Noninvasive Determination of the IDH Status of Gliomas Using MRI and MRI-Based Radiomics: Impact on Diagnosis and Prognosis
Abstract
1. Introduction
2. Correlation of Conventional Magnetic Resonance Imaging (cMRI) Findings with IDH Mutation Status and The Prognosis of Gliomas
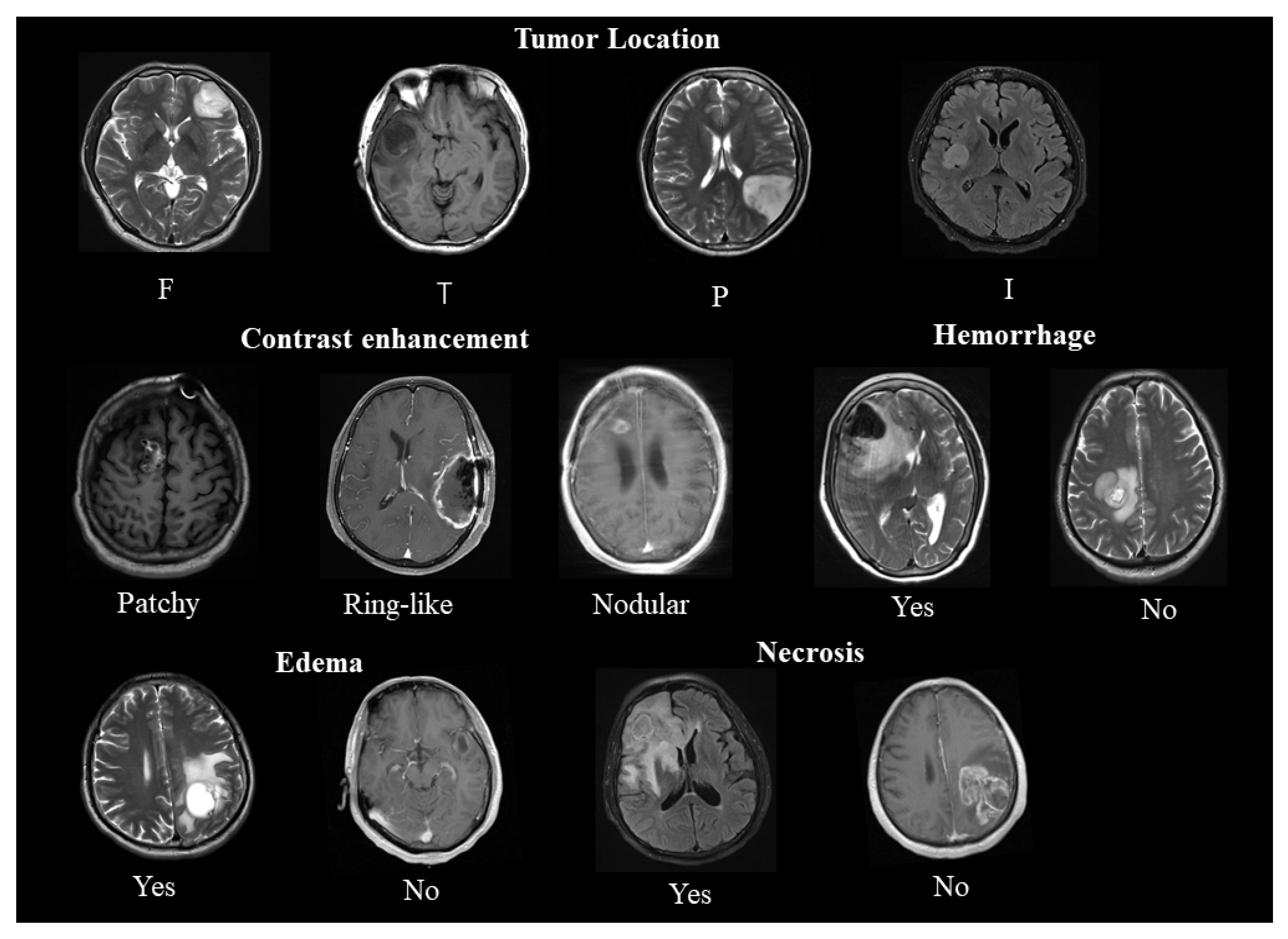
2.1. Location
2.2. Enhancement
2.3. Edema, Necrosis, and Hemorrhage
3. Correlation of Advanced Magnetic Resonance Imaging (aMRI) Findings with IDH Mutation Status and The Prognosis of Gliomas
3.1. Perfusion Weighted Imaging (PWI)
3.2. Diffusion Weighted Imaging (DWI)
3.3. Diffusion Tensor Imaging (DTI)
3.4. Diffusion Kurtosis Imaging (DKI)
| Variable | First Author, Year | Classification | IDH1 Status | p-Value | ||
|---|---|---|---|---|---|---|
| Total | Mutant | Wildtype | ||||
| Age | Song et al. (2014) [26] | Astrocytoma | 36.5 | 32.7 | 42.5 | <0.001 |
| Xiong et al. (2016) [57] | Oligodendroglial tumor | 37.9 ± 10.2 | 44.5 ± 12.1 | 0.038 | ||
| Lasocki et al. (2017) [23] | Glioblastoma | 50.6 ± 11.2 | 64.9 ± 12.0 | 0.014 | ||
| Xing et al. (2017) [50] | Astrocytoma | 35.76 ± 9.13 | 45.96 ± 18.36 | 0.041 | ||
| Xing et al. (2019) [49] | Glioblastoma | 40.7 ± 10.77 | 52.23 ± 12.71 | 0.008 | ||
| Tan et al. (2020) [64] | Astrocytoma | 43.23 ± 11.87 | 55.97 ± 11.74 | 0.001 | ||
| Frontal lobe location (Yes/no) (cMRI) | Song et al. (2014) [26] | Astrocytoma | 66/127 | 53/64 | 13/63 | NA |
| Wang et al. (2015) [33] | Anaplastic glioma | 50/34 | 45/22 | 5/12 | 0.005 | |
| Lasocki et al. (2017) [23] | Glioblastoma | 58/95 | 2/3 | 56/92 | 1 | |
| Xing et al. (2017) [50] | Astrocytoma | 13/29 | 9/8 | 4/2 | 0.006 | |
| Xing et al. (2019) [49] | Glioblastoma | 32/43 | 9/1 | 23/42 | 0.002 | |
| Contrast enhancement (Yes/no) (cMRI) | Song et al. (2014) [26] | Astrocytoma | 96/97 | 43/74 | 53/23 | <0.001 |
| Wang et al. (2015) [33] | Anaplastic glioma | 173/43 | 57/27 | 116/16 | <0.001 | |
| Xiong et al. (2016) [57] | Oligodendroglial tumor | 43/41 | 38/29 | 5/12 | 0.013 | |
| Wang et al. (2016) [34] | Glioblastoma | 256/24 | 33/12 | 223/12 | <0.001 | |
| Xing et al. (2017) [50] | Astrocytoma | 19/23 | 6/11 | 13/12 | 0.286 | |
| Tan et al. (2020) [64] | Astrocytoma | 52/10 | 21/9 | 31/1 | 0.002 | |
| Edema (Yes/no) (cMRI) | Song et al. (2014) [26] | Astrocytoma | 84/109 | 46/71 | 38/38 | 0.181 |
| Xiong et al. (2016) [57] | Oligodendroglial tumor | 57/27 | 47/20 | 10/7 | 0.372 | |
| Wang et al. (2016) [34] | Glioblastoma | 213/67 | 32/13 | 181/54 | 0.395 | |
| Xing et al. (2017) [50] | Astrocytoma | 10/32 | 3/14 | 7/18 | 0.746 | |
| Tan et al. (2020) [64] | Astrocytoma | 39/23 | 11/19 | 28/4 | 0.000 | |
| Hemorrhage (Yes/no) (cMRI) | Xiong et al. (2016) [57] | Oligodendroglial tumor | 6/78 | 3/64 | 3/14 | 0.06 |
| Xing et al. (2019) [49] | Glioblastoma | 58/17 | 9/1 | 49/16 | 0.439 | |
| Tan et al. (2020) [64] | Astrocytoma | 12/50 | 4/26 | 8/24 | 0.249 | |
| Necrosis (Yes/no) (cMRI) | Xiong et al. (2016) [57] | Oligodendroglial tumor | 42/42 | 37/30 | 5/12 | 0.162 |
| Xing et al. (2019) [49] | Glioblastoma | 62/13 | 8/2 | 54/11 | 1.000 | |
| Tan et al. (2020) [64] | Astrocytoma | 45/17 | 21/9 | 24/8 | 0.697 | |
3.5. Magnetic Resonance Spectroscopy (MRS)
3.6. Amide Proton Transfer (APT)
| First Author, Year | Classification | Variable | IDG Status | p-Value | |
|---|---|---|---|---|---|
| Mutant | Wildtype | ||||
| Xiong et al. (2016) [57] | Oligodendroglial tumor | Maximal FA (DTI) | 0.23 ± 0.10 | 0.30 ± 0.07 | 0.009 |
| Ratio of maximal FA (DTI) | 0.33 ± 0.15 | 0.44 ± 0.11 | 0.004 | ||
