A Novel Fatty Acid Metabolism-Associated Risk Model for Prognosis Prediction in Acute Myeloid Leukaemia
Abstract
1. Introduction
2. Materials and Methods
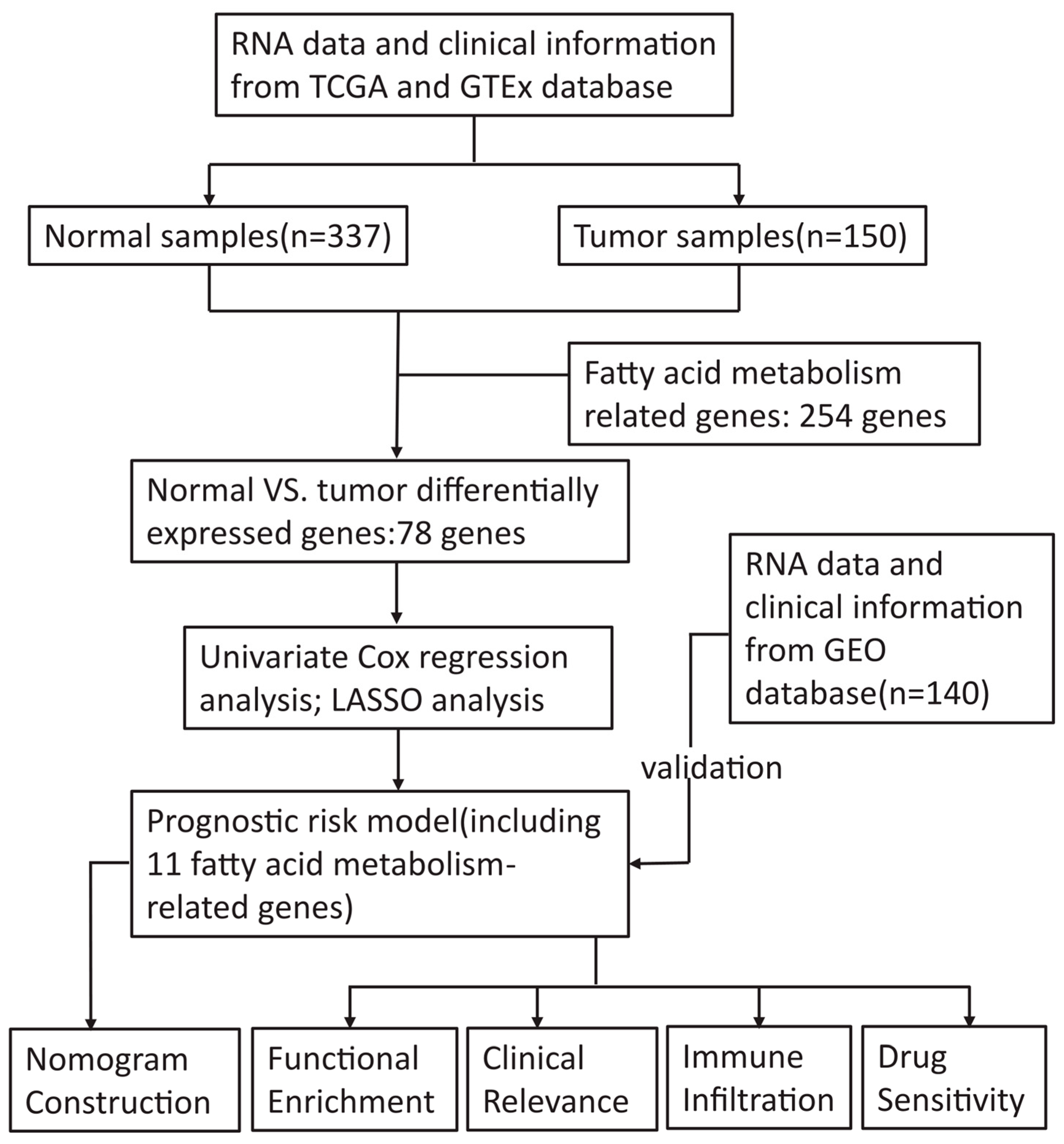
2.1. Data Collection
2.2. Identification and Enrichment Analysis of the Differentially Expressed Genes (DEGs)
2.3. Construction and Validation of the Prognostic Signature Associated with FAM
2.4. Comprehensive Analysis of the Prognostic Risk Score and Clinicopathological Parameters of AML Patients
2.5. Development and Assessment of the Nomogram for AML Patients
2.6. Gene Set Enrichment Analysis
2.7. Immune Infiltration Level Analysis
2.8. Drug Sensitivity Analysis
2.9. Protein-Protein Interaction (PPI) Network
2.10. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
3. Results
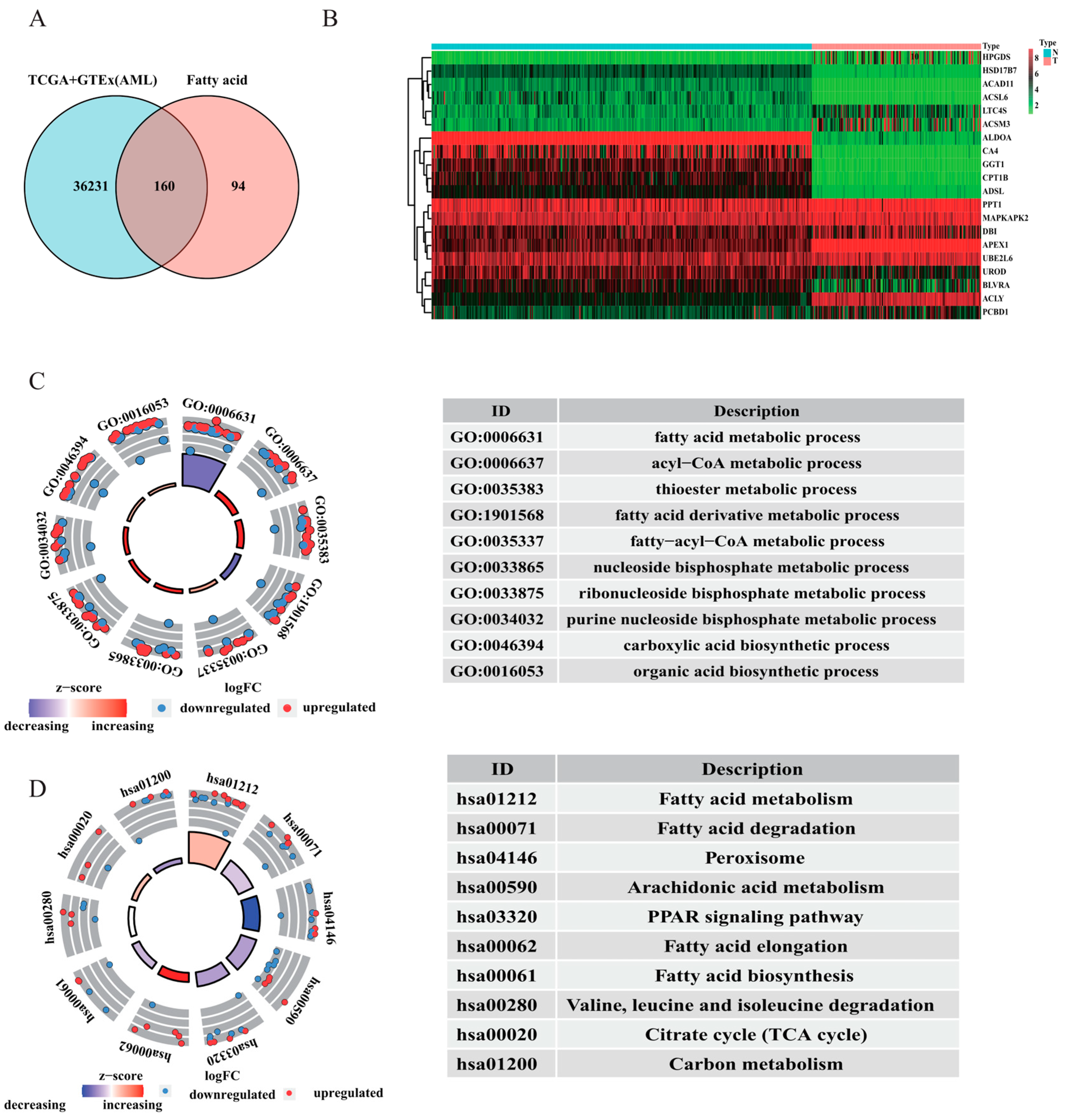
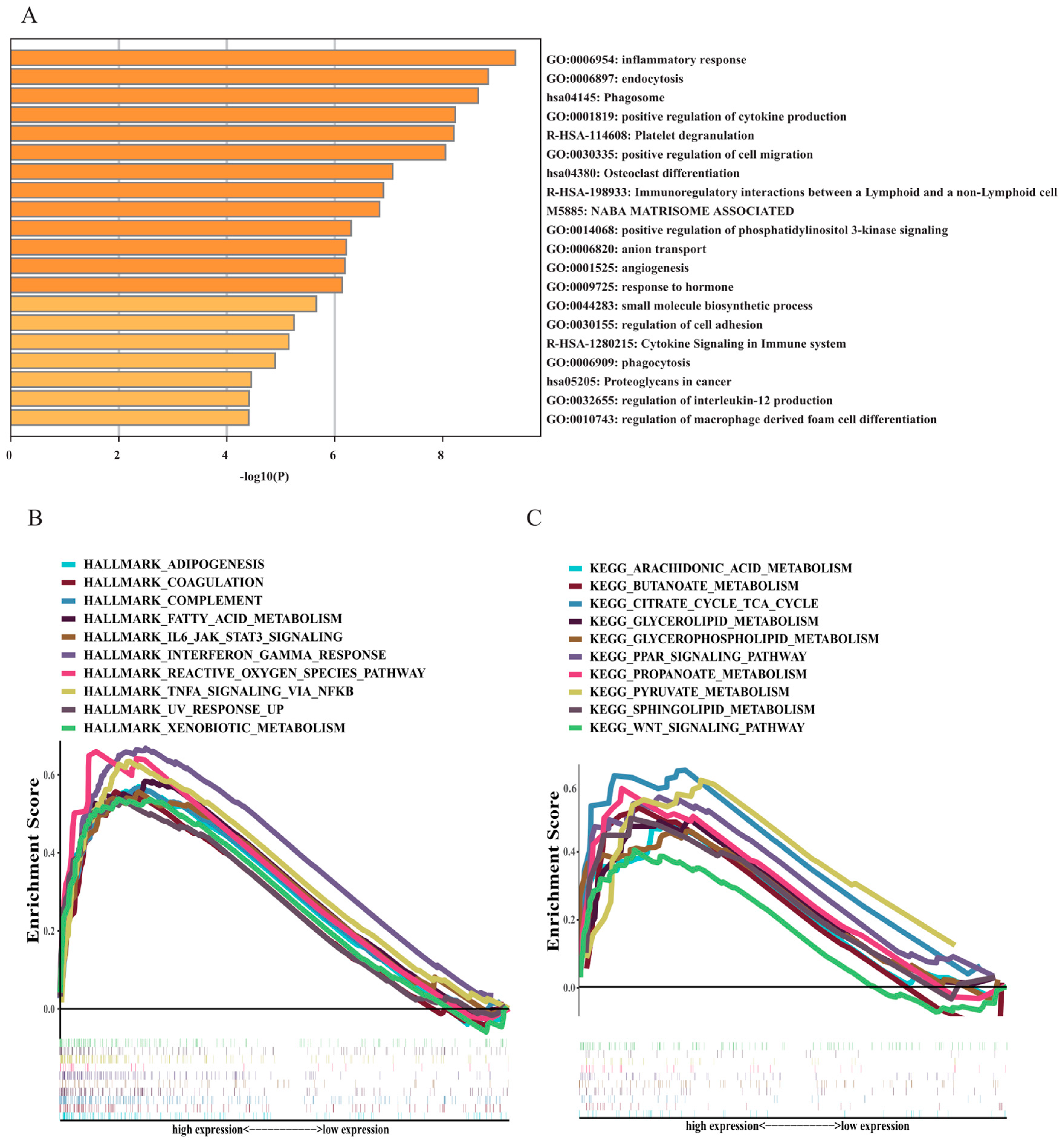
3.1. Enrichment Analysis of AML Patient Samples
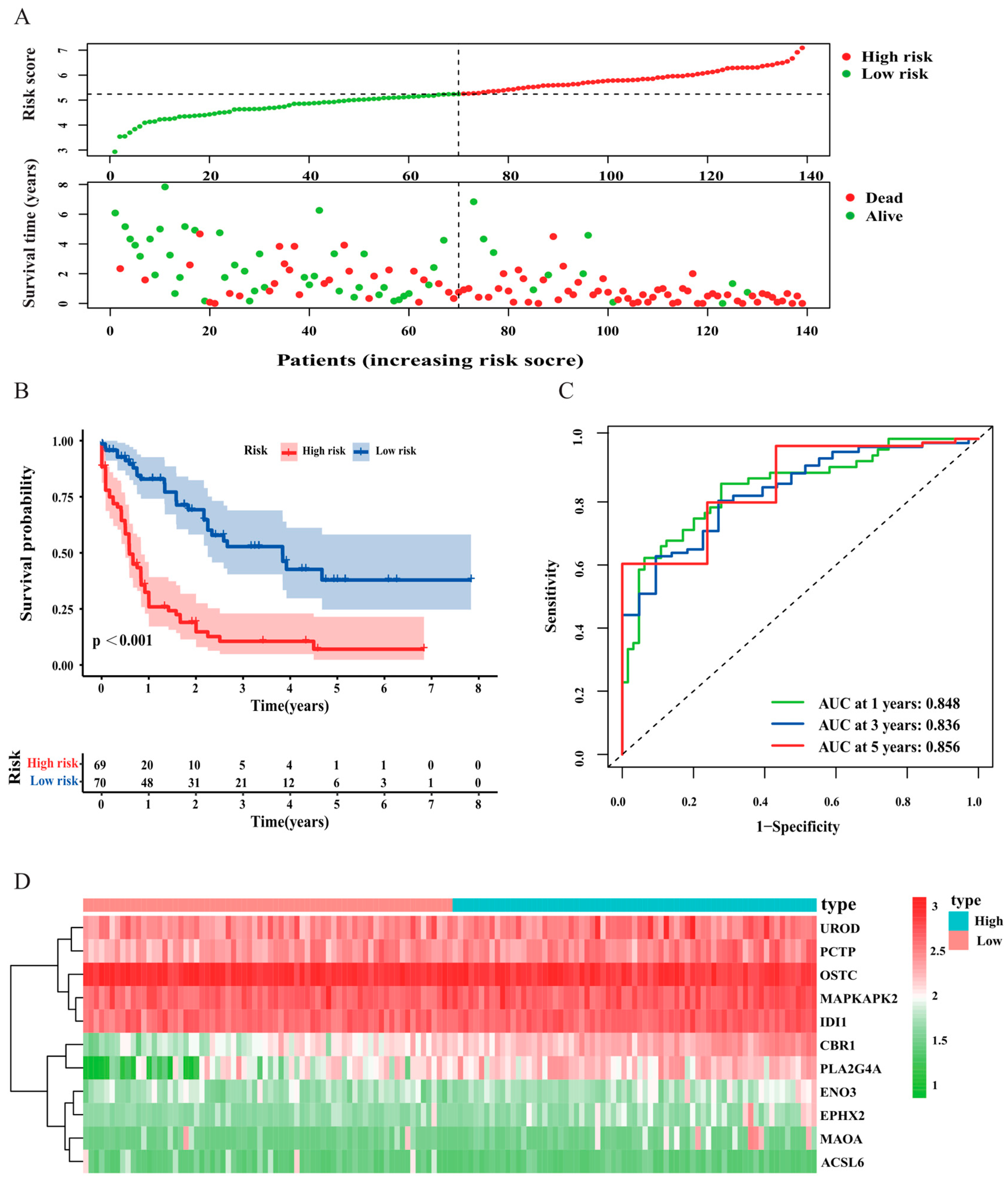
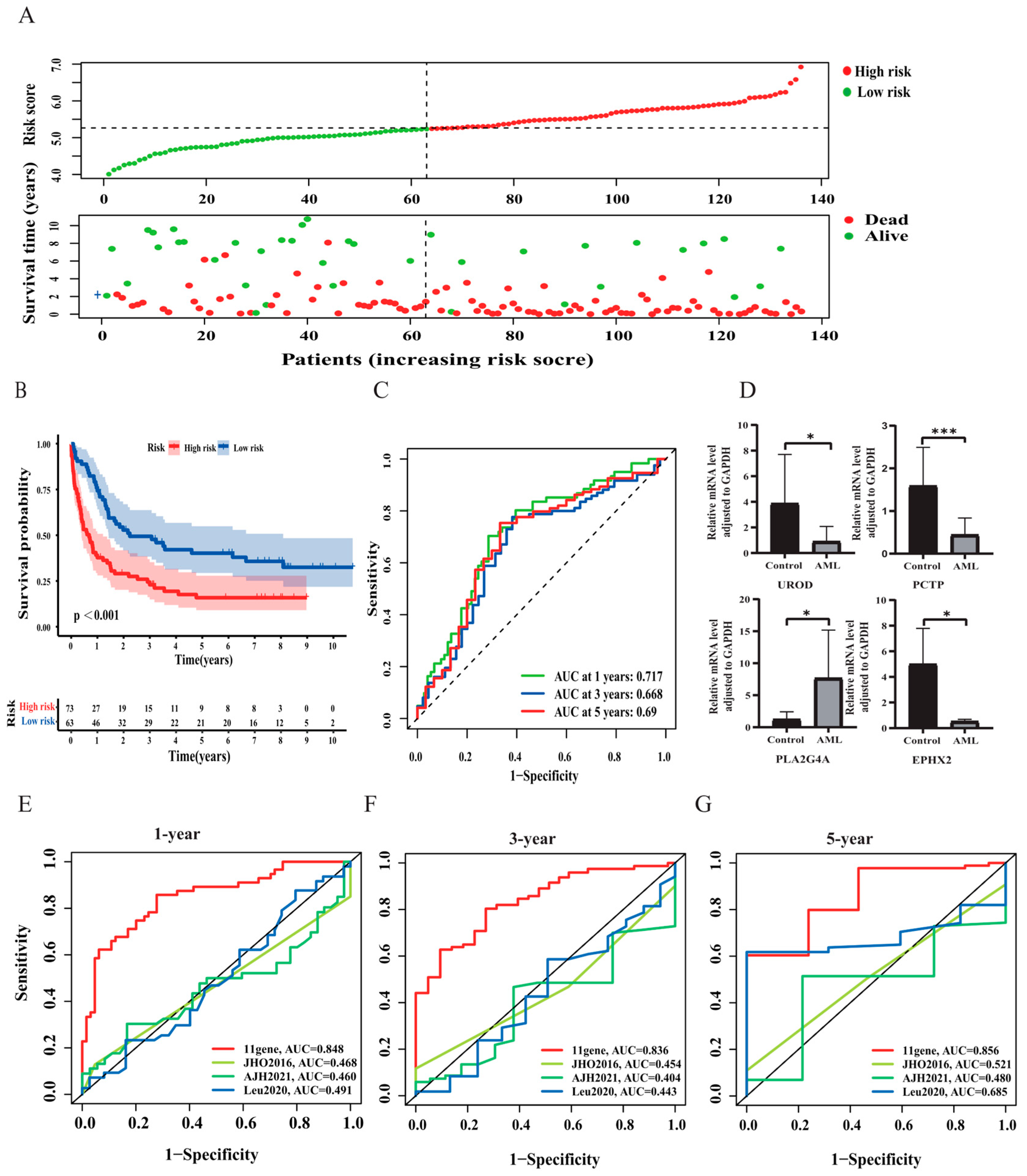
3.2. Construction and Validation of the Risk Signature
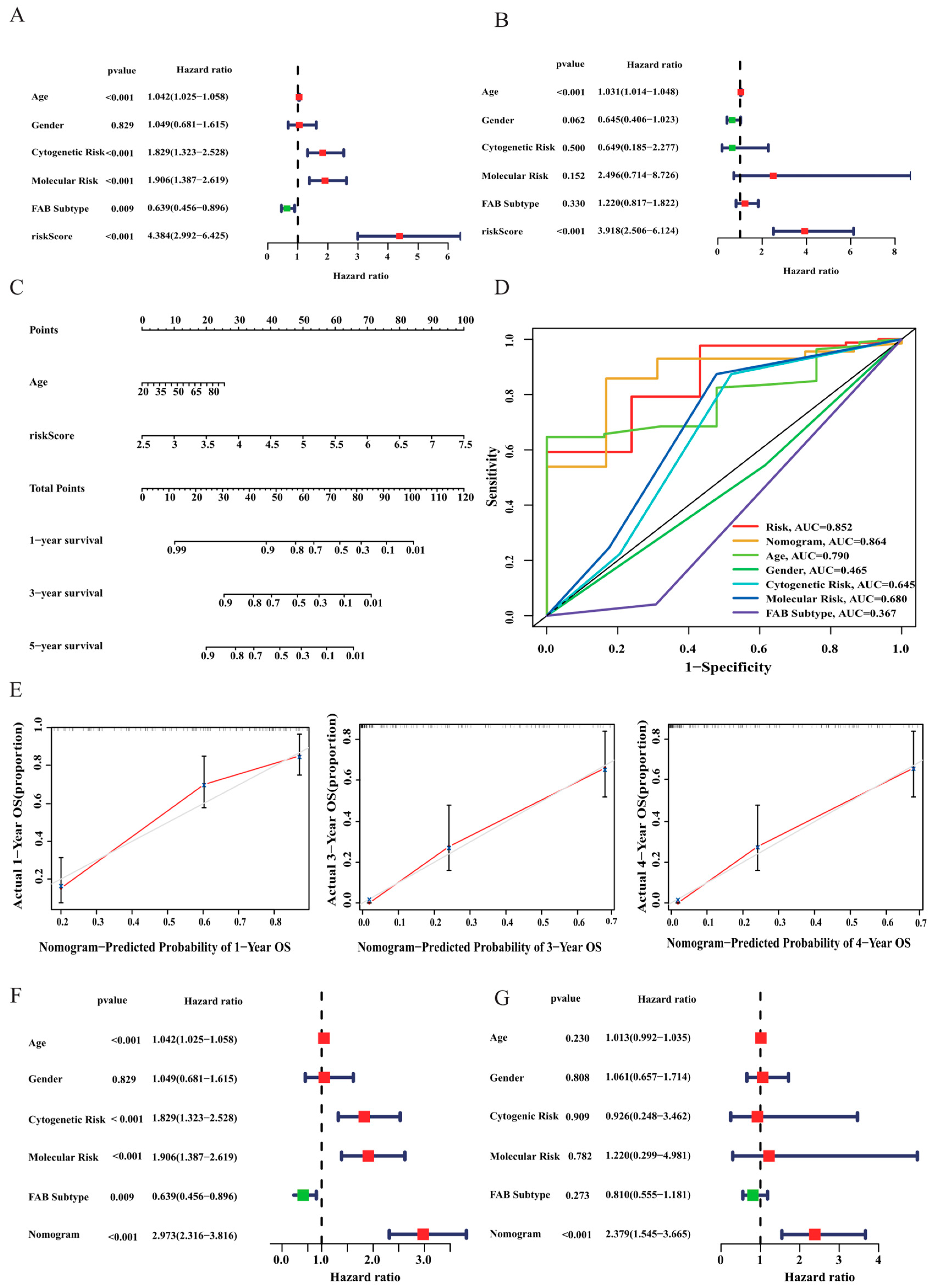
3.3. Correlation Analysis between Risk Score and Clinicopathological Features
3.4. Construction of a Nomogram for AML Patients
3.5. Functional and Annotation Analyses
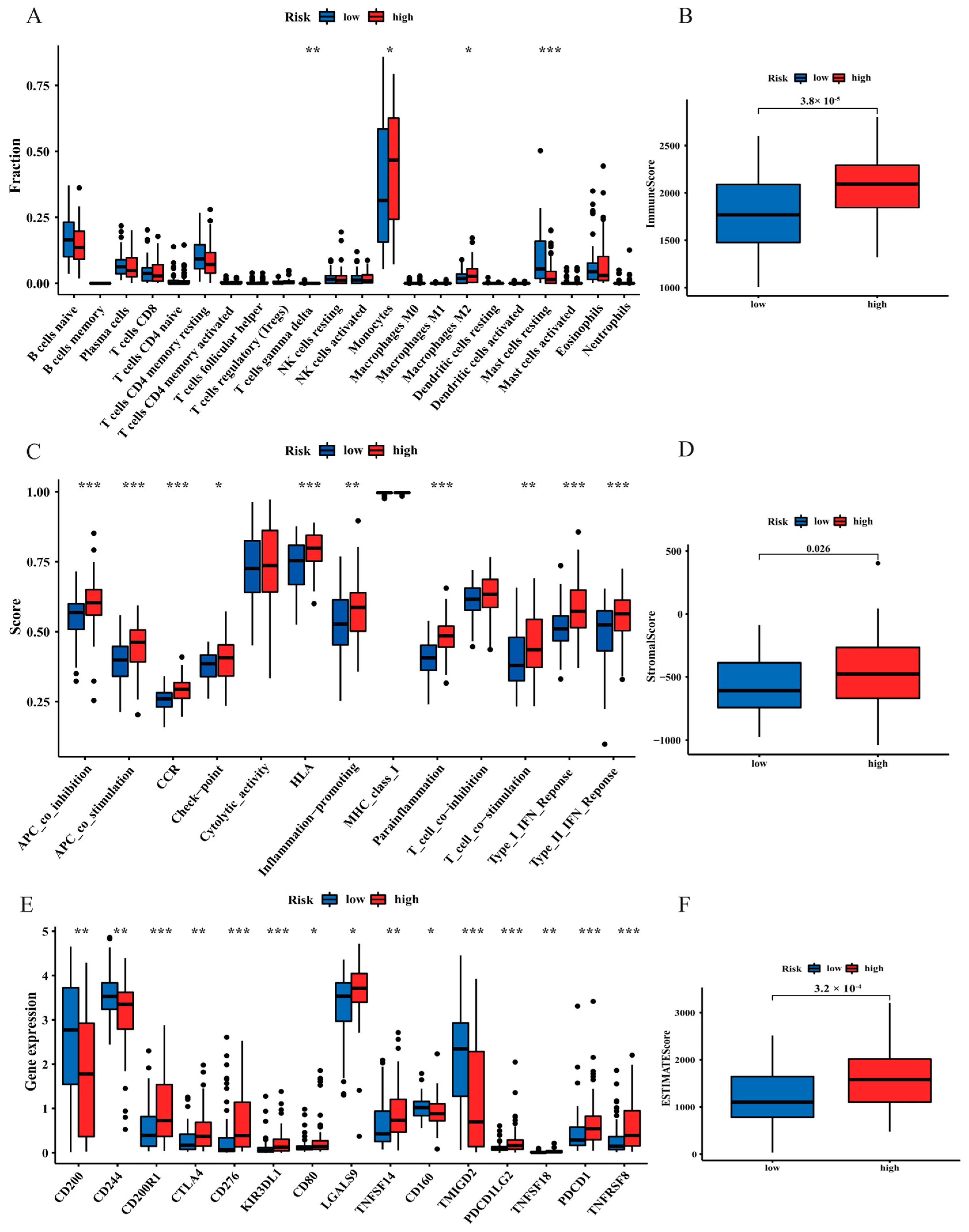
3.6. The Landscape of the Tumour Microenvironment (TME) and Immune Cell Infiltration in AML Patients
3.7. Analysis of Drug Sensitivity in the Two Risk Groups
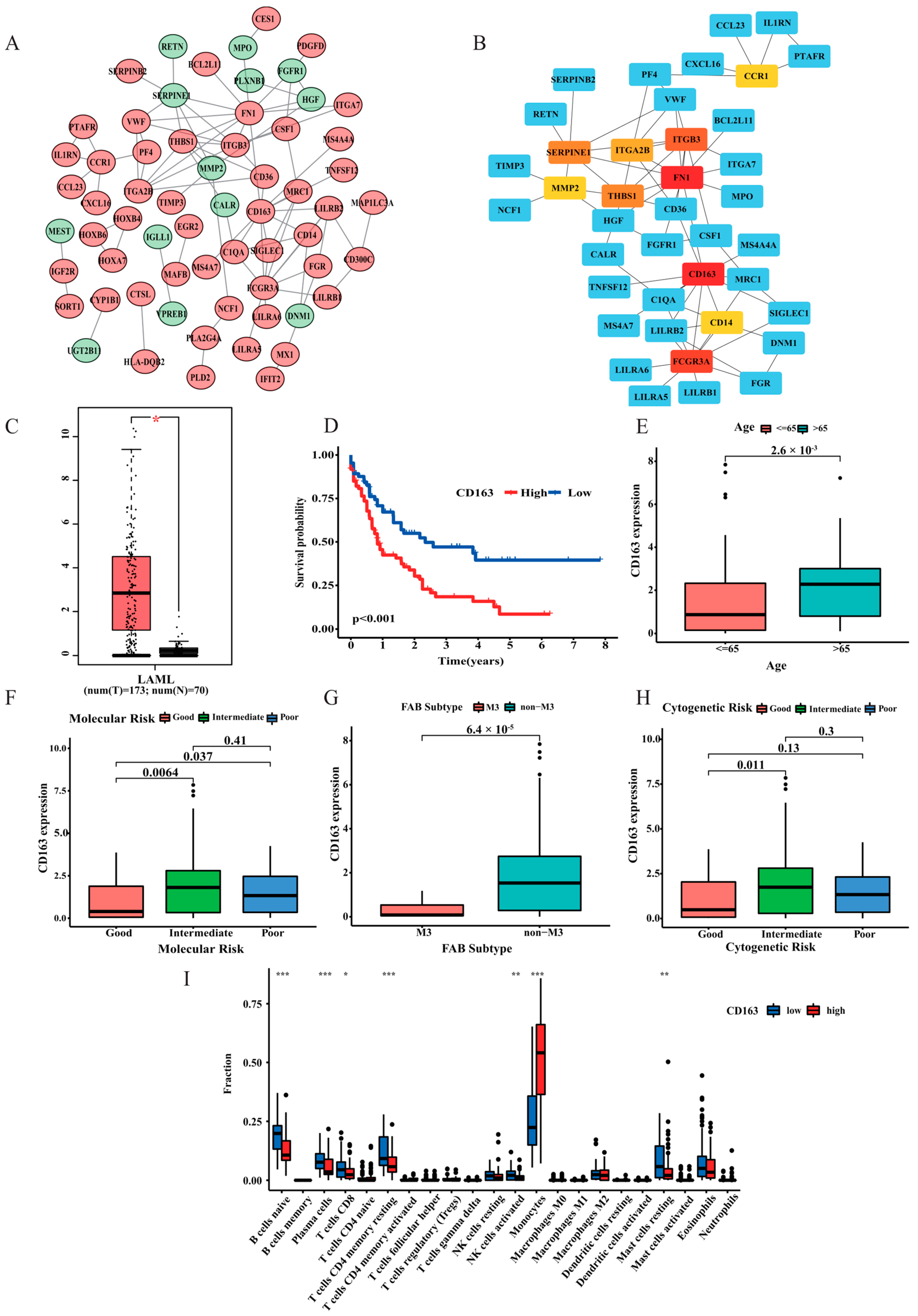
3.8. PPI Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Heuser, M.; Ofran, Y.; Boissel, N.; Brunet Mauri, S.; Craddock, C.; Janssen, J.; Wierzbowska, A.; Buske, C.; ESMO Guidelines Committee. Acute myeloid leukaemia in adult patients: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2020, 31, 697–712. [Google Scholar] [CrossRef] [PubMed]
