Reclamation of Marine Chitinous Materials for Chitosanase Production via Microbial Conversion by Paenibacillus macerans
Abstract
:1. Introduction
2. Results and Discussion
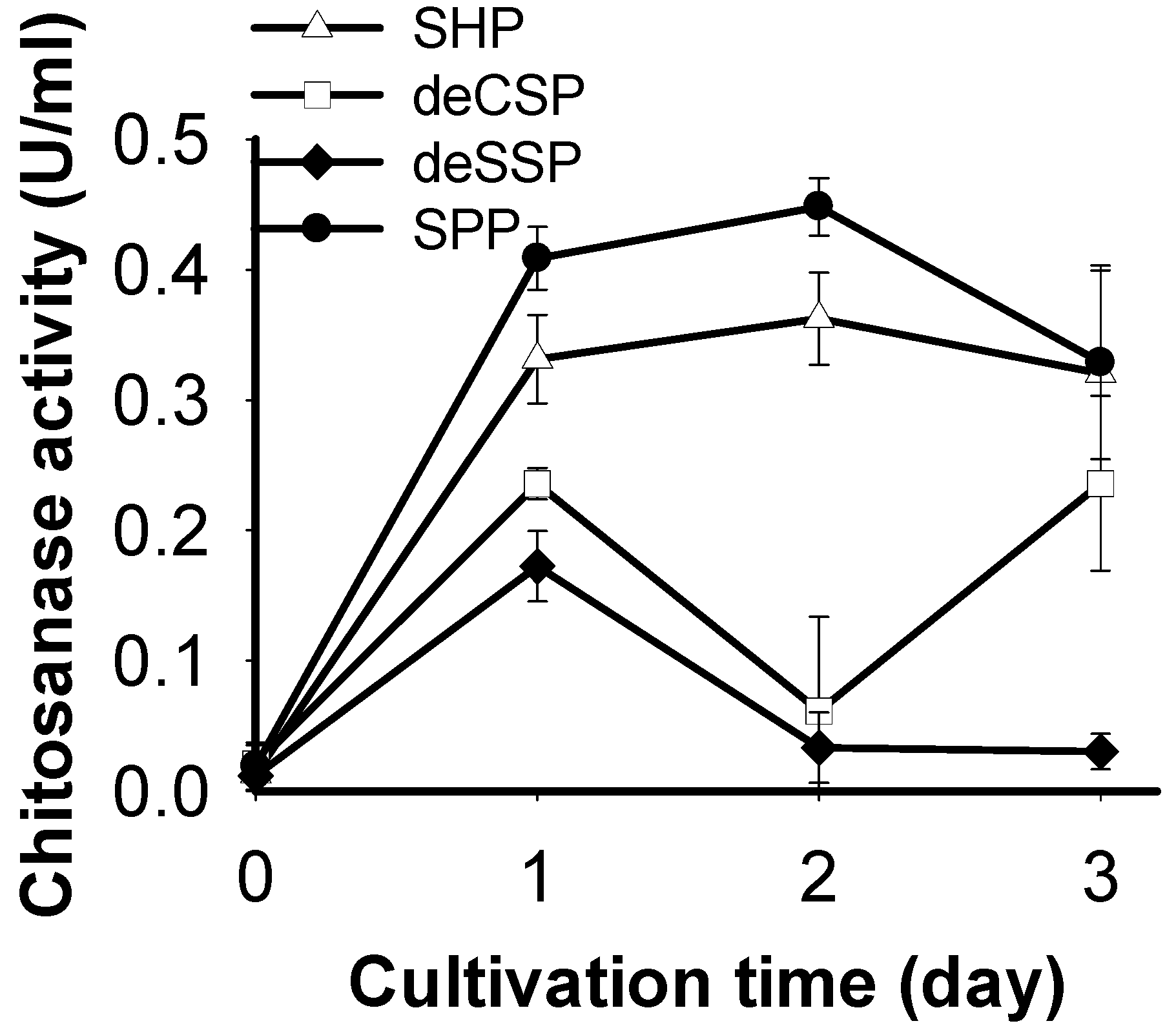
2.1. Screening of Chitinous Materials as Sole C/N Source for Chitosanase Production
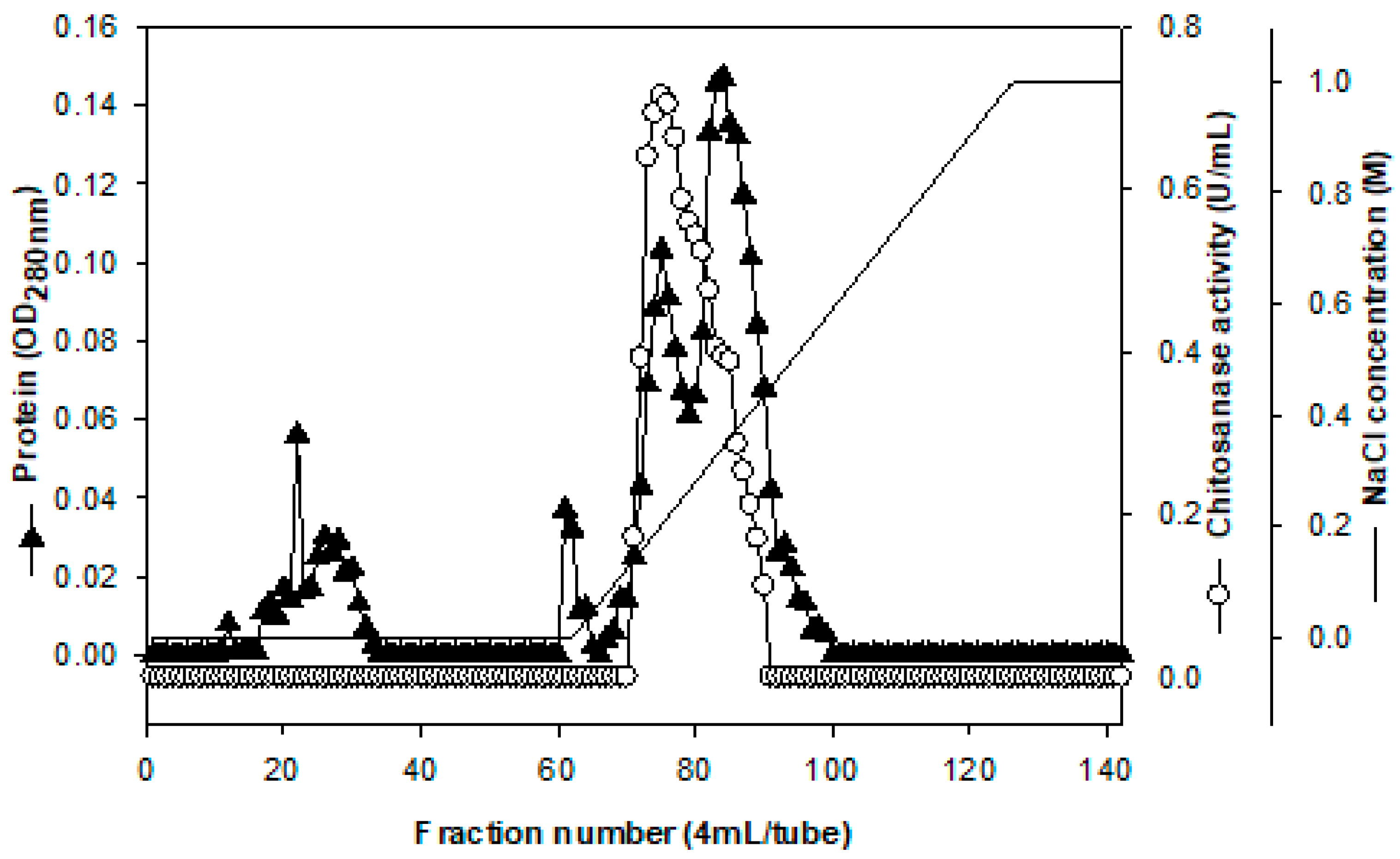
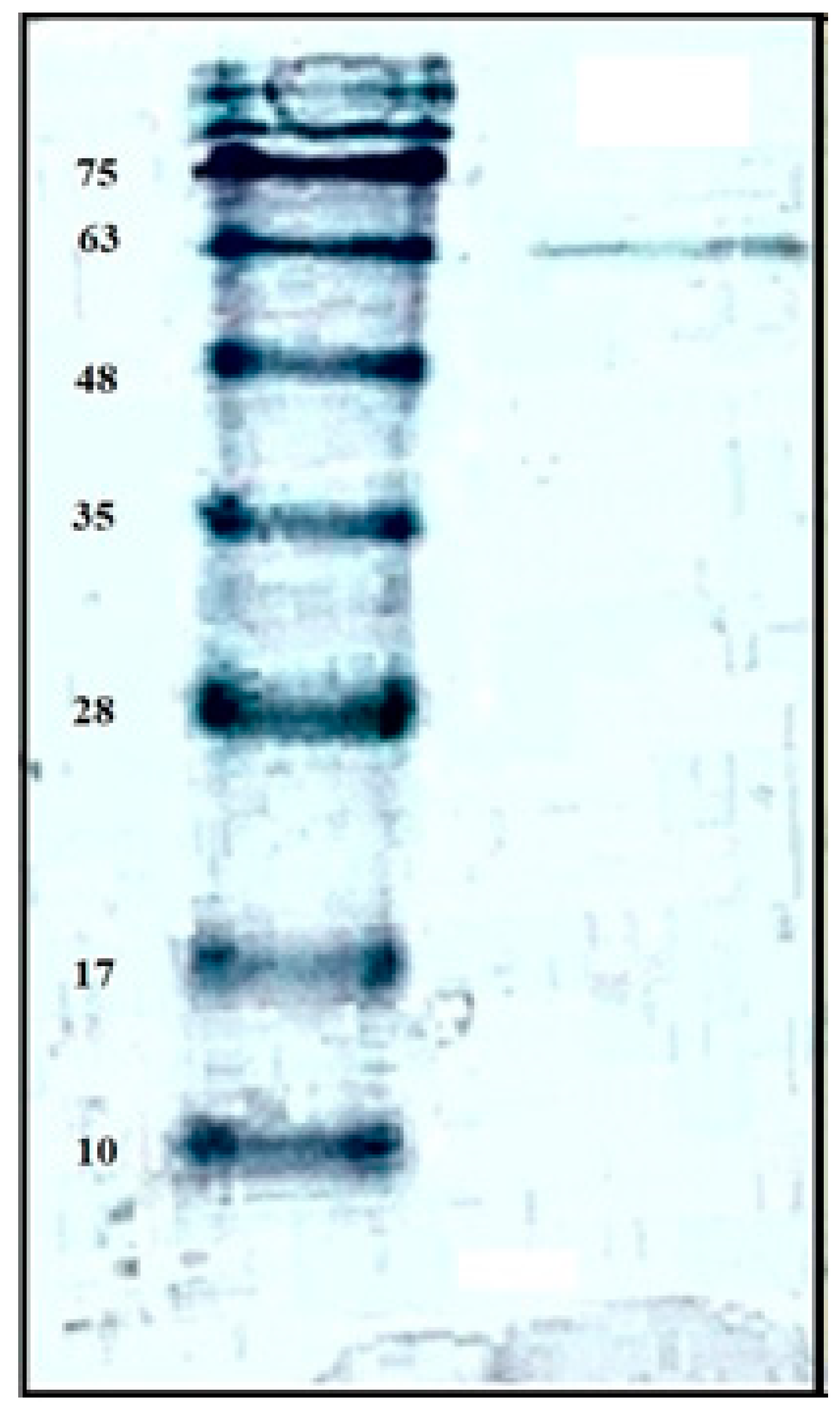
2.2. Purification and Characterization of Chitosanase
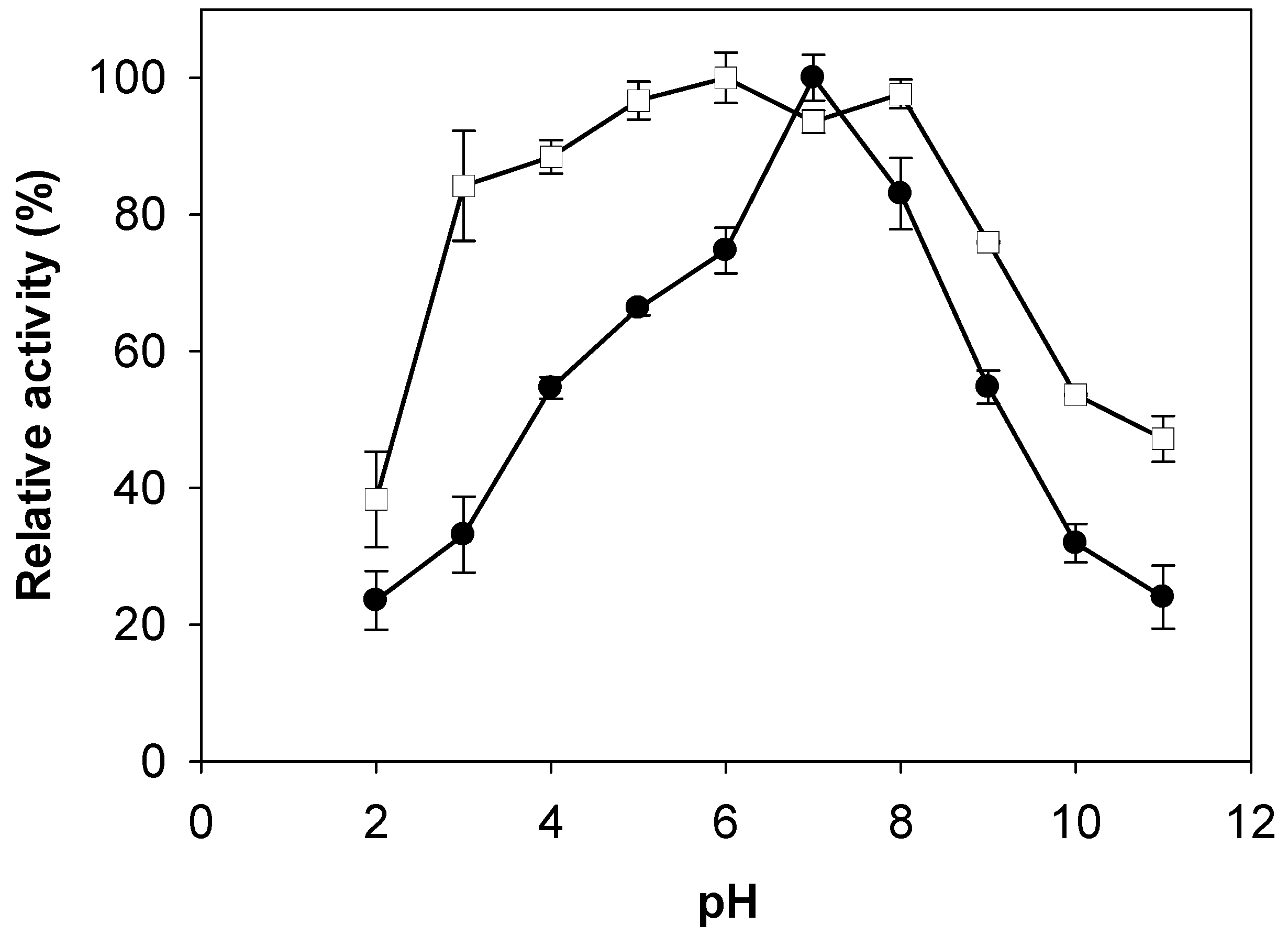
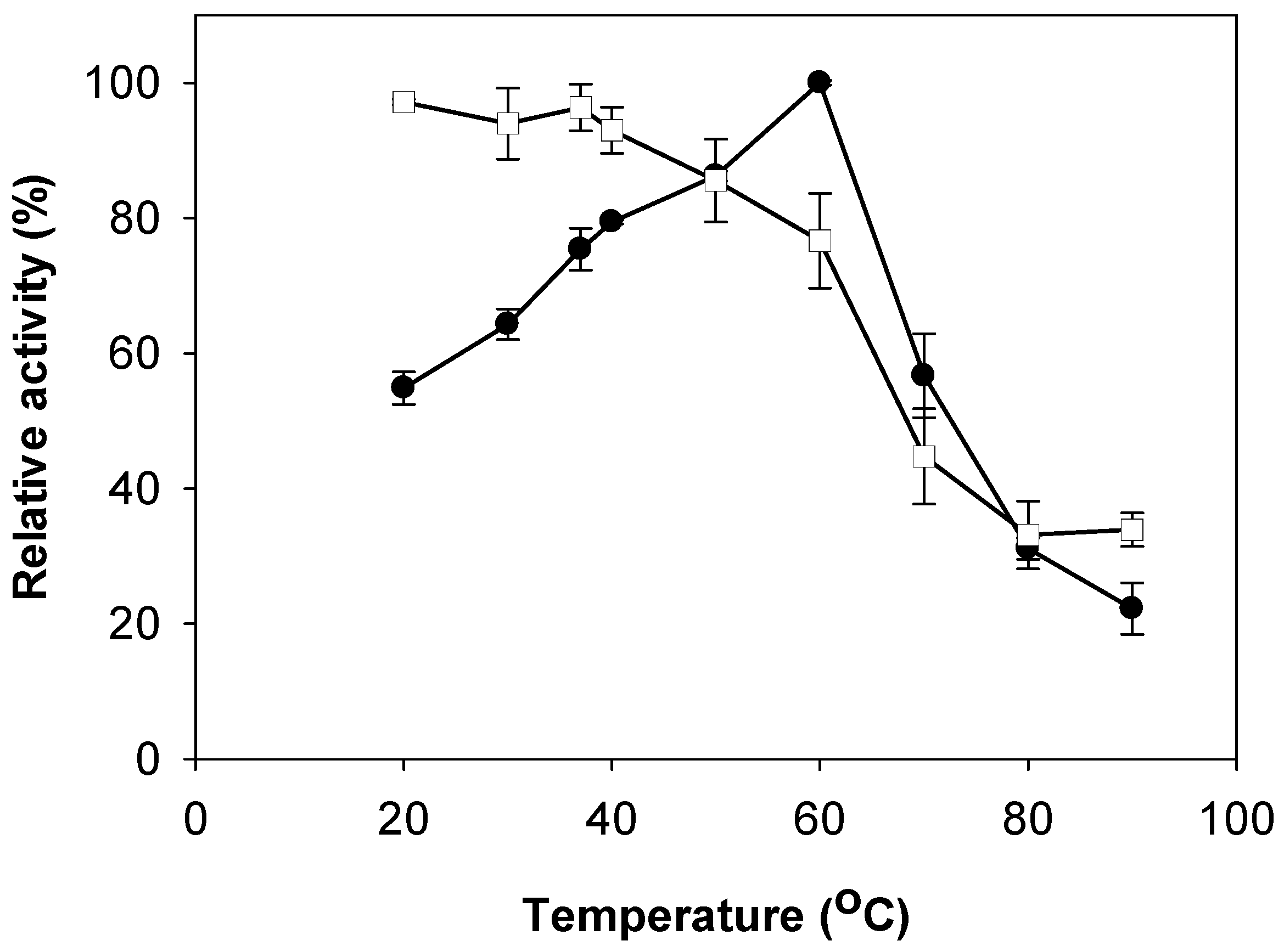
2.3. Effects of pH and Temperature on Activity and Stability of Chitosanase
2.4. Effect of Metal Ions on Activity of Chitosanase
2.5. Substrate Specificity of Chitosanase
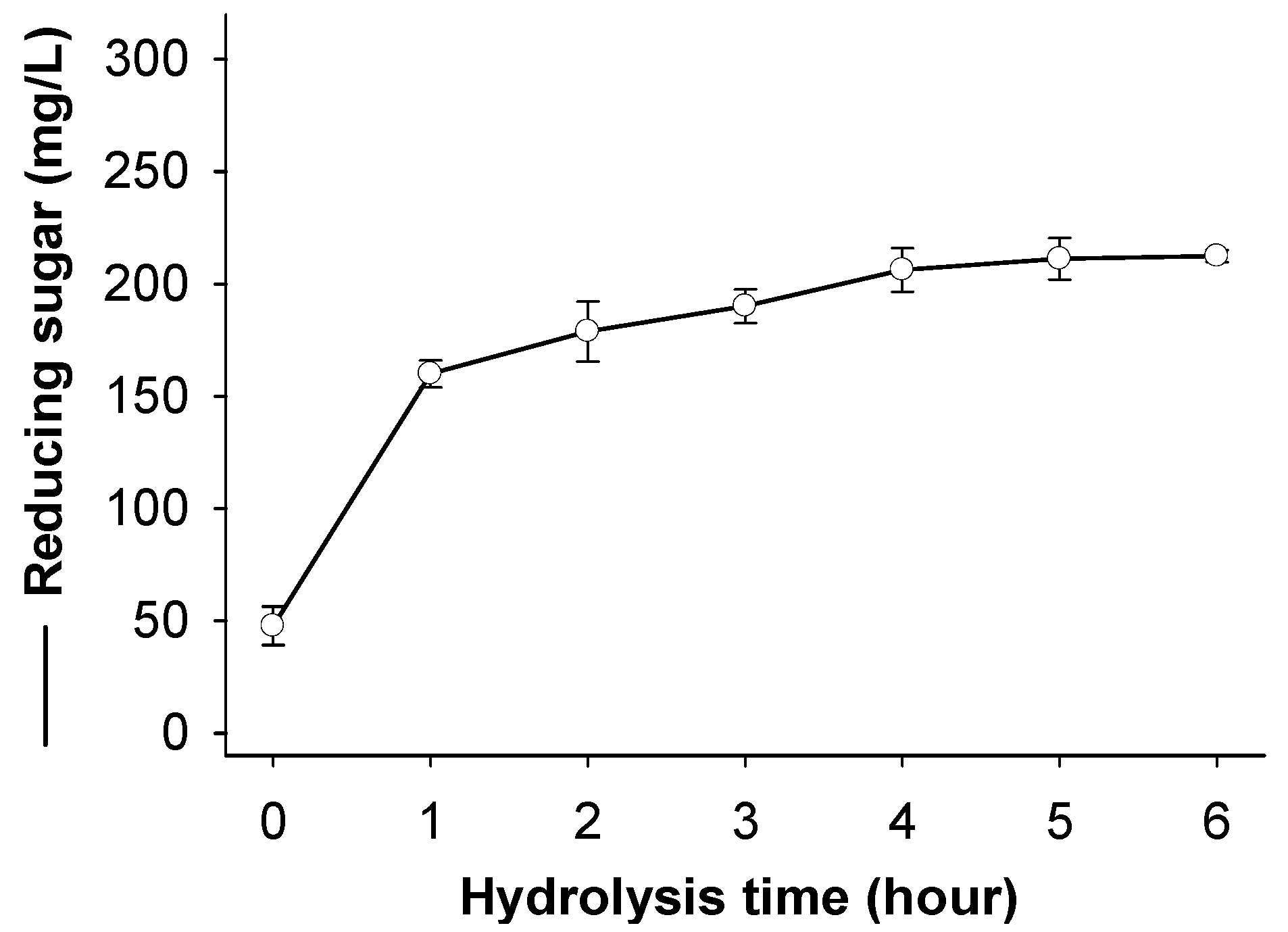
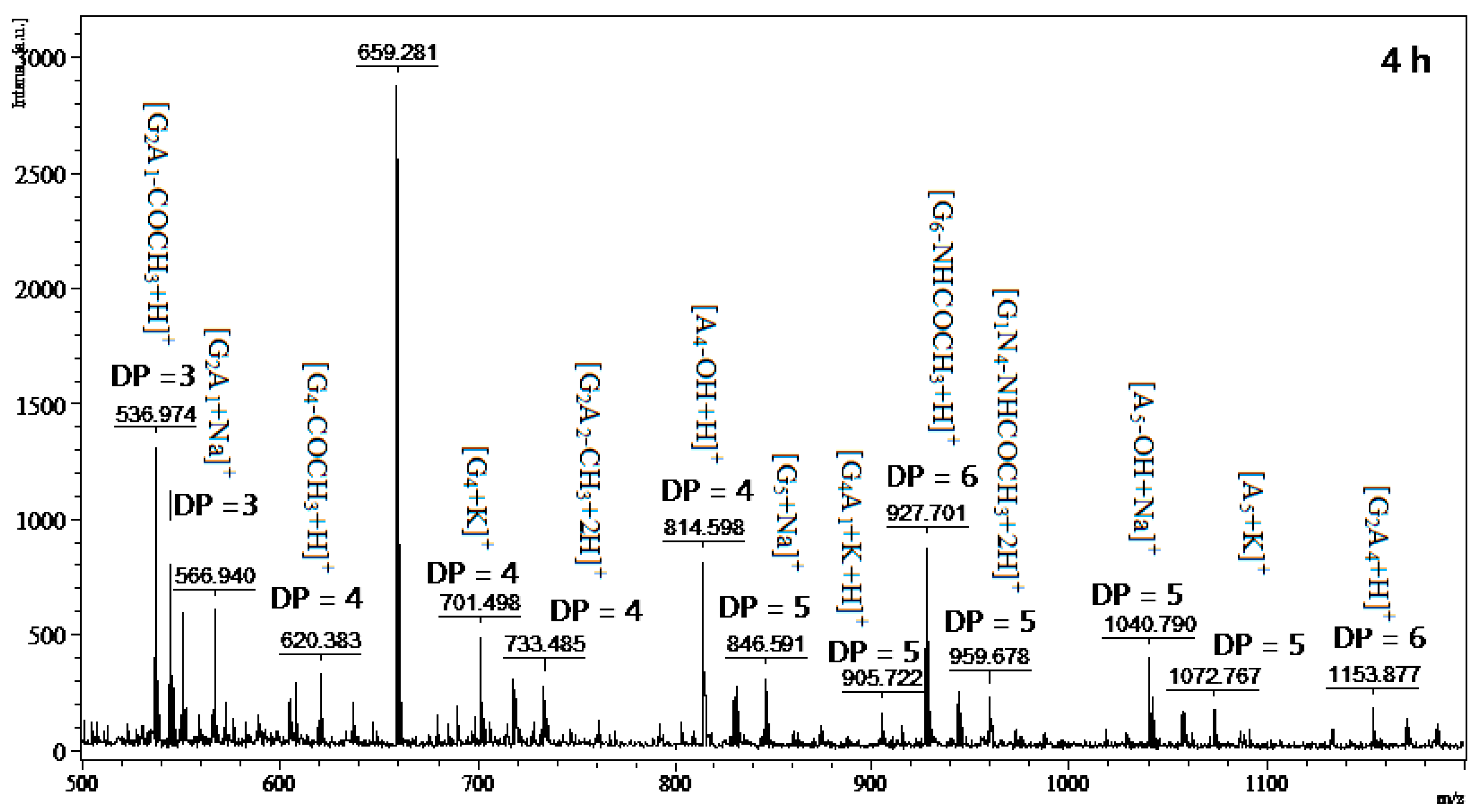
2.6. Chitosan Hydrolysis and COS Production
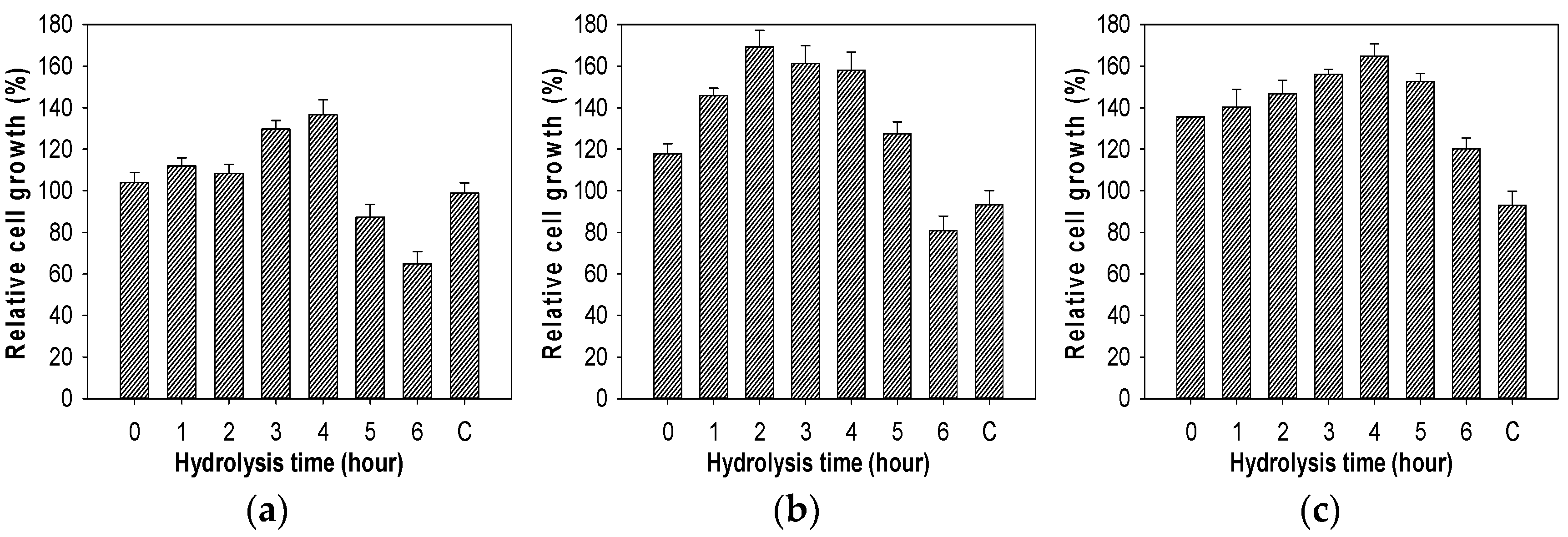
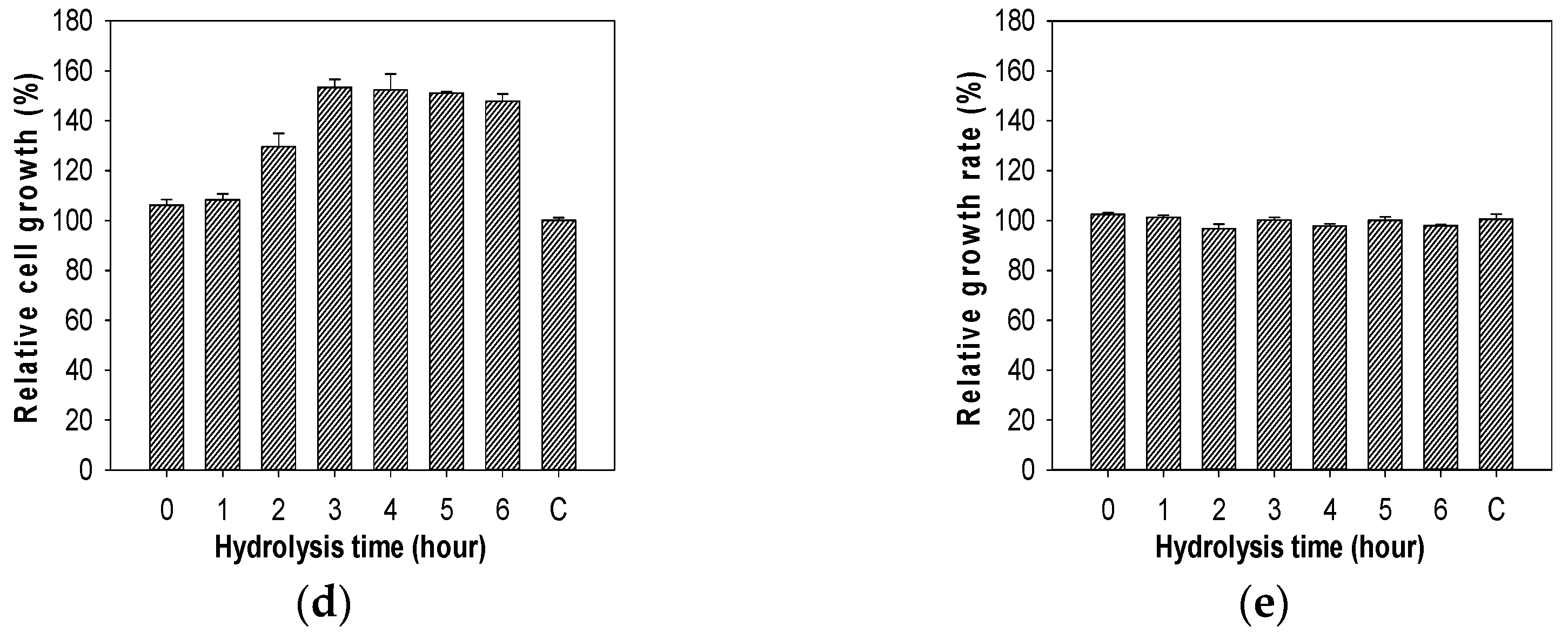
2.7. Evaluation of Growth Enhancing Effect of COS on Lactic Acid Bacteria
3. Materials and Methods
3.1. Materials
3.2. Measurement of Chitosanase Activity
3.3. Screening of Chitinous Materials as Sole C/N Source for Enzyme Activity
3.4. Purification of Chitosanae
3.5. Effects of pH and Temperature on Activity and Stability of Chitosanase
3.6. Effect of Metal Ions on Chitosanase Activity
3.7. Substrate Specificity
3.8. MALDI-TOF MS Analysis
3.9. Growth Enhancing Effect of COS on Lactic Acid Bacteria Test