| Oedema FA (DTI) | 0.26 ± 0.14 | 0.21 ± 0.08 | 0.138 | ||
| Ratio of oedema FA (DTI) | 0.37 ± 0.20 | 0.30 ± 0.11 | 0.15 | ||
| Normal FA (DTI) | 0.71 ± 0.03 | 0.69 ± 0.03 | 0.122 | ||
| Minimal ADC (×10−3 mm2/s) (DWI) | 1.10 ± 0.22 | 0.81 ± 0.16 | 0.001 | ||
| Ratio of minimal ADC (DWI) | 1.40 ± 0.32 | 1.13 ± 0.23 | 0.002 | ||
| Oedema ADC (×10−3 mm2/s) (DWI) | 1.20 ± 0.30 | 1.37 ± 0.30 | 0.036 | ||
| Ratio of oedema ADC (DWI) | 1.67 ± 0.43 | 1.91 ± 0.42 | 0.034 | ||
| Normal ADC (DWI) | 0.72 ± 0.03 | 0.72 ± 0.03 | 0.746 | ||
| Tan et al. (2019) [64] | Astrocytoma | MK (DKI) | 0.48 ± 0.16 | 0.67 ± 0.13 | <0.001 |
| Kr (DKI) | 0.45 ± 0.18 | 0.68 ± 0.19 | <0.001 | ||
| Ka (DKI) | 0.53 ± 0.17 | 0.66 ± 0.14 | 0.002 | ||
| MD (DKI) | 1.49 ± 0.41 | 1.22 ± 0.26 | 0.005 | ||
| FA (DTI) | 0.18 ± 0.17 | 0.20 ± 0.09 | 0.408 | ||
| Xing et al. (2017) [50] | Astrocytoma | ADCmin (×10−3 mm2/s) (DWI) | 1.21 ± 0.27 | 0.87 ± 0.18 | <0.001 |
| rADC (DWI) | 1.88 ± 0.41 | 1.37 ± 0.31 | <0.001 | ||
| rCBVmax (PWI) | 1.41 ± 0.50 | 3.47 ± 2.34 | 0.004 | ||
| Jiang et al. (2017) [74] | Glioma | Maximum APT | 0.99 ± 0.33 | 2.03 ± 0.72 | <0.001 |
| Minimum APT | 0.59 ± 0.32 | 0.99 ± 0.47 | 0.02 | ||
| Mean APT | 0.93 ± 0.44 | 1.39 ± 0.49 | 0.03 | ||
3.7. Physiological MRI of Oxygen Metabolism
4. MRI-Based Radiomics Could Predict IDH Status and Clinical Outcome in Patients with Gliomas
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Weller, M.; dan der Bent, M.; Tonn, J.C.; Stupp, R.; Preusser, M.; Cohen-Jonathan-Moyal, E.; Henriksson, R.; Le Rhun, E.; Balana, C.; Chinot, O.; et al. European Association for Neuro-Oncology (EANO) guideline on the diagnosis and treatment of adult astrocytic and oligodendroglial gliomas. Lancet Oncol. 2017, 18, e315–e329. [Google Scholar] [CrossRef]
- Weller, M.; Wick, W.; Aldape, K.; Brada, M.; Berger, M.; Pfister, S.M.; Nishikawa, R.; Rosenthal, M.; Wen, P.Y.; Stupp, R.; et al. Glioma. Nat. Rev. Dis. Primers 2015, 15017. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestler, O.D.; Cavenee, W.K.; Burger, P.C.; Jouvet, A.; Scheithauer, B.W.; Kleihues, P. The 2007 WHO Classification of Tumours of the Central Nervous System. Acta Neuropathol. 2007, 114, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.-H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.-M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Dang, L.; Jin, S.; Su, S.M. IDH mutations in glioma and acute myeloid leukemia. Trends Mol. Med. 2010, 16, 387–397. [Google Scholar] [CrossRef]
- Reitman, Z.J.; Yan, H. Isocitrate Dehydrogenase 1 and 2 Mutations in Cancer: Alterations at a Crossroads of Cellular Metabolism. JNCI J. Natl. Cancer Inst. 2010, 102, 932–941. [Google Scholar] [CrossRef]
- Krell, D.; Mulholland, P.; E Frampton, A.; Krell, J.; Stebbing, J.; Bardella, C. IDH mutations in tumorigenesis and their potential role as novel therapeutic targets. Futur. Oncol. 2013, 9, 1923–1935. [Google Scholar] [CrossRef]
- Zhao, S.; Lin, Y.; Xu, W.; Jiang, W.; Zha, Z.; Wang, P.; Yu, W.; Li, Z.; Gong, L.; Peng, Y.; et al. Glioma-Derived Mutations in IDH1 Dominantly Inhibit IDH1 Catalytic Activity and Induce HIF-1α. Science 2009, 324, 261–265. [Google Scholar] [CrossRef] [PubMed]
- Tsukada, Y.-I.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2005, 439, 811–816. [Google Scholar] [CrossRef]
- Tahiliani, M.; Koh, K.P.; Shen, Y.; Pastor, W.A.; Bandukwala, H.; Brudno, Y.; Agarwal, S.; Iyer, L.M.; Liu, D.R.; Aravind, L.; et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324, 930–935. [Google Scholar] [CrossRef]