- Guieze, R.; Liu, V.M.; Rosebrock, D.; Jourdain, A.A.; Hernandez-Sanchez, M.; Martinez Zurita, A.; Sun, J.; Ten Hacken, E.; Baranowski, K.; Thompson, P.A.; et al. Mitochondrial Reprogramming Underlies Resistance to BCL-2 Inhibition in Lymphoid Malignancies. Cancer Cell 2019, 36, 369–384.e13. [Google Scholar] [CrossRef]
- Tallman, M.S.; Wang, E.S.; Altman, J.K.; Appelbaum, F.R.; Bhatt, V.R.; Bixby, D.; Coutre, S.E.; De Lima, M.; Fathi, A.T.; Fiorella, M.; et al. Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Canc. Netw. 2019, 17, 721–749. [Google Scholar] [CrossRef]
- Madak-Erdogan, Z.; Band, S.; Zhao, Y.C.; Smith, B.P.; Kulkoyluoglu-Cotul, E.; Zuo, Q.; Santaliz Casiano, A.; Wrobel, K.; Rossi, G.; Smith, R.L.; et al. Free Fatty Acids Rewire Cancer Metabolism in Obesity-Associated Breast Cancer via Estrogen Receptor and mTOR Signaling. Cancer Res. 2019, 79, 2494–2510. [Google Scholar] [CrossRef]
- He, D.; Cai, L.; Huang, W.; Weng, Q.; Lin, X.; You, M.; Liao, S. Prognostic value of fatty acid metabolism-related genes in patients with hepatocellular carcinoma. Aging 2021, 13, 17847–17863. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.N.; Zeng, Z.L.; Lu, J.; Wang, Y.; Liu, Z.X.; He, M.M.; Zhao, Q.; Wang, Z.X.; Li, T.; Lu, Y.X.; et al. CPT1A-mediated fatty acid oxidation promotes colorectal cancer cell metastasis by inhibiting anoikis. Oncogene 2018, 37, 6025–6040. [Google Scholar] [CrossRef] [PubMed]
- Tabe, Y.; Konopleva, M.; Andreeff, M. Fatty Acid Metabolism, Bone Marrow Adipocytes, and AML. Front. Oncol. 2020, 10, 155. [Google Scholar] [CrossRef]