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Azuma, K.; Osaki, T.; Minami, S.; Okamoto, Y. Anticancer and anti-inflammatory properties of chitin and chitosan oligosaccharides. J. Funct. Biomater. 2015, 6, 33–49. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Chen, W.T.; Lin, Z.H.; Kuo, Y.H.; Nguyen, A.D.; Pan, P.S.; Wang, S.L. An amphiprotic novel chitosanase from Bacillus mycoides and its application in the production of chitooligomers with their antioxidant and anti-inflammatory evaluation. Int. J. Mol. Sci. 2017, 17, 1302. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Chen, Y.J.; Yen, Y.H.; Wang, S.L. The antitumor activity of the hydrolysates of chitinous materials hydrolyzed by crude enzyme from Bacillus amyloliquefaciens V656. Process Biochem. 2007, 42, 527–534. [Google Scholar] [CrossRef]
- Sun, T.; Qin, Y.; Xu, H.; Xie, J.; Hu, D.; Xue, B.; Hua, X. Antibacterial activities and preservative effect of chitosan oligosaccharide Maillard reaction products on Penaeus vannamei. Int. J. Biol. Macromol. 2017, 105, 764–768. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Liu, C.P.; Wu, C.; Wang, S.L. Applied development of crude enzyme from Bacillus cereus in prebiotics and microbial community changes in soil. Carbohydr. Polym. 2013, 92, 2141–2148. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Park, Y.S.; Jung, J.S.; Shin, W.S. Chitosan oligosaccharides, dp 2–8, have prebiotic effect on the Bifidobacterium bifidium and Lactobacillus sp. Anaerobe 2002, 8, 319–324. [Google Scholar] [CrossRef]