- Xu, W.; Yang, H.; Liu, Y.; Yang, Y.; Wang, P.; Kim, S.-H.; Ito, S.; Yang, C.; Wang, P.; Xiao, M.-T.; et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell 2011, 19, 17–30. [Google Scholar] [CrossRef] [PubMed]
- van den Bent, M.J.; Dubbink, H.J.; Marie, Y.; Brandes, A.A.; Taphoorn, M.J.; Wesseling, P.; Frenay, M.; Tijssen, C.C.; Lacombe, D.; Idbaih, A.; et al. IDH1 and IDH2 mutations are prognostic but not predictive for outcome in anaplastic oligodendroglial tumors: A report of the European Organization for Research and Treatment of Cancer Brain Tumor Group. Clin. Cancer Res. 2010, 16, 1597–1604. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wei, W.; Liu, Z.; Liang, Y.; Liu, X.; Li, Y.; Tang, Z.; Jiang, T.; Tian, J. Predicting the Type of Tumor-Related Epilepsy in Patients With Low-Grade Gliomas: A Radiomics Study. Front. Oncol. 2020, 10, 235. [Google Scholar] [CrossRef]
- Vives, K.; Piepmeier, J. Complications and expected outcome of glioma surgery. J. Neuro-Oncol. 1999, 42, 289–302. [Google Scholar] [CrossRef]
- Parker, N.R.; Khong, P.; Parkinson, J.F.; Howell, V.M.; Wheeler, H.R. Molecular heterogeneity in glioblastoma: Potential clinical implications. Front. Oncol. 2015, 5, 55. [Google Scholar] [CrossRef]
- Patel, A.P.; Tirosh, I.; Trombetta, J.J.; Shalek, A.K.; Gillespie, S.M.; Wakimoto, H.; Cahill, D.P.; Nahed, B.V.; Curry, W.T.; Martuza, R.L.; et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014, 344, 1396–1401. [Google Scholar] [CrossRef]
- Jackson, R.J.; Fuller, G.N.; Abi-Said, D.; Lang, F.F.; Gokaslan, Z.L.; Shi, W.M.; Wildrick, D.M.; Sawaya, R. Limitations of stereotactic biopsy in the initial management of gliomas. Neuro-oncology 2001, 3, 193–200. [Google Scholar] [CrossRef]
- Muragaki, Y.; Chernov, M.; Maruyama, T.; Ochiai, T.; Taira, T.; Kubo, O.; Nakamura, R.; Iseki, H.; Hori, T.; Takakura, K. Low-grade glioma on stereotactic biopsy: How often is the diagnosis accurate? Minim. Invasive Neurosurg. 2008, 51, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Abrams, D.A.; Hanson, J.A.; Brown, J.M.; Hsu, F.P.; Delashaw, J.B., Jr.; Bota, D.A. Timing of surgery and bevacizumab therapy in neurosurgical patients with recurrent high grade glioma. J. Clin. Neurosci. 2015, 22, 35–39. [Google Scholar] [CrossRef][Green Version]
- Lasocki, A.; Tsui, A.; Tacey, M.; Drummond, K.J.; Field, K.M.; Gaillard, F. MRI Grading versus Histology: Predicting Survival of World Health Organization Grade II-IV Astrocytomas. Am. J. Neuroradiol. 2014, 36, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Shofty, B.; Artzi, M.; Ben Bashat, D.; Liberman, G.; Haim, O.; Kashanian, A.; Bokstein, F.; Blumenthal, D.T.; Ram, Z.; Shahar, T. MRI radiomics analysis of molecular alterations in low-grade gliomas. Int. J. Comput. Assist. Radiol. Surg. 2017, 13, 563–571. [Google Scholar] [CrossRef] [PubMed]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.P.M.; Granton, P.; Zegers, C.M.L.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Qi, S.; Yu, L.; Li, H.; Ou, Y.; Qiu, X.; Ding, Y.; Han, H.; Zhang, X. Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol. Lett. 2014, 7, 1895–1902. [Google Scholar] [CrossRef]
- Metellus, P.; Coulibaly, B.; Colin, C.; de Paula, A.M.; Vasiljevic, A.; Taieb, D.; Barlier, A.; Boisselier, B.; Mokhtari, K.; Wang, X.W.; et al. Absence of IDH mutation identifies a novel radiologic and molecular subtype of WHO grade II gliomas with dismal prognosis. Acta Neuropathol. 2010, 120, 719–729. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, T.; Li, S.; Fan, X.; Ma, J.; Wang, L.; Jiang, T. Anatomical localization of isocitrate dehydrogenase 1 mutation: A voxel-based radiographic study of 146 low-grade gliomas. Eur. J. Neurol. 2014, 22, 348–354. [Google Scholar] [CrossRef]