- Stevens, B.M.; Jones, C.L.; Pollyea, D.A.; Culp-Hill, R.; D’Alessandro, A.; Winters, A.; Krug, A.; Abbott, D.; Goosman, M.; Pei, S.; et al. Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat. Cancer 2020, 1, 1176–1187. [Google Scholar] [CrossRef]
- Han, S.; Wei, R.; Zhang, X.; Jiang, N.; Fan, M.; Huang, J.H.; Xie, B.; Zhang, L.; Miao, W.; Butler, A.C.; et al. CPT1A/2-Mediated FAO Enhancement-A Metabolic Target in Radioresistant Breast Cancer. Front. Oncol. 2019, 9, 1201. [Google Scholar] [CrossRef]
- Wu, Y.; Fabritius, M.; Ip, C. Chemotherapeutic sensitization by endoplasmic reticulum stress: Increasing the efficacy of taxane against prostate cancer. Cancer Biol. Ther. 2009, 8, 146–152. [Google Scholar] [CrossRef]
- Tcheng, M.; Roma, A.; Ahmed, N.; Smith, R.W.; Jayanth, P.; Minden, M.D.; Schimmer, A.D.; Hess, D.A.; Hope, K.; Rea, K.A.; et al. Very long chain fatty acid metabolism is required in acute myeloid leukemia. Blood 2021, 137, 3518–3532. [Google Scholar] [CrossRef] [PubMed]
- Geeleher, P.; Cox, N.; Huang, R.S. pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE 2014, 9, e107468. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Song, J.; Wang, W.; Bai, J.; Zhang, Y.; Shi, J.; Bai, J.; Zhou, Y. A novel 4-mRNA signature predicts the overall survival in acute myeloid leukemia. Am. J. Hematol. 2021, 96, 1385–1395. [Google Scholar] [CrossRef] [PubMed]
- Elsayed, A.H.; Rafiee, R.; Cao, X.; Raimondi, S.; Downing, J.R.; Ribeiro, R.; Fan, Y.; Gruber, T.A.; Baker, S.; Klco, J.; et al. A six-gene leukemic stem cell score identifies high risk pediatric acute myeloid leukemia. Leukemia 2020, 34, 735–745. [Google Scholar] [CrossRef] [PubMed]
- Wilop, S.; Chou, W.C.; Jost, E.; Crysandt, M.; Panse, J.; Chuang, M.K.; Brummendorf, T.H.; Wagner, W.; Tien, H.F.; Kharabi Masouleh, B. A three-gene expression-based risk score can refine the European LeukemiaNet AML classification. J. Hematol. Oncol. 2016, 9, 78. [Google Scholar] [CrossRef]
