The Down-Regulation of Circ_0059707 in Acute Myeloid Leukemia Promotes Cell Growth and Inhibits Apoptosis by Regulating miR-1287-5p
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. BALB/c Nude Mice
2.3. Cell Culture and Transduction
2.4. Cell Growth Assays
2.5. Cell Apoptosis Assays
2.6. RNA Isolation, Reverse Transcription, and Real-Time Quantitative PCR (qRT-PCR)
2.7. HE Staining
2.8. Bioinformatics Analysis
2.9. Statistical Analysis
3. Results
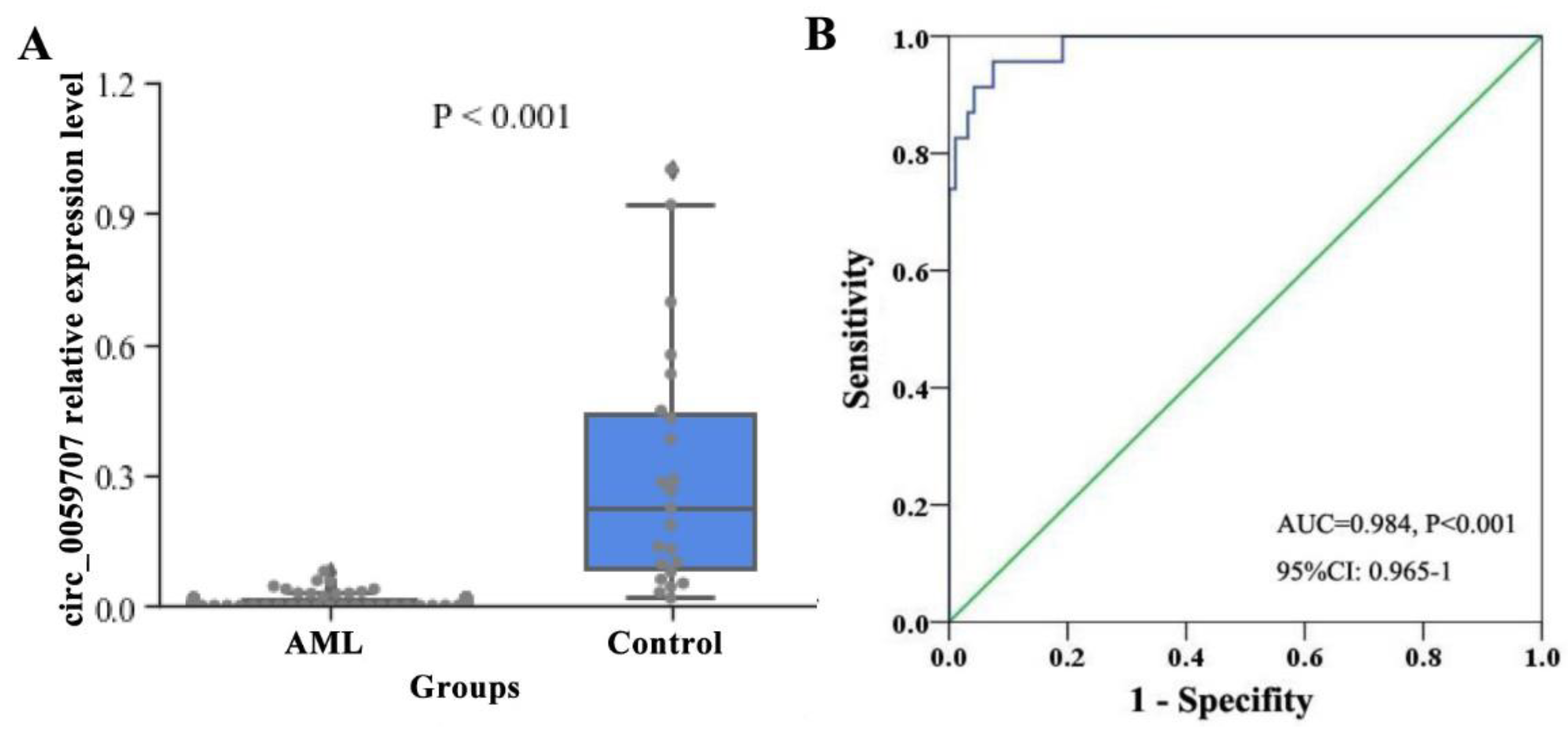
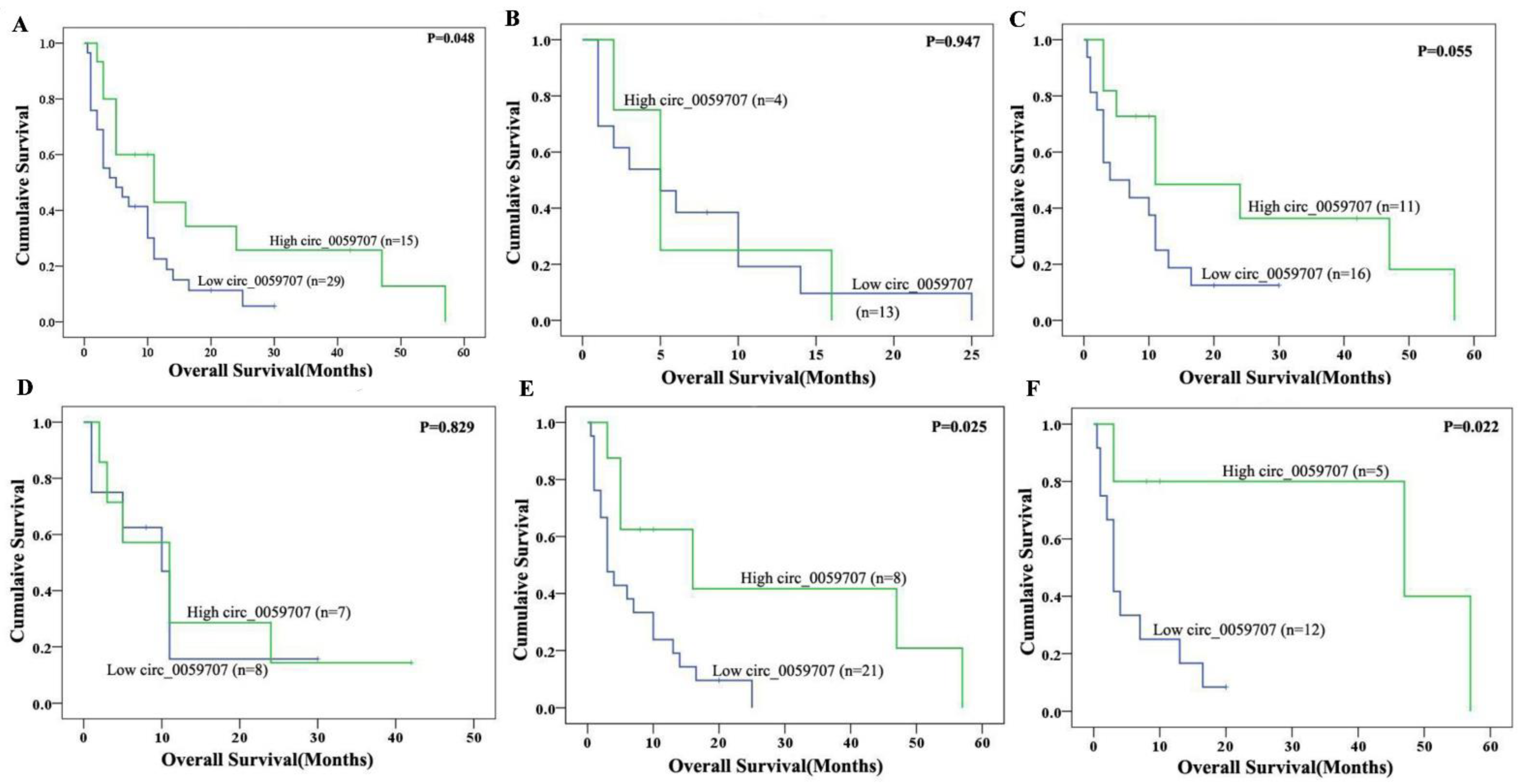
3.1. Relative Expression Levels of Circ_0059707 and Clinical Characteristics of AML
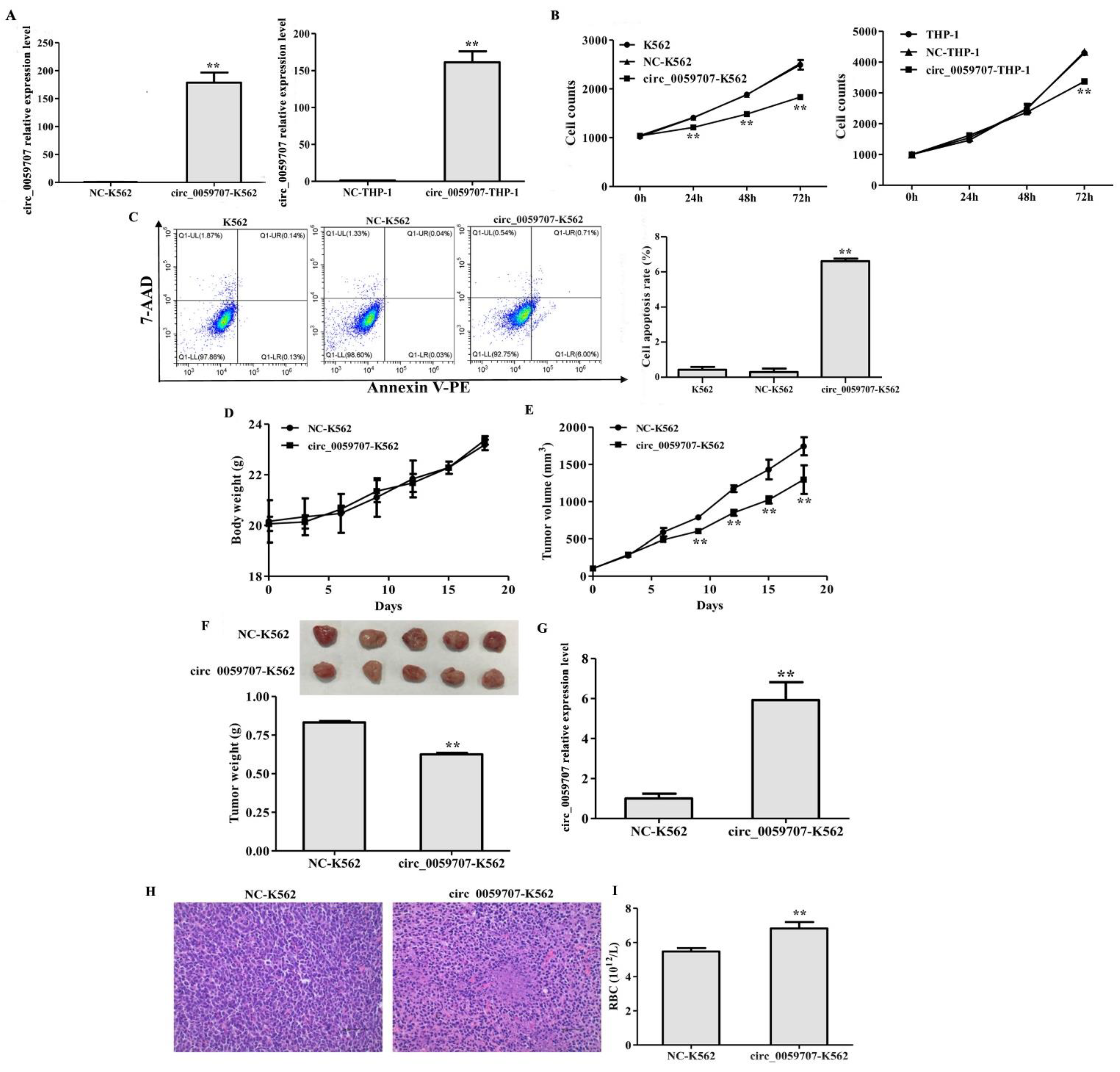
3.2. Circ_0059707 Inhibited the Growth of Leukemia Cells, Increased Apoptosis, and Inhibited Tumor Growth in Mice
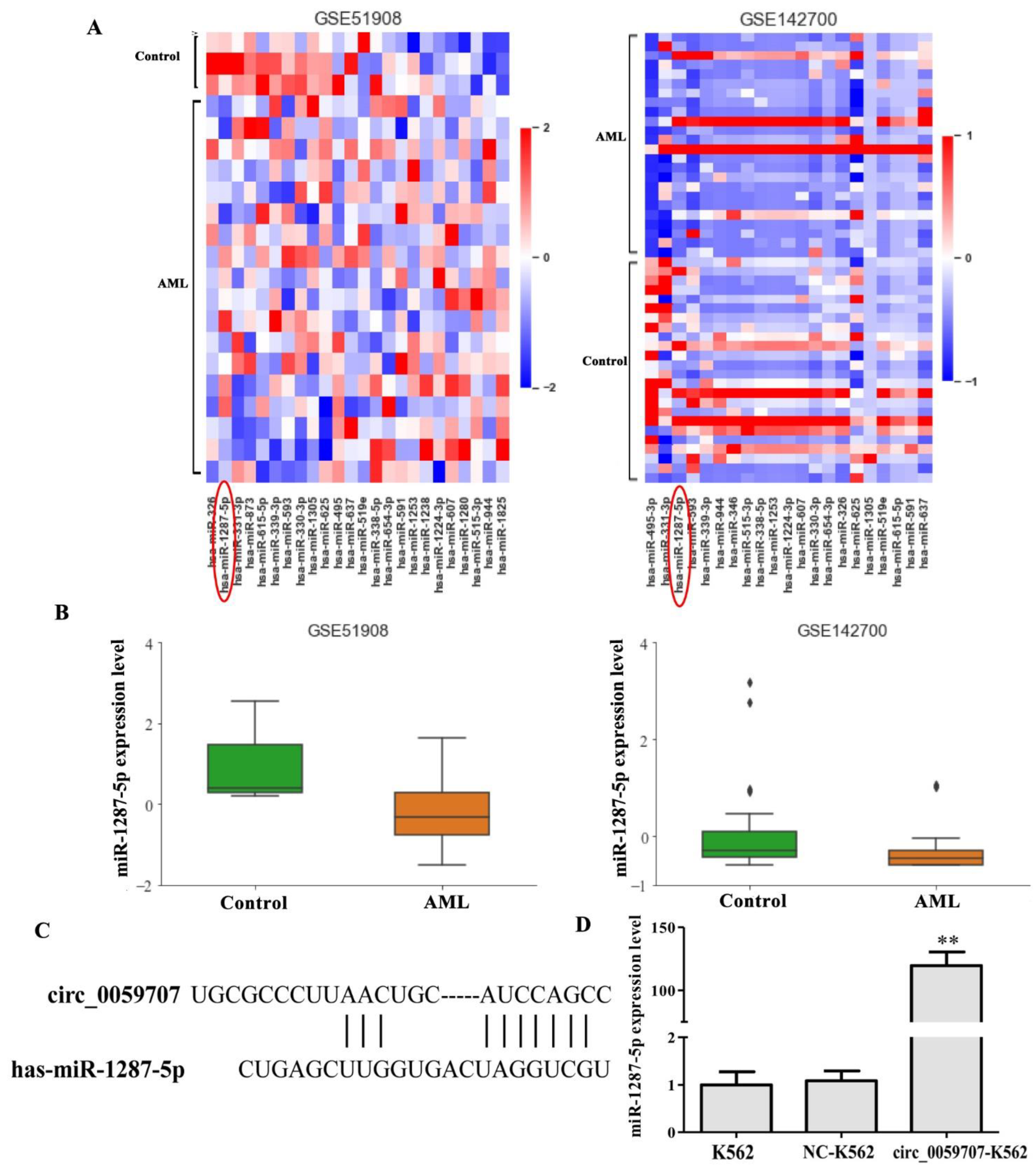
3.3. Mir-1287-5p Was Up-Regulated by Circ_0059707
4. Discussion
5. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shlush, L.I.; Zandi, S.; Mitchell, A.; Chen, W.C.; Brandwein, J.M.; Gupta, V.; Kennedy, J.A.; Schimmer, A.D.; Schuh, A.C.; Yee, K.W.; et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature 2014, 506, 328–333. [Google Scholar] [CrossRef]
- Khwaja, A.; Bjorkholm, M.; Gale, R.E.; Levine, R.L.; Jordan, C.T.; Ehninger, G.; Bloomfield, C.D.; Estey, E.; Burnett, A.; Cornelissen, J.J.; et al. Acute myeloid leukaemia. Nat. Rev. Dis. Primers 2016, 2, 16010. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Wang, P.; Han, G.; Zhang, T.; Chang, J.; Yin, R.; Shan, Y.; Wen, J.; Xie, X.; et al. Leukemogenic Chromatin Alterations Promote AML Leukemia Stem Cells via a KDM4C-ALKBH5-AXL Signaling Axis. Cell Stem Cell 2020, 27, 81–97.e88. [Google Scholar] [CrossRef] [PubMed]