- Zhang, C.B.; Bao, Z.S.; Wang, H.J.; Yan, W.; Liu, Y.W.; Li, M.Y.; Zhang, W.; Chen, L.; Jiang, T. Correlation of IDH1/2 mutation with clinicopathologic factors and prognosis in anaplastic gliomas: A report of 203 patients from China. J. Cancer Res. Clin. Oncol. 2014, 140, 45–51. [Google Scholar] [CrossRef]
- Lai, A.; Kharbanda, S.; Pope, W.B.; Tran, A.; Solis, O.E.; Peale, F.; Forrest, W.F.; Pujara, K.; Carrillo, J.A.; Pandita, A.; et al. Evidence for Sequenced Molecular Evolution of IDH1 Mutant Glioblastoma From a Distinct Cell of Origin. J. Clin. Oncol. 2011, 29, 4482–4490. [Google Scholar] [CrossRef]
- Persson, A.I.; Petritsch, C.; Swartling, F.J.; Itsara, M.; Sim, F.J.; Auvergne, R.; Goldenberg, D.D.; Vandenberg, S.R.; Nguyen, K.N.; Yakovenko, S.; et al. Non-Stem Cell Origin for Oligodendroglioma. Cancer Cell 2010, 18, 669–682. [Google Scholar] [CrossRef]
- Merkle, F.T.; Fuentealba, L.C.; A Sanders, T.; Magno, L.; Kessaris, N.; Alvarez-Buylla, A. Adult neural stem cells in distinct microdomains generate previously unknown interneuron types. Nat. Neurosci. 2013, 17, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, K.; Li, S.; Wang, J.; Ma, J.; Jiang, T.; Dai, J. Patterns of Tumor Contrast Enhancement Predict the Prognosis of Anaplastic Gliomas with IDH1 Mutation. Am. J. Neuroradiol. 2015, 36, 2023–2029. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Y.; Fan, X.; Wang, J.; Li, G.; Ma, J.; Ma, J.; Jiang, T.; Dai, J. Radiological features combined with IDH1 status for predicting the survival outcome of glioblastoma patients. Neuro Oncol. 2016, 18, 589–597. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.B.; Sayre, J.; Perlina, A.; Villablanca, J.P.; Mischel, P.S.; Cloughesy, T.F. MR Imaging Correlates of Survival in Patients with High-Grade Gliomas. Am. J. Neuroradiol. 2005, 26, 2466–2474. [Google Scholar] [PubMed]
- Lacerda, S.; Law, M. Magnetic Resonance Perfusion and Permeability Imaging in Brain Tumors. Neuroimaging Clin. N. Am. 2009, 19, 527–557. [Google Scholar] [CrossRef]
- Van Meter, T.; Dumur, C.; Hafez, N.; Garrett, C.; Fillmore, H.; Broaddus, W. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn. Mol. Pathol. Am. J. Surg. Pathol. Part B 2006, 15, 195–205. [Google Scholar] [CrossRef]
- Diehn, M.; Nardini, C.; Wang, D.S.; McGovern, S.; Jayaraman, M.; Liang, Y.; Aldape, K.; Cha, S.; Kuo, M.D. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc. Natl. Acad. Sci. USA 2008, 105, 5213–5218. [Google Scholar] [CrossRef]
- Dong, Y.; Dimopoulos, G. Anopheles Fibrinogen-related Proteins Provide Expanded Pattern Recognition Capacity against Bacteria and Malaria Parasites. J. Biol. Chem. 2009, 284, 9835–9844. [Google Scholar] [CrossRef]
- Suchorska, B.; Schüller, U.; Biczok, A.; Lenski, M.; Albert, N.L.; Giese, A.; Kreth, F.-W.; Ertl-Wagner, B.; Tonn, J.-C.; Ingrisch, M. Contrast enhancement is a prognostic factor in IDH1/2 mutant, but not in wild-type WHO grade II/III glioma as confirmed by machine learning. Eur. J. Cancer 2019, 107, 15–27. [Google Scholar] [CrossRef]
- Voss, M.; Franz, K.; Steinbach, J.P.; Fokas, E.; Forster, M.-T.; Filipski, K.; Hattingen, E.; Wagner, M.; Breuer, S. Contrast enhancing spots as a new pattern of late onset pseudoprogression in glioma patients. J. Neuro-Oncol. 2019, 142, 161–169. [Google Scholar] [CrossRef]