- Nepstad, I.; Hatfield, K.J.; Gronningsaeter, I.S.; Reikvam, H. The PI3K-Akt-mTOR Signaling Pathway in Human Acute Myeloid Leukemia (AML) Cells. Int. J. Mol. Sci. 2020, 21, 2907. [Google Scholar] [CrossRef]
- Espinoza-Sanchez, N.A.; Gotte, M. Role of cell surface proteoglycans in cancer immunotherapy. Semin. Cancer Biol. 2020, 62, 48–67. [Google Scholar] [CrossRef]
- Deng, X.; Lin, D.; Zhang, X.; Shen, X.; Yang, Z.; Yang, L.; Lu, X.; Yu, L.; Zhang, N.; Lin, J. Profiles of immune-related genes and immune cell infiltration in the tumor microenvironment of diffuse lower-grade gliomas. J. Cell. Physiol. 2020, 235, 7321–7331. [Google Scholar] [CrossRef]
- Jiang, X.; Yan, Q.; Xie, L.; Xu, S.; Jiang, K.; Huang, J.; Wen, Y.; Yan, Y.; Zheng, J.; Tang, S.; et al. Construction and Validation of a Ferroptosis-Related Prognostic Model for Gastric Cancer. J. Oncol. 2021, 2021, 6635526. [Google Scholar] [CrossRef]
- Stone, R.M.; Mandrekar, S.J.; Sanford, B.L.; Laumann, K.; Geyer, S.; Bloomfield, C.D.; Thiede, C.; Prior, T.W.; Dohner, K.; Marcucci, G.; et al. Midostaurin plus Chemotherapy for Acute Myeloid Leukemia with a FLT3 Mutation. N. Engl. J. Med. 2017, 377, 454–464. [Google Scholar] [CrossRef]
- Short, N.J.; Rytting, M.E.; Cortes, J.E. Acute myeloid leukaemia. Lancet 2018, 392, 593–606. [Google Scholar] [CrossRef]
- Currie, E.; Schulze, A.; Zechner, R.; Walther, T.C.; Farese, R.V., Jr. Cellular fatty acid metabolism and cancer. Cell Metab. 2013, 18, 153–161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Sun, Z.K.; Zhong, F.M.; Yao, F.Y.; Liu, J.; Zhang, J.; Zhang, N.; Lin, J.; Li, S.Q.; Li, M.Y.; et al. A novel fatty acid metabolism-related signature identifies features of the tumor microenvironment and predicts clinical outcome in acute myeloid leukemia. Lipids Health Dis. 