- Martelli, M.P.; Rossi, R.; Venanzi, A.; Meggendorfer, M.; Perriello, V.M.; Martino, G.; Spinelli, O.; Ciurnelli, R.; Varasano, E.; Brunetti, L.; et al. Novel NPM1 exon 5 mutations and gene fusions leading to aberrant cytoplasmic nucleophosmin in AML. Blood 2021, 138, 2696–2701. [Google Scholar] [CrossRef] [PubMed]
- McNeer, N.A.; Philip, J.; Geiger, H.; Ries, R.E.; Lavallee, V.P.; Walsh, M.; Shah, M.; Arora, K.; Emde, A.K.; Robine, N.; et al. Genetic mechanisms of primary chemotherapy resistance in pediatric acute myeloid leukemia. Leukemia 2019, 33, 1934–1943. [Google Scholar] [CrossRef] [PubMed]
- Crunkhorn, S. Epigenetic therapy prevents AML. Nat. Rev. Drug Discov. 2020, 19, 168. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, Z.; Pang, Y.; Cui, L.; Qian, T.; Quan, L.; Zhao, H.; Shi, J.; Ke, X.; Fu, L. Role of microRNAs, circRNAs and long noncoding RNAs in acute myeloid leukemia. J. Hematol. Oncol. 2019, 12, 51. [Google Scholar] [CrossRef]
- Bhayadia, R.; Krowiorz, K.; Haetscher, N.; Jammal, R.; Emmrich, S.; Obulkasim, A.; Fiedler, J.; Schwarzer, A.; Rouhi, A.; Heuser, M.; et al. Endogenous Tumor Suppressor microRNA-193b: Therapeutic and Prognostic Value in Acute Myeloid Leukemia. J. Clin. Oncol. 2018, 36, 1007–1016. [Google Scholar] [CrossRef]
- Benetatos, L.; Vartholomatos, G. Enhancer DNA methylation in acute myeloid leukemia and myelodysplastic syndromes. Cell. Mol. Life Sci. 2018, 75, 1999–2009. [Google Scholar] [CrossRef]
- Sun, L.; Wang, W.; Han, C.; Huang, W.; Sun, Y.; Fang, K.; Zeng, Z.; Yang, Q.; Pan, Q.; Chen, T.; et al. The oncomicropeptide APPLE promotes hematopoietic malignancy by enhancing translation initiation. Mol. Cell 2021, 81, 4493–4508.e9. [Google Scholar] [CrossRef]