- Siu, A.; Wind, J.J.; Iorgulescu, J.B.; Chan, T.A.; Yamada, Y.; Sherman, J.H. Radiation necrosis following treatment of high grade glioma—A review of the literature and current understanding. Acta Neurochir. 2011, 154, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Carrillo, J.A.; Lai, A.; Nghiemphu, P.L.; Kim, H.J.; Phillips, H.S.; Kharbanda, S.; Moftakhar, P.; Lalaezari, S.; Yong, W.; Ellingson, B.M.; et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am. J. Neuroradiol. 2012, 33, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Matsushima, S.; Fukasawa, N.; Akasaki, Y.; Mori, R.; Ojiri, H. Differentiating between glioblastomas with and without isocitrate dehydrogenase gene mutation by findings on conventional magnetic resonance images. J. Clin. Neurosci. 2020, 76, 140–144. [Google Scholar] [CrossRef] [PubMed]
- Losman, J.A.; Looper, R.E.; Koivunen, P.; Lee, S.; Schneider, R.K.; McMahon, C.; Cowley, G.S.; Root, D.E.; Ebert, B.L.; Kaelin, W.G., Jr. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 2013, 339, 1621–1625. [Google Scholar] [CrossRef]
- Kickingereder, P.; Sahm, F.; Radbruch, A.; Wick, W.; Heiland, S.; Von Deimling, A.; Bendszus, M.; Wiestler, B. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci. Rep. 2015, 5, 16238. [Google Scholar] [CrossRef]
- Hong, E.K.; Choi, S.H.; Shin, D.J.; Jo, S.W.; Yoo, R.E.; Kang, K.M.; Yun, T.J.; Kim, J.H.; Sohn, C.H.; Park, S.H.; et al. Radiogenomics correlation between MR imaging features and major genetic profiles in glioblastoma. Eur. Radiol. 2018, 28, 4350–4361. [Google Scholar] [CrossRef]
- Yamashita, K.; Hiwatashi, A.; Togao, O.; Kikuchi, K.; Hatae, R.; Yoshimoto, K.; Mizoguchi, M.; Suzuki, S.; Yoshiura, T.; Honda, H. MR Imaging–Based Analysis of Glioblastoma Multiforme: Estimation of IDH1 Mutation Status. Am. J. Neuroradiol. 2015, 37, 58–65. [Google Scholar] [CrossRef]
- Xing, Z.; Zhang, H.; She, D.; Lin, Y.; Zhou, X.; Zeng, Z.; Cao, D. IDH genotypes differentiation in glioblastomas using DWI and DSC-PWI in the enhancing and peri-enhancing region. Acta Radiol. 2019, 60, 1663–1672. [Google Scholar] [CrossRef]
- Xing, Z.; Yang, X.; She, D.; Lin, Y.; Zhang, Y.; Cao, D. Noninvasive Assessment of IDH Mutational Status in World Health Organization Grade II and III Astrocytomas Using DWI and DSC-PWI Combined with Conventional MR Imaging. AJNR Am. J. Neuroradiol. 2017, 38, 1138–1144. [Google Scholar] [CrossRef]
- Law, M.; Young, R.J.; Babb, J.S.; Peccerelli, N.; Chheang, S.; Gruber, M.L.; Miller, D.C.; Golfinos, J.G.; Zagzag, D.; Johnson, G. Gliomas: Predicting Time to Progression or Survival with Cerebral Blood Volume Measurements at Dynamic Susceptibility-weighted Contrast-enhanced Perfusion MR Imaging. Radiology 2008, 247, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.L.; Huang, W.Y.; Yin, B.; Xiong, J.; Wu, J.S.; Geng, D.Y. Can Diffusion Tensor Imaging Noninvasively Detect IDH1 Gene Mutations in Astrogliomas? A Retrospective Study of 112 Cases. Am. J. Neuroradiol. 2014, 35, 920–927. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.; Lee, S.; Agid, R.; Bae, J.; Keller, A.; Terbrugge, K. Preoperative Grading of Presumptive Low-Grade Astrocytomas on MR Imaging: Diagnostic Value of Minimum Apparent Diffusion Coefficient. Am. J. Neuroradiol. 2008, 29, 1872–1877. [Google Scholar] [CrossRef] [PubMed]
- Higano, S.; Yun, X.; Kumabe, T.; Watanabe, M.; Mugikura, S.; Umetsu, A.; Sato, A.; Yamada, T.; Takahashi, S. Malignant Astrocytic Tumors: Clinical Importance of Apparent Diffusion Coefficient in Prediction of Grade and Prognosis. Radiology 2006, 241, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Feraco, P.; Bacci, A.; Ferrazza, P.; Hauwe, L.V.D.; Pertile, R.; Girlando, S.; Barbareschi, M.; Gagliardo, C.; Morganti, A.G.; Petralia, B. Magnetic Resonance Imaging Derived Biomarkers of IDH Mutation Status and Overall Survival in Grade III Astrocytomas. Diagnostics 2020, 10, 247. [Google Scholar] [CrossRef]