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Plebuch, M.; Soldan, M.; Hungerer, C.; Koch, L.; Maser, E. Increased resistance of tumor cells to daunorubicin after transfection of cDNAs coding for anthracycline inactivating enzymes. Cancer Lett. 2007, 255, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Olson, L.E.; Bedja, D.; Alvey, S.J.; Cardounel, A.J.; Gabrielson, K.L.; Reeves, R.H. Protection from doxorubicin-induced cardiac toxicity in mice with a null allele of carbonyl reductase 1. Cancer Res. 2003, 63, 6602–6606. [Google Scholar]
- Varatharajan, S.; Abraham, A.; Zhang, W.; Shaji, R.V.; Ahmed, R.; Abraham, A.; George, B.; Srivastava, A.; Chandy, M.; Mathews, V.; et al. Carbonyl reductase 1 expression influences daunorubicin metabolism in acute myeloid leukemia. Eur. J. Clin. Pharmacol. 2012, 68, 1577–1586. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, X.; Zhang, L.; Geng, D.; Wang, Y.; Sun, D.; Sui, P.; Zhao, X.; Xin, C.; Jiang, J.; et al. Inhibition of PLA2G4A Reduces the Expression of Lung Cancer-Related Cytokines. DNA Cell Biol. 2018, 37, 1076–1081. [Google Scholar] [CrossRef]
- Zhan, Y.; Zheng, L.; Liu, J.; Hu, D.; Wang, J.; Liu, K.; Guo, J.; Zhang, T.; Kong, D. PLA2G4A promotes right-sided colorectal cancer progression by inducing CD39+gammadelta Treg polarization. JCI Insight 2021, 6, e148028. [Google Scholar] [CrossRef]
- Bai, H.; Zhou, M.; Zeng, M.; Han, L. PLA2G4A Is a Potential Biomarker Predicting Shorter Overall Survival in Patients with Non-M3/NPM1 Wildtype Acute Myeloid Leukemia. DNA Cell Biol. 2020, 39, 700–708. [Google Scholar] [CrossRef]
