, Z.; et al. Prediction of KRAS, NRAS and BRAF status in colorectal cancer patients with liver metastasis using a deep artificial neural network based on radiomics and semantic features. Am. J. Cancer Res. 2020, 10, 4513–4526. [Google Scholar]
- Chiti, G.; Grazzini, G.; Flammia, F.; Matteuzzi, B.; Tortoli, P.; Bettarini, S.; Pasqualini, E.; Granata, V.; Busoni, S.; Messserini, L.; et al. Gastroenteropancreatic neuroendocrine neoplasms (GEP-NENs): A radiomic model to predict tumor grade. Radiol. Med. 2022, 127, 928–938. [Google Scholar] [CrossRef]
- Wang, F.-H.; Zheng, H.-L.; Li, J.-T.; Li, P.; Zheng, C.-H.; Chen, Q.-Y.; Huang, C.-M.; Xie, J.-W. Prediction of recurrence-free survival and adjuvant therapy benefit in patients with gastrointestinal stromal tumors based on radiomics features. Radiol. Med. 2022, 127, 1085–1097. [Google Scholar] [CrossRef]
- Masci, G.M.; Ciccarelli, F.; Mattei, F.I.; Grasso, D.; Accarpio, F.; Catalano, C.; Laghi, A.; Sammartino, P.; Iafrate, F. Role of CT texture analysis for predicting peritoneal metastases in patients with gastric cancer. Radiol. Med. 2022, 127, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Palatresi, D.; Fedeli, F.; Danti, G.; Pasqualini, E.; Castiglione, F.; Messerini, L.; Massi, D.; Bettarini, S.; Tortoli, P.; Busoni, S.; et al. Correlation of CT radiomic features for GISTs with pathological classification and molecular subtypes: Preliminary and monocentric experience. Radiol. Med. 2022, 127, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Yu, N.; Yu, Y.; He, T.; Duan, X. Performance of CT radiomics in predicting the overall survival of patients with stage III clear cell renal carcinoma after radical nephrectomy. Radiol. Med. 2022, 127, 837–847. [Google Scholar] [CrossRef]
- Xue, K.; Liu, L.; Liu, Y.; Guo, Y.; Zhu, Y.; Zhang, M. Radiomics model based on multi-sequence MR images for predicting preoperative immunoscore in rectal cancer. Radiol. Med. 2022, 127, 702–713. [Google Scholar] [CrossRef]
- Chiloiro, G.; Cusumano, D.; de Franco, P.; Lenkowicz, J.; Boldrini, L.; Carano, D.; Barbaro, B.; Corvari, B.; Dinapoli, N.; Giraffa, M.; et al. Does restaging MRI radiomics analysis improve pathological complete response prediction in rectal cancer patients? A prognostic model development. Radiol. Med. 2022, 127, 11–20. [Google Scholar] [CrossRef]
- Cusumano, D.; Meijer, G.; Lenkowicz, J.; Chiloiro, G.; Boldrini, L.; Masciocchi, C.; Dinapoli, N.; Gatta, R.; Casà, C.; Damiani, A.; et al. A field strength independent MR radiomics model to predict pathological complete response in locally advanced rectal cancer. Radiol. Med. 2021, 126, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Chiloiro, G.; Rodriguez-Carnero, P.; Lenkowicz, J.; Casà, C.; Masciocchi, C.; Boldrini, L.; Cusumano, D.; Dinapoli, N.; Meldolesi, E.; Carano, D.; et al. Delta Radiomics Can Predict Distant Metastasis in Locally Advanced Rectal Cancer: The Challenge to Personalize the Cure. Front. Oncol. 2020, 10, 595012. [Google Scholar] [CrossRef]
- Chen, H.; Shi, L.; Nguyen, K.N.B.; Monjazeb, A.M.; Matsukuma, K.E.; Loehfelm, T.W.; Huang, H.; Qiu, J.; Rong, Y. MRI Radiomics for Prediction of Tumor Response and Downstaging in Rectal Cancer Patients after Preoperative Chemoradiation. Adv. Radiat. Oncol. 2020, 5, 1286–1295. [Google Scholar] [CrossRef]
- Cozzi, D.; Bicci, E.; Cavigli, E.; Danti, G.; Bettarini, S.; Tortoli, P.; Mazzoni, L.N.; Busoni, S.; Pradella, S.; Miele, V. Radiomics in pulmonary neuroendocrine tumours (NETs). Radiol. Med. 2022, 127, 609–615. [Google Scholar] [CrossRef]