- Price, S.J.; Jena, R.; Burnet, N.G.; Carpenter, T.A.; Pickard, J.D.; Gillard, J.H. Predicting patterns of glioma recurrence using diffusion tensor imaging. Eur. Radiol. 2007, 17, 1675–1684. [Google Scholar] [CrossRef]
- Xiong, J.; Tan, W.-L.; Pan, J.-W.; Wang, Y.; Yin, B.; Zhang, J.; Geng, D. Detecting isocitrate dehydrogenase gene mutations in oligodendroglial tumors using diffusion tensor imaging metrics and their correlations with proliferation and microvascular density. J. Magn. Reson. Imaging 2016, 43, 45–54. [Google Scholar] [CrossRef]
- Aliotta, E.; Batchala, P.P.; Schiff, D.; Lopes, B.M.; Druzgal, J.T.; Mukherjee, S.; Patel, S.H. Increased intratumoral infiltration in IDH wild-type lower-grade gliomas observed with diffusion tensor imaging. J. Neuro-Oncol. 2019, 145, 257–263. [Google Scholar] [CrossRef]
- Ko, S.-F.; Lee, T.-Y.; Huang, C.-C.; Cheng, Y.-F.; Ng, S.-H.; Kuo, Y.-L.; Lin, M.-C.; Liu, J.-W.; Yang, K.D.; Chen, M.-C.; et al. Severe Acute Respiratory Syndrome: Prognostic Implications of Chest Radiographic Findings in 52 Patients. Radiology 2004, 233, 173–181. [Google Scholar] [CrossRef]
- Hui, E.S.; Fieremans, E.; Jensen, J.H.; Tabesh, A.; Feng, W.; Bonilha, L.; Spampinato, M.V.; Adams, R.; Helpern, J.A. Stroke Assessment With Diffusional Kurtosis Imaging. Stroke 2012, 43, 2968–2973. [Google Scholar] [CrossRef]
- Zheng, W.; Wu, C.; Huang, L.; Wu, R. Diffusion Kurtosis Imaging of Microstructural Alterations in the Brains of Paediatric Patients with Congenital Sensorineural Hearing Loss. Sci. Rep. 2017, 7, 1–8. [Google Scholar] [CrossRef]
- Van Cauter, S.; De Keyzer, F.; Sima, D.M.; Sava, A.C.; D’Arco, F.; Veraart, J.; Peeters, R.R.; Leemans, A.; Van Gool, S.; Wilms, G.; et al. Integrating diffusion kurtosis imaging, dynamic susceptibility-weighted contrast-enhanced MRI, and short echo time chemical shift imaging for grading gliomas. Neuro Oncol. 2014, 16, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Raab, P.; Hattingen, E.; Franz, K.; Zanella, F.; Lanfermann, H. Cerebral gliomas: Diffusional kurtosis imaging analysis of microstructural differences. Radiology 2010, 254, 876–881. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, H.; Wang, X.; Qin, J.; Wang, L.; Yang, G.; Yan, H. Comparing the value of DKI and DTI in detecting isocitrate dehydrogenase genotype of astrocytomas. Clin. Radiol. 2019, 74, 314–320. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.-L.; Li, X.-B.; Hu, M.-S.; Li, Z.-H.; Song, Y.-K.; Wang, J.-Y.; Tian, Y.-S.; Liu, D.-W.; Yan, X.; et al. Comparative analysis of the diffusion kurtosis imaging and diffusion tensor imaging in grading gliomas, predicting tumour cell proliferation and IDH-1 gene mutation status. J. Neuro-Oncol. 2018, 141, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.; Ganji, S.K.; DeBerardinis, R.J.; Hatanpaa, K.J.; Rakheja, D.; Kovacs, Z.; Yang, X.-L.; Mashimo, T.; Raisanen, J.M.; Marin-Valencia, I.; et al. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat. Med. 2012, 18, 624–629. [Google Scholar] [CrossRef]
- Andronesi, O.C.; Kim, G.S.; Gerstner, E.; Batchelor, T.; Tzika, A.A.; Fantin, V.R.; Vander Heiden, M.G.; Sorensen, A.G. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci. Transl. Med. 2012, 4, 116ra4. [Google Scholar] [CrossRef]