- Zhu, G.; Luo, H.; Feng, Y.; Guryanova, O.A.; Xu, J.; Chen, S.; Lai, Q.; Sharma, A.; Xu, B.; Zhao, Z.; et al. HOXBLINC long non-coding RNA activation promotes leukemogenesis in NPM1-mutant acute myeloid leukemia. Nat. Commun. 2021, 12, 1956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Nguyen, L.X.T.; Zhao, D.; Frankhouser, D.E.; Wang, H.; Hoang, D.H.; Qiao, J.; Abundis, C.; Brehove, M.; Su, Y.L.; et al. Treatment-induced arteriolar revascularization and miR-126 enhancement in bone marrow niche protect leukemic stem cells in AML. J. Hematol. Oncol. 2021, 14, 122. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pan, J.; Huang, S.; Li, F.; Huang, J.; Li, X.; Ling, Q.; Ye, W.; Wang, Y.; Yu, W.; et al. Development and validation of a novel circular RNA as an independent prognostic factor in acute myeloid leukemia. BMC Med. 2021, 19, 28. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Sun, Y.M.; Wang, W.T.; Zeng, Z.C.; Chen, T.Q.; Han, C.; Pan, Q.; Huang, W.; Fang, K.; Sun, L.Y.; Zhou, Y.F.; et al. circMYBL2, a circRNA from MYBL2, regulates FLT3 translation by recruiting PTBP1 to promote FLT3-ITD AML progression. Blood 2019, 134, 1533–1546. [Google Scholar] [CrossRef]
- Wang, X.; Jin, P.; Zhang, Y.; Wang, K. CircSPI1 acts as an oncogene in acute myeloid leukemia through antagonizing SPI1 and interacting with microRNAs. Cell Death Dis. 2021, 12, 297. [Google Scholar] [CrossRef]
- Liu, X.; Liu, X.; Cai, M.; Luo, A.; He, Y.; Liu, S.; Zhang, X.; Yang, X.; Xu, L.; Jiang, H. CircRNF220, not its linear cognate gene RNF220, regulates cell growth and is associated with relapse in pediatric acute myeloid leukemia. Mol. Cancer 2021, 20, 139. [Google Scholar] [CrossRef]
- Hasskarl, J.; Duensing, S.; Manuel, E.; Munger, K. The helix-loop-helix protein ID1 localizes to centrosomes and rapidly induces abnormal centrosome numbers. Oncogene 2004, 23, 1930–1938. [Google Scholar] [CrossRef]