- Liu, K.; Zheng, X.; Lu, D.; Tan, Y.; Hou, C.; Dai, J.; Shi, W.; Jiang, B.; Yao, Y.; Lu, Y.; et al. A multi-institutional study to predict the benefits of DEB-TACE and molecular targeted agent sequential therapy in unresectable hepatocellular carcinoma using a radiological-clinical nomogram. Radiol. Med. 2023. [Google Scholar] [CrossRef]
- Tsuchiya, M.; Masui, T.; Terauchi, K.; Yamada, T.; Katyayama, M.; Ichikawa, S.; Noda, Y.; Goshima, S. MRI-based radiomics analysis for differentiating phyllodes tumors of the breast from fibroadenomas. Eur. Radiol. 2022, 32, 4090–4100. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, A.; Fusco, R.; Di Bernardo, E.; Petrosino, T.; Barretta, M.L.; Porto, A.; Granata, V.; Di Bonito, M.; Fanizzi, A.; Massafra, R.; et al. Prediction of Breast Cancer Histological Outcome by Radiomics and Artificial Intelligence Analysis in Contrast-Enhanced Mammography. Cancers 2022, 14, 2132. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.; Wang, W.; Zou, C.; Li, M.; Chen, H.; Meng, F.; Dong, G.; Wang, J.; Yu, Q.; Sun, M.; et al. Predicting Tumor Mutation Burden and EGFR Mutation Using Clinical and Radiomic Features in Patients with Malignant Pulmonary Nodules. J. Pers. Med. 2022, 13, 16. [Google Scholar] [CrossRef]
- Gangil, T.; Sharan, K.; Rao, B.D.; Palanisamy, K.; Chakrabarti, B.; Kadavigere, R. Utility of adding Radiomics to clinical features in predicting the outcomes of radiotherapy for head and neck cancer using machine learning. PLoS ONE 2022, 17, e0277168. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Ai, Q.Y.H.; Wong, L.M.; Green, C.; Qamar, S.; So, T.Y.; Vlantis, A.C.; King, A.D. Radiomics for Discriminating Benign and Malignant Salivary Gland Tumors; Which Radiomic Feature Categories and MRI Sequences Should Be Used? Cancers 2022, 14, 5804. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Yin, J. Radiomics of dynamic contrast-enhanced magnetic resonance imaging parametric maps and apparent diffusion coefficient maps to predict Ki-67 status in breast cancer. Front. Oncol. 2022, 12, 847880. [Google Scholar] [CrossRef]
- Wang, X.; Xie, T.; Luo, J.; Zhou, Z.; Yu, X.; Guo, X. Radiomics predicts the prognosis of patients with locally advanced breast cancer by reflecting the heterogeneity of tumor cells and the tumor microenvironment. Breast Cancer Res. 2022, 24, 20. [Google Scholar] [CrossRef]
- Helmreich, J.E. Regression modeling strategies with applications to linear models, logistic and ordinal regression and survival analysis (2nd edition). J. Stat. Softw. 2016, 70, 1–3. [Google Scholar] [CrossRef]
- Thimansson, E.; Bengtsson, J.; Baubeta, E.; Engman, J.; Flondell-Sité, D.; Bjartell, A.; Zackrisson, S. Deep learning algorithm performs similarly to radiologists in the assessment of prostate volume on MRI. Eur. Radiol. 2022, 33, 2519–2528. [Google Scholar] [CrossRef]
- Autorino, R.; Gui, B.; Panza, G.; Boldrini, L.; Cusumano, D.; Russo, L.; Nardangeli, A.; Persiani, S.; Campitelli, M.; Ferrandina, G.; et al. Radiomics-based prediction of two-year clinical outcome in locally advanced cervical cancer patients undergoing neoadjuvant chemoradiotherapy. Radiol. Med. 2022, 127, 498–506. [Google Scholar] [CrossRef]
- Danala, G.; Maryada, S.K.; Islam, W.; Faiz, R.; Jones, M.; Qiu, Y.; Zheng, B. A Comparison of Computer-Aided Diagnosis Schemes Optimized Using Radiomics and Deep Transfer Learning Methods. Bioengineering 2022, 9, 256. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, Y.; Zhan, C.; Zhang, Q.; Wu, Y.; Ai, T. A nomogram based on radiomics signature and deep-learning signature for preoperative prediction of axillary lymph node metastasis in breast cancer. Front. Oncol. 2022, 12, 940655. [Google Scholar] [CrossRef] [PubMed]