- Liang, T.W.; Chen, Y.Y.; Pan, P.S.; Wang, S.L. Purification of chitinase/chitosanase from Bacillus cereus and discovery of an enzyme inhibitor. Int. J. Biol. Macromol. 2014, 63, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Lo, B.C.; Wang, S.L. Chitinolytic bacteria-assisted conversion of squid pen and its effect on dyes and adsorption. Mar. Drugs 2015, 13, 4576–4593. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.L.; Su, J.W.; Liang, T.W.; Nguyen, A.D.; Wang, S.L. Production, purification and characterization of a chitosanase from Bacillus cereus. Res. Chem. Intermed. 2014, 40, 2237–2248. [Google Scholar] [CrossRef]
- Wang, S.L.; Wu, P.C.; Liang, T.W. Utilization of squid pen for the efficient production of chitosanase and antioxidants through prolonged autoclave treatment. Carbohydr. Res. 2009, 244, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Jen, S.N.; Nguyen, A.D.; Wang, S.L. Application of chitinous materials in production and purification of a poly (L-lactic acid) depolymerase from Pseudomonas tamsuii TKU015. Polymers 2016, 8, 98. [Google Scholar] [CrossRef]
- Wang, S.L.; Chen, S.J.; Liang, T.W.; Lin, Y.D. A novel nattokinase produced by Pseudomonas sp. TKU015 using shrimp shells as substrate. Process Biochem. 2009, 44, 70–76. [Google Scholar] [CrossRef]
- Wang, S.L.; Hsu, W.H.; Liang, T.W. Conversion of squid pen by Pseudomonas aeruginosa K-187 fermentation for the production of N-acetyl chitooligosaccharides and biofertilizers. Carbohydr. Res. 2010, 345, 880–885. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.H.; Liang, T.W.; Liu, K.C.; Hsu, Y.W.; Hsu, H.; Wang, S.L. Isolation and identification of a novel antioxidant with antitumor activity from Serratia ureilytica using squid pen as fermentation substrate. Mar. Biotechnol. 2011, 13, 451–461. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Wang, C.Y.; Yen, Y.H.; Liang, T.W.; Chen, S.Y.; Chen, C.H. Enhanced production of insecticidal prodigiosin from Serratia marcescens TKU011 in media containing squid pen. Process Biochem. 2012, 47, 1684–1690. [Google Scholar] [CrossRef]
- Liang, T.W.; Wu, C.C.; Cheng, W.T.; Chen, Y.C.; Wang, C.L.; Wang, I.L.; Wang, S.L. Exopolysaccharides and antimicrobial biosurfactants produced by Paenibacillus macerans TKU029. Appl. Biochem. Biotechnol. 2014, 172, 933–950. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Tseng, S.C.; Wang, S.L. Production and characterization of antioxidant properties of exopolysaccharides from Paenibacillus mucilaginosus TKU032. Mar. Drugs 2016, 14, 40. [Google Scholar] [CrossRef] [PubMed]
- Liang, T.W.; Wang, S.L. Recent advances in exopolysaccharides from Paenibacillus spp.: Production, isolation, structure, and bioactivities. Mar. Drugs 2015, 13, 1847–1863. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Utilization of fishery processing by product squid pens for Paenibacillus sp. fermentation on producing potent α-glucosidase inhibitors. Mar. Drugs 2017, 15, 274. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.B.; Wang, S.L. Reclamation of marine chitinous materials for the production of α-glucosidase inhibitors via microbial conversion. Mar. Drugs 2017, 15, 350. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.L.; Li, H.T.; Zhang, L.J.; Lin, Z.H.; Kuo, Y.H. Conversion of squid pen to homogentisic acid via Paenibacillus sp. TKU036 and the antioxidant and anti-inflammatory activities of homogentisic acid. Mar. Drugs 2016, 14, 183. [Google Scholar] [CrossRef] [PubMed]
- Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.L. Conversion of squid pens to chitosanases and proteases via Paenibacillus sp. TKU042. Mar. Drugs 2018, 16, 83. [Google Scholar] [CrossRef] [PubMed]
- Loni, P.P.; Patil, J.U.; Phugare, S.S.; Bajekal, S.S. Purification and characterization of alkaline chitinase from Paenibacillus pasadenensis NCIM 5434. J. Basic Microbiol. 2014, 54, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Zitouni, M.; Fortin, M.; Scheerle, R.K.; Letzel, T.; Matteau, D.; Rodrigue, S.; Brzezinski, R. Biochemical and molecular characterization of a thermostable chitosanase produced by the strain Paenibacillus sp. 1794 newly isolated from compost. Appl. Microbiol. Biotechnol. 2013, 97, 5801–5813. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Kurosawa, N. Characterization of an extracellular thermophilic chitinase from Paenibacillus thermoaerophilus strain TC22-2b isolated from compost. World J. Microbiol. Biotechnol. 2015, 31, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalez, E.; Poppinga, L.; Fünfhaus, A.; Hertlein, G.; Hedtke, K.; Jakubowska, A.; Genersch, E. Paenibacillus larvae chitin-degrading protein PlCBP49 is a key virulence factor in American foulbrood of honey bees. PLoS Pathogen. 2014, 10, e1004284. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Chhatpar, H.S. Purification and characterization of chitinase from Paenibacillus sp. D1. Appl. Biochem. Biotechnol. 