- Cook, P.J.; Thomas, R.; Kingsley, P.J.; Shimizu, F.; Montrose, D.C.; Marnett, L.J.; Tabar, V.S.; Dannenberg, A.J.; Benezra, R. Cox-2-derived PGE2 induces Id1-dependent radiation resistance and self-renewal in experimental glioblastoma. Neuro Oncol. 2016, 18, 1379–1389. [Google Scholar] [CrossRef]
- Yin, X.; Tang, B.; Li, J.H.; Wang, Y.; Zhang, L.; Xie, X.Y.; Zhang, B.H.; Qiu, S.J.; Wu, W.Z.; Ren, Z.G. ID1 promotes hepatocellular carcinoma proliferation and confers chemoresistance to oxaliplatin by activating pentose phosphate pathway. J. Exp. Clin. Cancer Res. 2017, 36, 166. [Google Scholar] [CrossRef]
- Valanti, E.K.; Dalakoura-Karagkouni, K.; Fotakis, P.; Vafiadaki, E.; Mantzoros, C.S.; Chroni, A.; Zannis, V.; Kardassis, D.; Sanoudou, D. Reconstituted HDL-apoE3 promotes endothelial cell migration through ID1 and its downstream kinases ERK1/2, AKT and p38 MAPK. Metabolism 2022, 127, 154954. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.D.; Yang, L.; Zhu, X.W.; Wen, X.M.; Yang, J.; Guo, H.; Chen, Q.; Yao, D.M.; Ma, J.C.; Lin, J.; et al. Clinical significance of up-regulated ID1 expression in Chinese de novo acute myeloid leukemia. Int. J. Clin. Exp. Pathol. 2015, 8, 5336–5344. [Google Scholar] [PubMed]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Bennett, J.M.; Catovsky, D.; Daniel, M.T.; Flandrin, G.; Galton, D.A.; Gralnick, H.R.; Sultan, C. Proposed revised criteria for the classification of acute myeloid leukemia. A report of the French-American-British Cooperative Group. Ann. Intern. Med. 1985, 103, 620–625. [Google Scholar] [CrossRef]
- Dohner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef]