- Granata, V.; Grassi, R.; Fusco, R.; Setola, S.V.; Belli, A.; Ottaiano, A.; Nasti, G.; La Porta, M.; Danti, G.; Cappabianca, S.; et al. Intrahepatic cholangiocarcinoma and its differential diagnosis at MRI: How radiologist should assess MR features. Radiol. Med. 2021, 126, 1584–1600. [Google Scholar] [CrossRef]
- Huynh, B.Q.; Li, H.; Giger, M.L. Digital mammographic tumor classification using transfer learning from deep convolutional neural networks. J. Med. Imaging 2016, 3, 34501. [Google Scholar] [CrossRef]
- Wu, K.; Miu, X.; Wang, H.; Li, X. A Bayesian optimization tunning integrated multi-stacking classifier framework for the prediction of radiodermatitis from 4D-CT of patients underwent breast cancer radiotherapy. Front. Oncol. 2023, 13, 1152020. [Google Scholar] [CrossRef]
- Zhu, Y.; Ma, Y.; Zhang, Y.; Liu, A.; Wang, Y.; Zhao, M.; Li, H.; He, N.; Wu, Y.; Ye, Z. Radiomics nomogram for predicting axillary lymph node metastasis-a potential method to address the limitation of axilla coverage in cone-beam breast CT: A bi-center retrospective study. Radiol. Med. 2023, 128, 1472–1482. [Google Scholar] [CrossRef]
- Hu, Q.; Giger, M.L. Clinical Artificial Intelligence Applications. Radiol. Clin. N. Am. 2021, 59, 1027–1043. [Google Scholar] [CrossRef] [PubMed]
- Huang, H. Editorial: The Application of Radiomics and Artificial Intelligence in Cancer Imaging. Front. Oncol. 2022, 12, 864940. [Google Scholar] [CrossRef]
- Candela-Leal, M.O.; Gutiérrez-Flores, E.A.; Presbítero-Espinosa, G.; Sujatha-Ravindran, A.; Ramírez-Mendoza, R.A.; Lozoya-Santos, J.d.J.; Ramírez-Moreno, M.A. Multi-Output Sequential Deep Learning Model for Athlete Force Prediction on a Treadmill Using 3D Markers. Appl. Sci. 2022, 12, 5424. [Google Scholar] [CrossRef]
- Ramlakhan, S.L.; Saatchi, R.; Sabir, L.; Ventour, D.; Shobayo, O.; Hughes, R.; Singh, Y. Building artificial intelligence and machine learning models: A primer for emergency physicians. Emerg. Med. J. 2022, 39, e1. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, Q.; Yin, W.; Yang, L.; Xiao, B.; Wang, J.; Yao, X. Development and validation of a radiopathomic model for predicting pathologic complete response to neoadjuvant chemotherapy in breast cancer patients. BMC Cancer 2023, 23, 431. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.A.; Islam, W.; Faiz, R.; Chen, X.; Zheng, B. Applying artificial intelligence technology to assist with breast cancer diagnosis and prognosis prediction. Front. Oncol. 2022, 12, 980793. [Google Scholar] [CrossRef] [PubMed]
- Ho, S.Y.; Phua, K.; Wong, L.; Bin Goh, W.W. Extensions of the External Validation for Checking Learned Model Interpretability and Generalizability. Patterns 2020, 1, 100129. [Google Scholar] [CrossRef] [PubMed]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Endocrine 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Hatt, M.; Vallieres, M.; Visvikis, D.; Zwanenburg, A. IBSI: An international community radiomics standardization initiative. J. Nucl. Med. 2018, 59, 287. [Google Scholar]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceeding of the IEEE International Joint Conference on Neural Networks (IEEE World Congress on Computational Intelligence), Hong Kong, China, 1–8 June 2008; pp. 1322–1328. [Google Scholar] [CrossRef]
- Kosolwattana, T.; Liu, C.; Hu, R.; Han, S.; Chen, H.; Lin, Y. A self-inspected adaptive SMOTE algorithm (SASMOTE) for highly imbalanced data classification in healthcare. BioData Min. 2023, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Coppola, F.; Faggioni, L.; Gabelloni, M.; De Vietro, F.; Mendola, V.; Cattabriga, A.; Cocozza, M.A.; Vara, G.; Piccinino, A.; Monaco, S.L.; et al. Human, All Too Human? An All-Around Appraisal of the “Artificial Intelligence Revolution” in Medical Imaging. Front. Psychol. 2021, 12, 710982. [Google Scholar] [CrossRef]