2011, 164, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Yan, Q.; Wang, J.; Yang, S.; Jiang, Z. Purification and biochemical characterization of novel acidic chitinase from Paenibacillus barengoltzii. Int. J. Biol. Macromol. 2016, 91, 973–979. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.H.; Park, S.K.; Hur, J.Y.; Kim, Y.C. Purification and characterization of a major extracellular chitinase from a biocontrol bacterium, Paenibacillus elgii HOA73. Plant Pathol. J. 2017, 33, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Xu, P.; Zong, M.; Lou, W. Purification and characterization of alkaline chitinase from Paenibacillus pasadenensis CS0611. Chin. J. Catalys. 2017, 38, 665–672. [Google Scholar] [CrossRef]
- Kimoto, H.; Kusaoke, H.; Yamamoto, I.; Fujii, Y.; Onodera, T.; Taketo, A. Biochemical and genetic properties of Paenibacillus glycosyl hydrolase having chitosanase activity and discoidin domain. J. Biol. Chem. 2002, 277, 14695–14702. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Sugimoto, I.; Hibi, T.; Suzuki, F.; Matsuo, K.; Fujii, Y.; Taketo, A.; Kimoto, H. Overexpression, purification, and characterization of Paenibacillus cell surface-expressed chitinase ChiW with two catalytic domains. Biosci. Biotechnol. Biochem. 2014, 78, 624–634. [Google Scholar] [CrossRef] [PubMed]
- Jung, W.J.; Kuk, J.K.; Kim, K.Y.; Kim, T.H.; Park, R.D. Purification and characterization of chitinase from Paenibacillus illinoisensis KJA-424. J. Microbiol. Biotechnol. 2005, 15, 274–280. [Google Scholar]