- Baldazzi, C.; Luatti, S.; Marzocchi, G.; Grassi, A.; Cavo, M.; Testoni, N. t(5;12)(q31;p13)/ETV6::ACSL6 and t(6;9)(p23;q34)/DEK::NUP214 concurrence in acute myeloid leukemia: An unusual association of two rare abnormalities. Cancer Genet. 2022, 262–263, 35–39. [Google Scholar] [CrossRef]
- Soni, S.; Anand, P.; Padwad, Y.S. MAPKAPK2: The master regulator of RNA-binding proteins modulates transcript stability and tumor progression. J. Exp. Clin. Cancer Res. 2019, 38, 121. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pu, T.; Yin, L.; Li, Q.; Liao, C.P.; Wu, B.J. MAOA-mediated reprogramming of stromal fibroblasts promotes prostate tumorigenesis and cancer stemness. Oncogene 2020, 39, 3305–3321. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Bai, Y.; Liao, S.; Chen, H.; Kuang, L.; Luo, Q.; Lv, L.; Qiu, L.; Mei, Z. Identification and validation of EPHX2 as a prognostic biomarker in hepatocellular carcinoma. Mol. Med. Rep. 2021, 24, 650. [Google Scholar] [CrossRef] [PubMed]
- Berk, L.S.; Webb, G.; Imperio, N.C.; Nehlsen-Cannarella, S.L.; Eby, W.C. Simple, rapid 125I-labeled cyclosporine double antibody/polyethylene glycol radioimmunoassay used in a pediatric cardiac transplant program. Ther. Drug Monit. 1986, 8, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Lu, H.; Zhang, S.; Wang, T.; Zhao, H.; Guan, F.; Zeng, P. Leveraging Methylation Alterations to Discover Potential Causal Genes Associated With the Survival Risk of Cervical Cancer in TCGA Through a Two-Stage Inference Approach. Front. Genet. 2021, 12, 667877. [Google Scholar] [CrossRef] [PubMed]
- Ito, E.; Yue, S.; Moriyama, E.H.; Hui, A.B.; Kim, I.; Shi, W.; Alajez, N.M.; Bhogal, N.; Li, G.; Datti, A.; et al. Uroporphyrinogen decarboxylase is a radiosensitizing target for head and neck cancer. Sci. Transl. Med. 2011, 3, 67ra7. [Google Scholar] [CrossRef]