- Ding, L.; Zhao, Y.; Dang, S.; Wang, Y.; Li, X.; Yu, X.; Li, Z.; Wei, J.; Liu, M.; Li, G. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol. Cancer 2019, 18, 45. [Google Scholar] [CrossRef]
- Chen, L.Y.; Wang, L.; Ren, Y.X.; Pang, Z.; Liu, Y.; Sun, X.D.; Tu, J.; Zhi, Z.; Qin, Y.; Sun, L.N.; et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1alpha translation. Mol. Cancer 2020, 19, 164. [Google Scholar] [CrossRef]
- Shangguan, H.; Feng, H.; Lv, D.; Wang, J.; Tian, T.; Wang, X. Circular RNA circSLC25A16 contributes to the glycolysis of non-small-cell lung cancer through epigenetic modification. Cell Death Dis. 2020, 11, 437. [Google Scholar] [CrossRef]
- Jiang, T.; Xia, Y.; Lv, J.; Li, B.; Li, Y.; Wang, S.; Xuan, Z.; Xie, L.; Qiu, S.; He, Z.; et al. A novel protein encoded by circMAPK1 inhibits progression of gastric cancer by suppressing activation of MAPK signaling. Mol. Cancer 2021, 20, 66. [Google Scholar] [CrossRef]
- Jiao, J.; Zhang, T.; Jiao, X.; Huang, T.; Zhao, L.; Ma, D.; Cui, B. hsa_circ_0000745 promotes cervical cancer by increasing cell proliferation, migration, and invasion. J. Cell Physiol. 2020, 235, 1287–1295. [Google Scholar] [CrossRef]
- Liu, F.; Zhang, H.; Xie, F.; Tao, D.; Xiao, X.; Huang, C.; Wang, M.; Gu, C.; Zhang, X.; Jiang, G. Hsa_circ_0001361 promotes bladder cancer invasion and metastasis through miR-491-5p/MMP9 axis. Oncogene 2020, 39, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.; Zheng, H.; Wu, Z.; Chen, M.; Huang, Y. Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 2020, 10, 3503–3517. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.Y.; Yi, J.; Zhu, X.; Zhang, J.; Zhou, J.; Tang, X.; Lin, J.; Wang, P.; Deng, Z.Q. Circular RNA of vimentin expression as a valuable predictor for acute myeloid leukemia development and prognosis. J. Cell Physiol. 