- Meena, S.; Gothwal, R.K.; Saxena, J.; Nehra, S.; Mohan, M.K.; Ghosh, P. Effect of metal ions and chemical compounds on chitinase produced by a newly isolated thermotolerant Paenibacillus sp. BISR-047 and its shelf-life. Int. J. Curr. Microbiol. Appl. Sci. 2015, 4, 872–881. [Google Scholar]
- Wang, S.L.; Liu, C.P.; Liang, T.W. Fermented and enzymatic production of chitin/chitosan oligosaccharides by extracellular chitinases from Bacillus cereus TKU027. Carbohydr. Polym. 2012, 90, 1305–1313. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.D.; Huang, C.C.; Liang, T.W.; Nguyen, V.B.; Pan, P.S.; Wang, S.L. Production and purification of a fungal chitosanase and chitooligomers from Penicillium janthinellum D4 and discovery of the enzyme activators. Carbohydr. Polym. 2014, 108, 331–337. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.G.; Kim, H.Y.; Kim, H.K.; Kim, K.H.; Hwang, H.J.; Cho, H.Y. Cloning and expression of thermostable chitosanase gene from Bacillus sp. KFB-C108. J. Microbiol. Biotechnol. 1999, 9, 631–636. [Google Scholar]
- Liang, T.W.; Hsieh, J.L.; Wang, S.L. Production and purification of a protease, a chitosanase, and chitin oligosaccharides by Bacillus cereus TKU022 fermentation. Carbohydr. Res. 2012, 362, 38–46. [Google Scholar] [CrossRef] [PubMed]
| Step | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Recovery (%) | Purification (Fold) |
|---|---|---|---|---|---|
| Culture supernatant | 1245.88 | 328.09 | 0.26 | 100.00 | 1.00 |
| EtOH precipitation | 60.31 | 83.90 | 1.39 | 25.57 | 5.28 |
| DEAE-Sepharose CL-6B | 10.45 | 63.17 | 6.04 | 19.25 | 22.95 |
| Macro-Prep DEAE | 1.43 | 34.48 | 24.19 | 10.51 | 91.87 |
| Relative Activity (%) | |
|---|---|