- Emir, U.E.; Larkin, S.J.; de Pennington, N.; Voets, N.; Plaha, P.; Stacey, R.; Al-Qahtani, K.; Mccullagh, J.; Schofield, C.J.; Clare, S.; et al. Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations. Cancer Res. 2016, 76, 43–49. [Google Scholar] [CrossRef]
- Leather, T.; Jenkinson, M.D.; Das, K.; Poptani, H. Magnetic Resonance Spectroscopy for Detection of 2-Hydroxyglutarate as a Biomarker for IDH Mutation in Gliomas. Metabolites 2017, 7, 29. [Google Scholar] [CrossRef]
- Choi, C.; Raisanen, J.M.; Ganji, S.K.; Zhang, S.; McNeil, S.S.; An, Z.; Madan, A.; Hatanpaa, K.J.; Vemireddy, V.; Sheppard, C.A.; et al. Prospective Longitudinal Analysis of 2-Hydroxyglutarate Magnetic Resonance Spectroscopy Identifies Broad Clinical Utility for the Management of Patients With IDH-Mutant Glioma. J. Clin. Oncol. 2016, 34, 4030–4039. [Google Scholar] [CrossRef]
- de la Fuente, M.I.; Young, R.J.; Rubel, J.; Rosenblum, M.; Tisnado, J.; Briggs, S.; Arevalo-Perez, J.; Cross, J.R.; Campos, C.; Straley, K.; et al. Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro Oncol. 2016, 18, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Ward, K.M.; Aletras, A.H.; Balaban, R.S. A new class of contrast agents for MRI based on proton chemical exchange dependent saturation transfer (CEST). J. Magn Reson. 2000, 14, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Payen, J.-F.; A Wilson, D.; Traystman, R.J.; Van Zijl, P.C.M. Using the amide proton signals of intracellular proteins and peptides to detect pH effects in MRI. Nat. Med. 2003, 9, 1085–1090. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zou, T.; Eberhart, C.G.; Villalobos, M.A.; Heo, H.-Y.; Zhang, Y.; Wang, Y.; Wang, X.; Yu, H.; Du, Y.; et al. Predicting IDH mutation status in grade II gliomas using amide proton transfer-weighted (APTw) MRI. Magn. Reson. Med. 2017, 78, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ke, C.; Liu, J.; Xu, S.; Han, L.; Yang, Y.; Qian, L.; Liu, X.; Zheng, H.; Lv, X.; et al. Diagnostic performance between MR amide proton transfer (APT) and diffusion kurtosis imaging (DKI) in glioma grading and IDH mutation status prediction at 3 T. Eur. J. Radiol. 2020, 134, 109466. [Google Scholar] [CrossRef]
- Togao, O.; Yoshiura, T.; Keupp, J.; Hiwatashi, A.; Yamashita, K.; Kikuchi, K.; Suzuki, Y.; Suzuki, S.O.; Iwaki, T.; Hata, N.; et al. Amide proton transfer imaging of adult diffuse gliomas: Correlation with histopathological grades. Neuro-Oncology 2013, 16, 441–448. [Google Scholar] [CrossRef]
- Joo, B.; Han, K.; Ahn, S.S.; Choi, Y.S.; Chang, J.H.; Kang, S.-G.; Kim, S.H.; Zhou, J.; Lee, S.-K. Amide proton transfer imaging might predict survival and IDH mutation status in high-grade glioma. Eur. Radiol. 2019, 29, 6643–6652. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Zimmermann, M.; Kitzwögerer, M.; Oberndorfer, S.; Rössler, K.; Dörfler, A.; Buchfelder, M.; Heinz, G. MR Imaging-derived Oxygen Metabolism and Neovascularization Characterization for Grading and IDH Gene Mutation Detection of Gliomas. Radiology 2017, 283, 799–809. [Google Scholar] [CrossRef]
- Pouratian, N.; Asthagiri, A.R.; Jagannathan, J.; Shaffrey, M.E.; Schiff, D. Surgery Insight: The role of surgery in the management of low-grade gliomas. Nat. Clin. Pr. Cardiovasc. Med. 2007, 3, 628–639. [Google Scholar] [CrossRef]
- Gevaert, O.; Xu, J.; Hoang, C.D.; Leung, A.N.; Xu, Y.; Quon, A.; Rubin, D.L.; Napel, S.; Plevritis, S.K. Non–Small Cell Lung Cancer: Identifying Prognostic Imaging Biomarkers by Leveraging Public Gene Expression Microarray Data—Methods and Preliminary Results. Radiology 2012, 264, 387–396. [Google Scholar] [CrossRef]