2019, 234, 3711–3719. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Liu, T.; Liu, J.; Feng, Y.; Wang, B.; Wang, J.; Bai, J.; Zhao, W.; Shen, Y.; Wang, X.; et al. Circ-ANAPC7 is Upregulated in Acute Myeloid Leukemia and Appears to Target the MiR-181 Family. Cell Physiol. Biochem. 2018, 47, 1998–2007. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Chen, W.M.; Wang, Z.H.; Wei, T.N.; Chen, Z.Z.; Wu, W.B. CircPAN3 mediates drug resistance in acute myeloid leukemia through the miR-153-5p/miR-183-5p-XIAP axis. Exp. Hematol. 2019, 70, 42–54.e43. [Google Scholar] [CrossRef]
- Lin, Y.; Huang, Y.; Liang, C.; Xie, S.; Xie, A. Silencing of circTASP1 inhibits proliferation and induces apoptosis of acute myeloid leukaemia cells through modulating miR-515-5p/HMGA2 axis. J. Cell Mol. Med. 2021, 25, 7367–7380. [Google Scholar] [CrossRef]
- Han, F.; Zhong, C.; Li, W.; Wang, R.; Zhang, C.; Yang, X.; Ji, C.; Ma, D. hsa_circ_0001947 suppresses acute myeloid leukemia progression via targeting hsa-miR-329-5p/CREBRF axis. Epigenomics 2020, 12, 935–953. [Google Scholar] [CrossRef]
- Schwarzenbacher, D.; Klec, C.; Pasculli, B.; Cerk, S.; Rinner, B.; Karbiener, M.; Ivan, C.; Barbano, R.; Ling, H.; Wulf-Goldenberg, A.; et al. MiR-1287-5p inhibits triple negative breast cancer growth by interaction with phosphoinositide 3-kinase CB, thereby sensitizing cells for PI3Kinase inhibitors. Breast Cancer Res. 2019, 21, 20. [Google Scholar] [CrossRef]
- Cui, G.; Zhao, H.; Li, L. Long noncoding RNA PRKCQ-AS1 promotes CRC cell proliferation and migration via modulating miR-1287-5p/YBX1 axis. J. Cell Biochem. 2020, 121, 4166–4175. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, L.; Wang, C.; Zhang, L.; Xu, W. MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through inhibiting GPX4. Free Radic. Res. 2021, 55, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