- Balagurunathan, Y.; Gu, Y.; Wang, H.; Kumar, V.; Grove, O.; Hawkins, S.; Kim, J.; Goldgof, D.B.; Hall, L.O.; Gatenby, R.A.; et al. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl. Oncol. 2014, 7, 72–87. [Google Scholar] [CrossRef]
- Yang, F.; Simpson, G.; Young, L.; Ford, J.; Dogan, N.; Wang, L. Impact of contouring variability on oncological PET radiomics features in the lung. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Young, L.A.; Johnson, P.B. Quantitative radiomics: Validating image textural features for oncological PET in lung cancer. Radiother. Oncol. 2018, 129, 209–217. [Google Scholar] [CrossRef]
- Avanzo, M.; Wei, L.; Stancanello, J.; Vallières, M.; Rao, A.; Morin, O.; Mattonen, S.A.; El Naqa, I. Machine and deep learning methods for radiomics. Med. Phys. 2020, 47, e185–e202. [Google Scholar] [CrossRef]
- Wagner, M.W.; Namdar, K.; Biswas, A.; Monah, S.; Khalvati, F.; Ertl-Wagner, B.B. Radiomics, machine learning, and artificial intelligence—What the neuroradiologist needs to know. Neuroradiology 2021, 63, 1957–1967. [Google Scholar] [CrossRef]
- Wu, G.; Jochems, A.; Refaee, T.; Ibrahim, A.; Yan, C.; Sanduleanu, S.; Woodruff, H.C.; Lambin, P. Structural and functional radiomics for lung cancer. Eur. J. Nucl. Med. 2021, 48, 3961–3974. [Google Scholar] [CrossRef] [PubMed]
- Rogers, W.; Seetha, S.T.; Refaee, T.A.G.; Lieverse, R.I.Y.; Granzier, R.W.Y.; Ibrahim, A.; Keek, S.A.; Sanduleanu, S.; Primakov, S.P.; Beuque, M.P.L.; et al. Radiomics: From qualitative to quantitative imaging. Br. J. Radiol. 2020, 93, 20190948. [Google Scholar] [CrossRef] [PubMed]
- Reig, B.; Heacock, L.; Geras, K.J.; Moy, L. Machine learning in breast MRI. J. Magn. Reson. Imaging 2020, 52, 998–1018. [Google Scholar] [CrossRef]
- Guiot, J.; Vaidyanathan, A.; Deprez, L.; Zerka, F.; Danthine, D.; Frix, A.; Lambin, P.; Bottari, F.; Tsoutzidis, N.; Miraglio, B.; et al. A review in radiomics: Making personalized medicine a reality via routine imaging. Med. Res. Rev. 2022, 42, 426–440. [Google Scholar] [CrossRef] [PubMed]
- Geras, K.J.; Mann, R.M.; Moy, L. Artificial Intelligence for Mammography and Digital Breast Tomosynthesis: Current Concepts and Future Perspectives. Radiology 2019, 293, 246–259. [Google Scholar] [CrossRef]
- Tsougos, I.; Vamvakas, A.; Kappas, C.; Fezoulidis, I.; Vassiou, K. Application of Radiomics and Decision Support Systems for Breast MR Differential Diagnosis. Comput. Math. Methods Med. 2018, 2018, 7417126. [Google Scholar] [CrossRef]
- Granata, V.; Fusco, R.; Avallone, A.; Catalano, O.; Filice, F.; Leongito, M.; Palaia, R.; Izzo, F.; Petrillo, A. Major and ancillary magnetic resonance features of LI-RADS to assess HCC: An overview and update. Infect. Agents Cancer 2017, 12, 23. [Google Scholar] [CrossRef]
- Tagliafico, A.S.; Piana, M.; Schenone, D.; Lai, R.; Massone, A.M.; Houssami, N. Overview of radiomics in breast cancer diagnosis and prognostication. Breast 2020, 49, 74–80. [Google Scholar] [CrossRef]
- Zheng, X.; Yao, Z.; Huang, Y.; Yu, Y.; Wang, Y.; Liu, Y.; Mao, R.; Li, F.; Xiao, Y.; Wang, Y.; et al. Deep learning radiomics can predict axillary lymph node status in early-stage breast cancer. Nat. Commun. 2020, 11, 1236. [Google Scholar] [CrossRef]
- Pesapane, F.; Rotili, A.; Agazzi, G.M.; Botta, F.; Raimondi, S.; Penco, S.; Dominelli, V.; Cremonesi, M.; Jereczek-Fossa, B.A.; Carrafiello, G.; et al. Recent Radiomics Advancements in Breast Cancer: Lessons and Pitfalls for the Next Future. Curr. Oncol. 2021, 28, 2351–2372. [Google Scholar] [CrossRef]