| Control | 100.00 ± 1.39 |
| Cu2+ | 63.42 ± 1.51 |
| Zn2+ | 99.75 ± 1.84 |
| Mg2+ | 64.31 ± 2.09 |
| Na+ | 154.22 ± 1.96 |
| Ba2+ | 76.99 ± 2.03 |
| Ca2+ | 108.81 ± 4.13 |
| Fe2+ | 133.23 ± 5.31 |
| EDTA | 68.10 ± 0.39 |
| Substrate | Relative Activity (%) |
|---|---|
| Chitosan | 100 ± 16.93 |
| Water soluble chitosan | 196.43 ± 15.55 |
| α-Chitin | 0 |
| β-Chitin | 12.30 ± 6.62 |
| Colloidal Chitin | 63.26 ± 4.08 |
| pNPG | 0 |
| Cellulose | 0 |
| Dextran | 0 |
| Starch | 0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doan, C.T.; Tran, T.N.; Nguyen, V.B.; Nguyen, A.D.; Wang, S.-L. Reclamation of Marine Chitinous Materials for Chitosanase Production via Microbial Conversion by Paenibacillus macerans. Mar. Drugs 2018, 16, 429. https://doi.org/10.3390/md16110429
Doan CT, Tran TN, Nguyen VB, Nguyen AD, Wang S-L. Reclamation of Marine Chitinous Materials for Chitosanase Production via Microbial Conversion by Paenibacillus macerans. Marine Drugs. 2018; 16(11):429. https://doi.org/10.3390/md16110429
Chicago/Turabian StyleDoan, Chien Thang, Thi Ngoc Tran, Van Bon Nguyen, Anh Dzung Nguyen, and San-Lang Wang. 2018. "Reclamation of Marine Chitinous Materials for Chitosanase Production via Microbial Conversion by Paenibacillus macerans" Marine Drugs 16, no. 11: 429. https://doi.org/10.3390/md16110429
APA StyleDoan, C. T., Tran, T. N., Nguyen, V. B., Nguyen, A. D., & Wang, S.-L. (2018). Reclamation of Marine Chitinous Materials for Chitosanase Production via Microbial Conversion by Paenibacillus macerans. Marine Drugs, 16(11), 429. https://doi.org/10.3390/md16110429