| Patient’s Parameters | Low (n = 62) | High (n = 32) | p Value |
|---|---|---|---|
| Sex, male/female | 37/25 | 19/13 | 0.847 |
| Age, median (range), years ∆ | 54 (15–81) | 59 (27–84) | 0.127 |
| WBC, median (range), ×109/L ∆ | 49.6 (0.8–528) | 55.8 (0.8–207.9) | 0.388 |
| Hemoglobin, median (range), g/L ∆ | 84.7 (42–138) | 93.3 (58–141) | 0.045 |
| Platelets, median (range), ×109/L ∆ | 46.6 (7–192) | 66.8 (5–382) | 0.103 |
| BM blasts, median (range), % ∆ | 46.1 (0–95.0) | 35.8 (0–80.0) | 0.050 |
| CR (+/−) | 27/22 | 8/13 | 0.297 |
| FAB classification | 0.433 | ||
| M0 | 1 | 0 | |
| M1 | 2 | 0 | |
| M2 | 24 | 17 | |
| M3 | 10 | 2 | |
| M4 | 14 | 5 | |
| M5 | 6 | 4 | |
| No data | 4 | 4 | |
| Risk classification | 0.273 | ||
| Low | 8 (13%) | 3 (9%) | |
| Intermediate | 37 (60%) | 23 (72%) | |
| High | 16 (26%) | 4 (13%) | |
| No data | 1 (2%) | 2 (6%) | |
| Karyotypes | 0.801 | ||
| normal | 29 (47%) | 17 (53%) | |
| t(8;21) | 5 (8%) | 2 (6%) | |
| t(16;16) | 1 (2%) | 0 (0%) | |
| t(15;17) | 10 (16%) | 2 (6%) | |
| t(9;22) | 1 (2%) | 0 (0%) | |
| +8 | 3 (5%) | 2 (6%) | |
| −7/7q- | 1 (2%) | 0 (0%) | |
| complex | 6 (10%) | 3 (9%) | |
| others | 5 (8%) | 4 (13%) | |
| No data | 1(2%) | 2 (6%) | |
| Gene mutations | |||
| CEBPA (+/−) | 3/49 | 2/23 | 0.361 |
| NPM1 (+/−) | 10/42 | 7/20 | 0.250 |
| FLT3-ITD (+/−) | 6/46 | 2/23 | 0.322 |
| c-KIT (+/−) | 2/50 | 1/24 | 0.494 |
| N/K-RAS (+/−) | 6/37 | 2/15 | 0.417 |
| IDH1/2 (+/−) | 2/41 | 1/16 | 0.431 |
| DNMT3A (+/−) | 4/48 | 2/23 | 0.486 |
| U2AF1 (+/−) | 0/43 | 0/17 | - |
| SRSF2 (+/−) | 0/43 | 1/16 | 0.060 |