- Su, G.-H.; Xiao, Y.; Jiang, L.; Zheng, R.-C.; Wang, H.; Chen, Y.; Gu, Y.-J.; You, C.; Shao, Z.-M. Radiomics features for assessing tumor-infiltrating lymphocytes correlate with molecular traits of triple-negative breast cancer. J. Transl. Med. 2022, 20, 471. [Google Scholar] [CrossRef]
- Vicini, S.; Bortolotto, C.; Rengo, M.; Ballerini, D.; Bellini, D.; Carbone, I.; Preda, L.; Laghi, A.; Coppola, F.; Faggioni, L. A narrative review on current imaging applications of artificial intelligence and radiomics in oncology: Focus on the three most common cancers. Radiol. Med. 2022, 127, 819–836. [Google Scholar] [CrossRef] [PubMed]
- Caruso, D.; Polici, M.; Rinzivillo, M.; Zerunian, M.; Nacci, I.; Marasco, M.; Magi, L.; Tarallo, M.; Gargiulo, S.; Iannicelli, E.; et al. CT-based radiomics for prediction of therapeutic response to Everolimus in metastatic neuroendocrine tumors. Radiol. Med. 2022, 127, 691–701. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Zhao, Z.; Wang, X.; Ai, H.; Yang, C.; Luo, Y.; Jiang, X. Radiomics for prediction of response to EGFR-TKI based on metastasis/brain parenchyma (M/BP)-interface. La Radiol. Med. 2022, 127, 1342–1354. [Google Scholar] [CrossRef] [PubMed]
- Satake, H.; Ishigaki, S.; Ito, R.; Naganawa, S. Radiomics in breast MRI: Current progress toward clinical application in the era of artificial intelligence. Radiol. Med. 2021, 127, 39–56. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, J.; Jin, W.; Li, R.; Xie, X.; Zhao, W.; Xia, S.; Han, D. Radiomics from dual-energy CT-derived iodine maps predict lymph node metastasis in head and neck squamous cell carcinoma. Radiol. Med. 2023, 1–16. [Google Scholar] [CrossRef]
- Song, F.; Ma, M.; Zeng, S.; Shao, F.; Huang, W.; Feng, Z.; Rong, P. CT enterography-based radiomics combined with body composition to predict infliximab treatment failure in Crohn’s disease. Radiol. Med. 2023, 1–13. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Guo, S.-M.; Lien, J.-J.J.; Lin, W.-T.; Liu, Y.-S.; Lai, C.-H.; Hsu, I.-L.; Chang, C.-C.; Tseng, Y.-L. Combined model integrating deep learning, radiomics, and clinical data to classify lung nodules at chest CT. Radiol. Med. 2023, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Zeng, S.; Li, F.; Liu, G. Utilizing grayscale ultrasound-based radiomics nomogram for preoperative identification of triple negative breast cancer. Radiol. Med. 2023, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Shur, J.D.; Doran, S.J.; Kumar, S.; ap Dafydd, D.; Downey, K.; O’connor, J.P.B.; Papanikolaou, N.; Messiou, C.; Koh, D.-M.; Orton, M.R. Radiomics in Oncology: A Practical Guide. RadioGraphics 2021, 41, 1717–1732. [Google Scholar] [CrossRef] [PubMed]
- Petrillo, A.; Fusco, R.; Barretta, M.L.; Granata, V.; Raso, M.M.; Porto, A.; Sorgente, E.; Fanizzi, A.; Massafra, R.; Lafranceschina, M.; et al. Radiomics and artificial intelligence analysis by T2-weighted imaging and dynamic contrast-enhanced magnetic resonance imaging to predict Breast Cancer Histological Outcome. Radiol. Med. 2023, 128, 1347–1371. [Google Scholar] [CrossRef]
- Shang, Y.; Chen, W.; Li, G.; Huang, Y.; Wang, Y.; Kui, X.; Li, M.; Zheng, H.; Zhao, W.; Liu, J. Computed Tomography-derived intratumoral and peritumoral radiomics in predicting EGFR mutation in lung adenocarcinoma. Radiol. Med. 2023, 128, 1483–1496. [Google Scholar] [CrossRef]
- Ma, X.; Qian, X.; Wang, Q.; Zhang, Y.; Zong, R.; Zhang, J.; Qian, B.; Yang, C.; Lu, X.; Shi, Y. Radiomics nomogram based on optimal VOI of multi-sequence MRI for predicting microvascular invasion in intrahepatic cholangiocarcinoma. Radiol. Med. 2023, 128, 1296–1309. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).