- Lochner, M.; Berod, L.; Sparwasser, T. Fatty acid metabolism in the regulation of T cell function. Trends Immunol. 2015, 36, 81–91. [Google Scholar] [CrossRef]
- Yao, J.; Chen, X.; Liu, X.; Li, R.; Zhou, X.; Qu, Y. Characterization of a ferroptosis and iron-metabolism related lncRNA signature in lung adenocarcinoma. Cancer Cell Int. 2021, 21, 340. [Google Scholar] [CrossRef]
- Liu, J.Y.; Peng, C.W.; Yang, G.F.; Hu, W.Q.; Yang, X.J.; Huang, C.Q.; Xiong, B.; Li, Y. Distribution pattern of tumor associated macrophages predicts the prognosis of gastric cancer. Oncotarget 2017, 8, 92757–92769. [Google Scholar] [CrossRef]
- Xu, Z.J.; Gu, Y.; Wang, C.Z.; Jin, Y.; Wen, X.M.; Ma, J.C.; Tang, L.J.; Mao, Z.W.; Qian, J.; Lin, J. The M2 macrophage marker CD206: A novel prognostic indicator for acute myeloid leukemia. Oncoimmunology 2020, 9, 1683347. [Google Scholar] [CrossRef]
- Yan, H.; Qu, J.; Cao, W.; Liu, Y.; Zheng, G.; Zhang, E.; Cai, Z. Identification of prognostic genes in the acute myeloid leukemia immune microenvironment based on TCGA data analysis. Cancer Immunol. Immunother. 2019, 68, 1971–1978. [Google Scholar] [CrossRef] [PubMed]
- Stilund, M.; Gjelstrup, M.C.; Petersen, T.; Moller, H.J.; Rasmussen, P.V.; Christensen, T. Biomarkers of inflammation and axonal degeneration/damage in patients with newly diagnosed multiple sclerosis: Contributions of the soluble CD163 CSF/serum ratio to a biomarker panel. PLoS ONE 2015, 10, e0119681. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.M.; Liu, K.; Liu, J.H.; Jiang, X.L.; Wang, X.L.; Chen, Y.Z.; Li, S.G.; Zou, H.; Pang, L.J.; Liu, C.X.; et al. CD163 as a marker of M2 macrophage, contribute to predicte aggressiveness and prognosis of Kazakh esophageal squamous cell carcinoma. Oncotarget 2017, 8, 21526–21538. [Google Scholar] [CrossRef] [PubMed]