| SETBP1 (+/−) | 1/34 | 0/16 | 0.263 |
| TP53 (+/−) | 1/12 | 1/8 | 0.420 |
| TET2 (+/−) | 0/8 | 1/4 | 0.134 |
| Univariate | Multivariate | |||
|---|---|---|---|---|
| Hazard Ratio (95% CI) | p Value | Hazard Ratio (95% CI) | p Value | |
| Circ_0059707 expression | 0.500 (0.240–1.044) | 0.065 | 0.441 (0.210–0.929) | 0.031 |
| Age | 1.841 (0.943–3.594) | 0.074 | 1.401 (0.707–2.778) | 0.334 |
| WBC | 2.008 (1.012–3.985) | 0.046 | 1.611 (0.756–3.433) | 0.009 |
| Risk classification | 1.579 (0.888–2.809) | 0.019 | 1.993 (1.199–3.311) | 0.008 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, J.; Wen, X.; Xu, Z.; Xia, P.; Jin, Y.; Lin, J.; Qian, J. The Down-Regulation of Circ_0059707 in Acute Myeloid Leukemia Promotes Cell Growth and Inhibits Apoptosis by Regulating miR-1287-5p. Curr. Oncol. 2022, 29, 6688-6699. https://doi.org/10.3390/curroncol29090525
Ma J, Wen X, Xu Z, Xia P, Jin Y, Lin J, Qian J. The Down-Regulation of Circ_0059707 in Acute Myeloid Leukemia Promotes Cell Growth and Inhibits Apoptosis by Regulating miR-1287-5p. Current Oncology. 2022; 29(9):6688-6699. https://doi.org/10.3390/curroncol29090525
Chicago/Turabian StyleMa, Jichun, Xiangmei Wen, Zijun Xu, Peihui Xia, Ye Jin, Jiang Lin, and Jun Qian. 2022. "The Down-Regulation of Circ_0059707 in Acute Myeloid Leukemia Promotes Cell Growth and Inhibits Apoptosis by Regulating miR-1287-5p" Current Oncology 29, no. 9: 6688-6699. https://doi.org/10.3390/curroncol29090525
APA StyleMa, J., Wen, X., Xu, Z., Xia, P., Jin, Y., Lin, J., & Qian, J. (2022). The Down-Regulation of Circ_0059707 in Acute Myeloid Leukemia Promotes Cell Growth and Inhibits Apoptosis by Regulating miR-1287-5p. Current Oncology, 29(9), 6688-6699. https://doi.org/10.3390/curroncol29090525