- Gutman, D.A.; Cooper, L.A.D.; Hwang, S.; Holder, C.A.; Gao, J.; Aurora, T.D.; Dunn, W.D.; Scarpace, L.; Mikkelsen, T.; Jain, R.; et al. MR Imaging Predictors of Molecular Profile and Survival: Multi-institutional Study of the TCGA Glioblastoma Data Set. Radiology 2013, 267, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.; Bai, H.X.; Zhou, H.; Su, C.; Bi, W.L.; Agbodza, E.; Kavouridis, V.K.; Senders, J.T.; Boaro, A.; Beers, A.; et al. Residual Convolutional Neural Network for the Determination ofIDHStatus in Low- and High-Grade Gliomas from MR Imaging. Clin. Cancer Res. 2018, 24, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Shi, Z.; Lian, Y.; Li, Z.; Liu, T.; Gao, Y.; Wang, Y.; Chen, L.; Mao, Y. Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur. Radiol. 2016, 27, 3509–3522. [Google Scholar] [CrossRef]
- Han, L.; Wang, S.; Miao, Y.; Shen, H.; Guo, Y.; Xie, L.; Shang, Y.; Dong, J.; Li, X.; Wang, W.; et al. MRI texture analysis based on 3D tumor measurement reflects the IDH1 mutations in gliomas – A preliminary study. Eur. J. Radiol. 2019, 112, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, Y.; Li, S.; Fan, X.; Sun, Z.; Yang, Z.; Wang, K.; Zhang, Z.; Jiang, T.; Liu, Y.; et al. IDH mutation-specific radiomic signature in lower-grade gliomas. Aging 2019, 11, 673–696. [Google Scholar] [CrossRef]
- Li, Z.-C.; Bai, H.; Sun, Q.; Zhao, Y.; Lv, Y.; Zhou, J.; Liang, C.; Chen, Y.; Liang, D.; Zheng, H. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018, 7, 5999–6009. [Google Scholar] [CrossRef]
- Kim, M.; Jung, S.Y.; Park, J.E.; Jo, Y.; Park, S.Y.; Nam, S.J.; Kim, J.H.; Kim, H.S. Diffusion- and perfusion-weighted MRI radiomics model may predict isocitrate dehydrogenase (IDH) mutation and tumor aggressiveness in diffuse lower grade glioma. Eur. Radiol. 2019, 30, 2142–2151. [Google Scholar] [CrossRef]
- Zhou, H.; Chang, K.; Bai, H.X.; Xiao, B.; Su, C.; Bi, W.L.; Zhang, P.J.; Senders, J.T.; Vallières, M.; Kavouridis, V.K.; et al. Machine learning reveals multimodal MRI patterns predictive of isocitrate dehydrogenase and 1p/19q status in diffuse low- and high-grade gliomas. J. Neuro-Oncol. 2019, 142, 299–307. [Google Scholar] [CrossRef]
- Peeken, J.C.; Hesse, J.; Haller, B.; Kessel, K.A.; Nüsslin, F.; Combs, S.E. Semantic imaging features predict disease progression and survival in glioblastoma multiforme patients. Strahlenther. Onkol. 2018, 194, 580–590. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Qin, Q.; Zhang, Y.; Cao, Y. Noninvasive Determination of the IDH Status of Gliomas Using MRI and MRI-Based Radiomics: Impact on Diagnosis and Prognosis. Curr. Oncol. 2022, 29, 6893-6907. https://doi.org/10.3390/curroncol29100542
Li Y, Qin Q, Zhang Y, Cao Y. Noninvasive Determination of the IDH Status of Gliomas Using MRI and MRI-Based Radiomics: Impact on Diagnosis and Prognosis. Current Oncology. 2022; 29(10):6893-6907. https://doi.org/10.3390/curroncol29100542
Chicago/Turabian StyleLi, Yurong, Qin Qin, Yumeng Zhang, and Yuandong Cao. 2022. "Noninvasive Determination of the IDH Status of Gliomas Using MRI and MRI-Based Radiomics: Impact on Diagnosis and Prognosis" Current Oncology 29, no. 10: 6893-6907. https://doi.org/10.3390/curroncol29100542
APA StyleLi, Y., Qin, Q., Zhang, Y., & Cao, Y. (2022). Noninvasive Determination of the IDH Status of Gliomas Using MRI and MRI-Based Radiomics: Impact on Diagnosis and Prognosis. Current Oncology, 29(10), 6893-6907. https://doi.org/10.3390/curroncol29100542