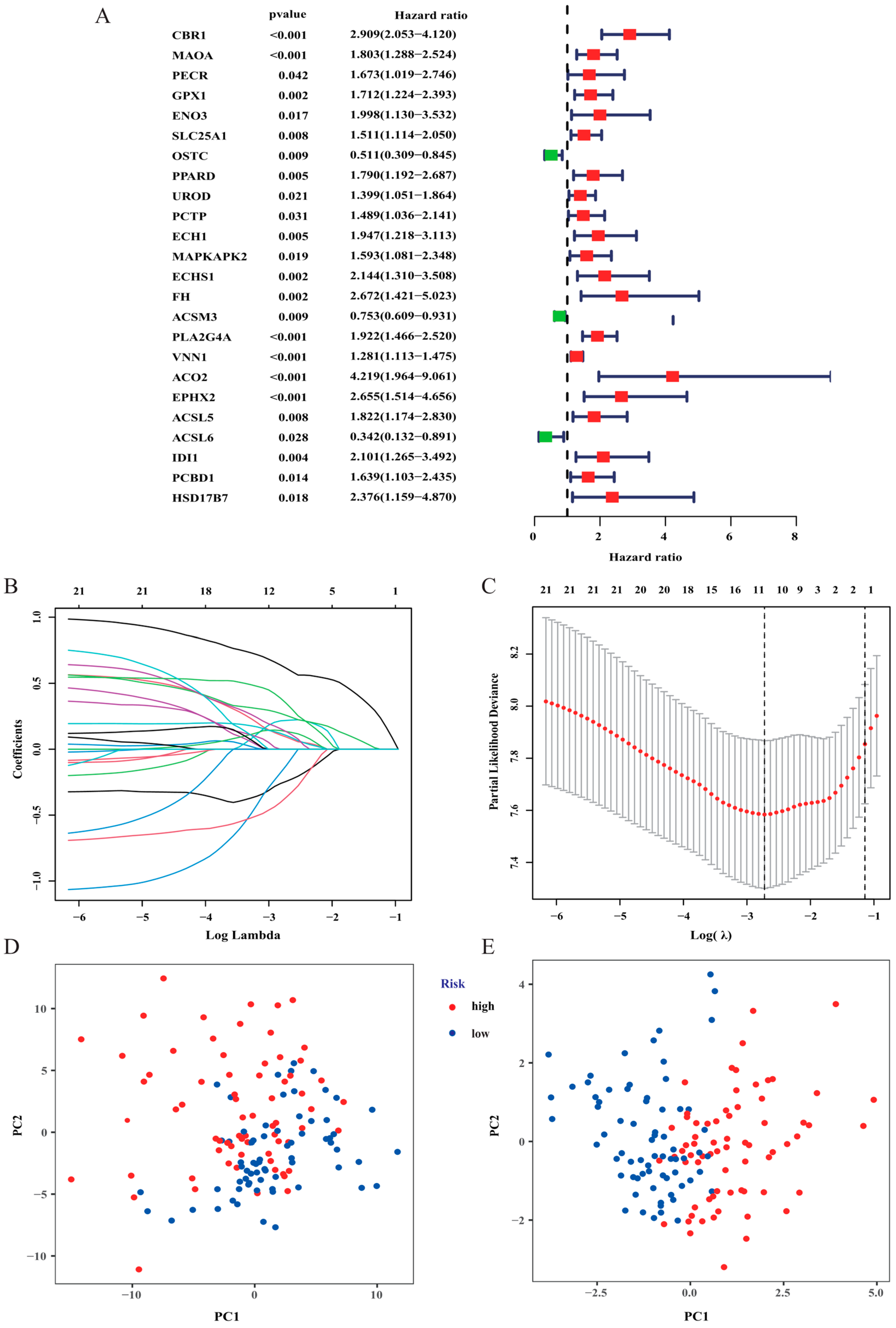

| Gene | Coefficient |
|---|---|
| CBR1 | 0.614533407779894 |
| MAOA | 0.0923271597975086 |
| ENO3 | 0.21850701264524 |
| OSTC | −0.243479390920753 |
| UROD | 0.134439432274683 |
| PCTP | −0.123309988656909 |
| MAPKAPK2 | 0.104413688378819 |
| PLA2G4A | 0.25685461709233 |
| EPHX2 | 0.133099892317426 |
| ACSL6 | −0.364168757158391 |
| IDI1 | 0.347710847872942 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, N.; Bai, X.; Wang, X.; Wang, D.; Ma, G.; Zhang, F.; Ye, J.; Lu, F.; Ji, C. A Novel Fatty Acid Metabolism-Associated Risk Model for Prognosis Prediction in Acute Myeloid Leukaemia. Curr. Oncol. 2023, 30, 2524-2542. https://doi.org/10.3390/curroncol30020193
Wang N, Bai X, Wang X, Wang D, Ma G, Zhang F, Ye J, Lu F, Ji C. A Novel Fatty Acid Metabolism-Associated Risk Model for Prognosis Prediction in Acute Myeloid Leukaemia. Current Oncology. 2023; 30(2):2524-2542. https://doi.org/10.3390/curroncol30020193
Chicago/Turabian StyleWang, Nana, Xiaoran Bai, Xinlu Wang, Dongmei Wang, Guangxin Ma, Fan Zhang, Jingjing Ye, Fei Lu, and Chunyan Ji. 2023. "A Novel Fatty Acid Metabolism-Associated Risk Model for Prognosis Prediction in Acute Myeloid Leukaemia" Current Oncology 30, no. 2: 2524-2542. https://doi.org/10.3390/curroncol30020193
APA StyleWang, N., Bai, X., Wang, X., Wang, D., Ma, G., Zhang, F., Ye, J., Lu, F., & Ji, C. (2023). A Novel Fatty Acid Metabolism-Associated Risk Model for Prognosis Prediction in Acute Myeloid Leukaemia. Current Oncology, 30(2), 2524-2542. https://doi.org/10.3390/curroncol30020193





