Antimicrobial Peptides in Reptiles
Abstract
:1. Introduction
| Peptide name | Sequence | APD Identified | Source Organism | Comment Reference | Activity (*) |
|---|---|---|---|---|---|
| Cathelicidin | |||||
| OH-CATH | KRFKKFFKKLKNSVKKRAKKFFKKPRVIGVSIPF | AP00895 | O. Hannah (Snake) | Derivatives: OH-CATH30; OH-CM6 [3,4] | G+, G− |
| Derivative OH-CATH30 | KFFKKLKNSVKKRAKKFFKKPRVIGVSIPF | [5,6] | G+, G− | ||
| Derivative OH-CM6 | KFFKKLKKAVKKGFKKFAKV | [3, 4] | G+, G− | ||
| BF-CATH | KRFKKFFKKLKKSVKKRAKKFFKKPRVIGVSIPF | AP00896 | Bungarus fasciatus (Snake) | [7] | G+, G−, F, Cancer cells |
| Derivative BF-30 | KFFRKLKKSVKKRAKEFFKKPRVIGVSIPF | AP01239 | B. fasciatus | [8,9,10] | G+, G− |
| Derivative BF-15 | KFFRKLKKSVVKRFK | B. fasciatus | [8] | G+, G− | |
| NA-CATH | KRFKKFFKKLKNSVKKRAKKFFKKPKVIGVTFPF | AP00897 | N. atra (Snake) | [3] | G+, G− |
| Waprin | |||||
| Omwaprin | KDRPKKPGLCPPRPQKPCVKECKNDDSCPGQQKCCNYGCKDECRDPIFVG | AP01589 | Oxyuranus microlepidotus (Snake) | 4S = S [11, 12] | G+ |
| Proline Rich | |||||
| Lethal peptide I/Waglerin | GGKPDLRPCHPPCHYIPRPKPR | AP00238 | Trimeresurus wagleri, Wagler’s pit viper (Snake) | P24335, [13] | Tx |
| β-Defensin or defensin-like | |||||
| TBD-1 (Turtle β-defensin 1) | YDLSKNCRLRGGICYIGKCPRRFFRSGSCSRGNVCCLRFG | AP01380 | Turtle | 3S = S [14] | G+, G−, F |
| Pelovaterin | DDTPSSRCGSGGWGPCLPIVDLLCIVHVTVGCSGGFGCCRIG | AP01381 | Turtle | 2JR3 BBBh2o [15, 16] | G− |
| TEWP (turtle egg-white protein) | EKKCPGRCTLKCGKHERPTLPYNCGKYICCVPVKVK | AP01382 | Turtle | 2B5B XXQ [17,18,19] | G-, V |
| Crotamine defensin-like toxin | |||||
| Crotamine | YKQCHKKGGHCFPKEKICLPPSSDFGKMDCRWRWKCCKKGSG | AP01650 | Snake Venom | 1Z99, 3S = S ZZP [20, 21] | G+, G−, F, P, Mammalian and Cancer cells |
2. Four Orders of Reptiles
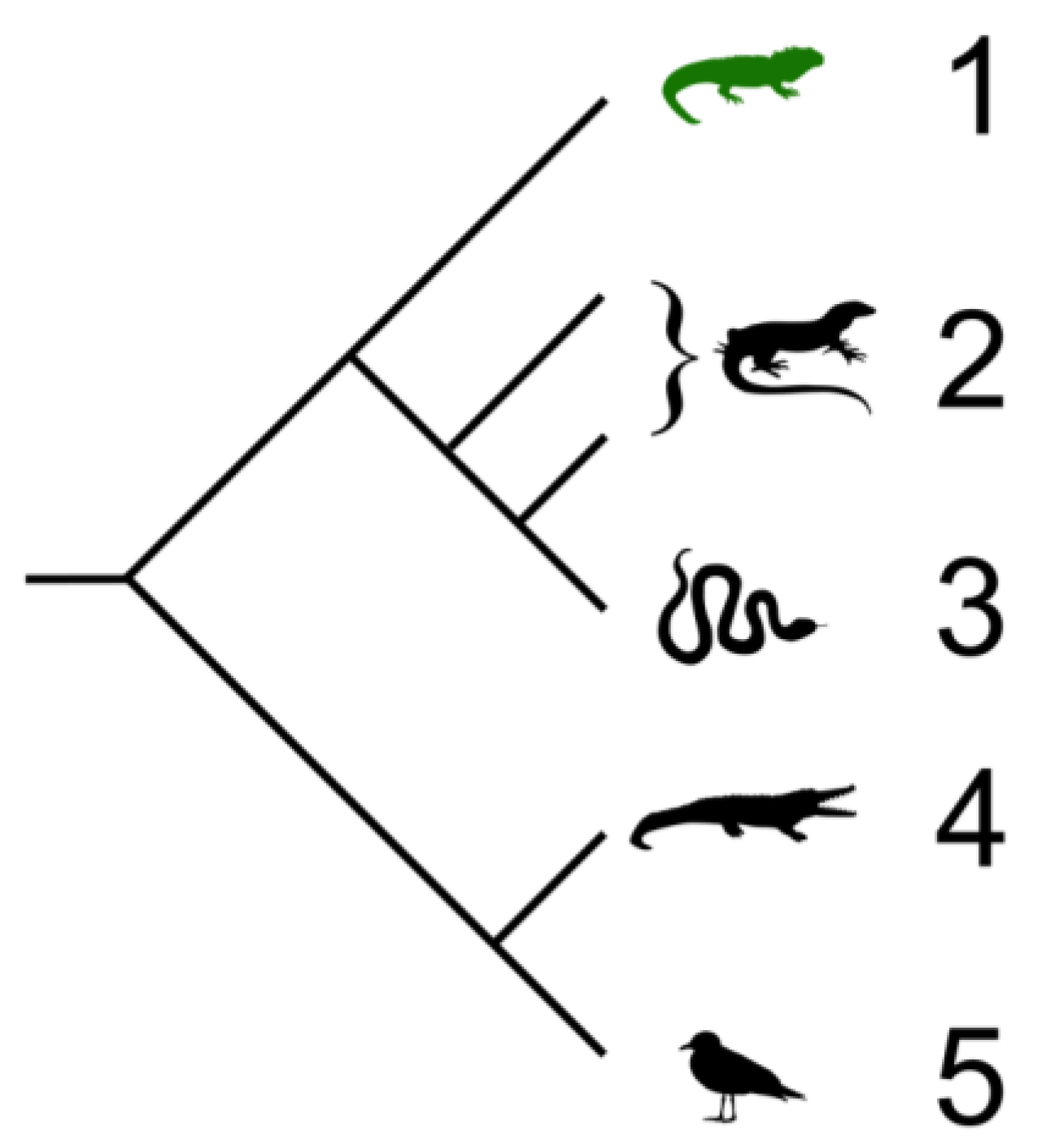
2.1. Testudines (Turtles and Tortises)

2.2. Sphenodontia (Tuataras)

2.3. Squamata (Snakes and Lizards)


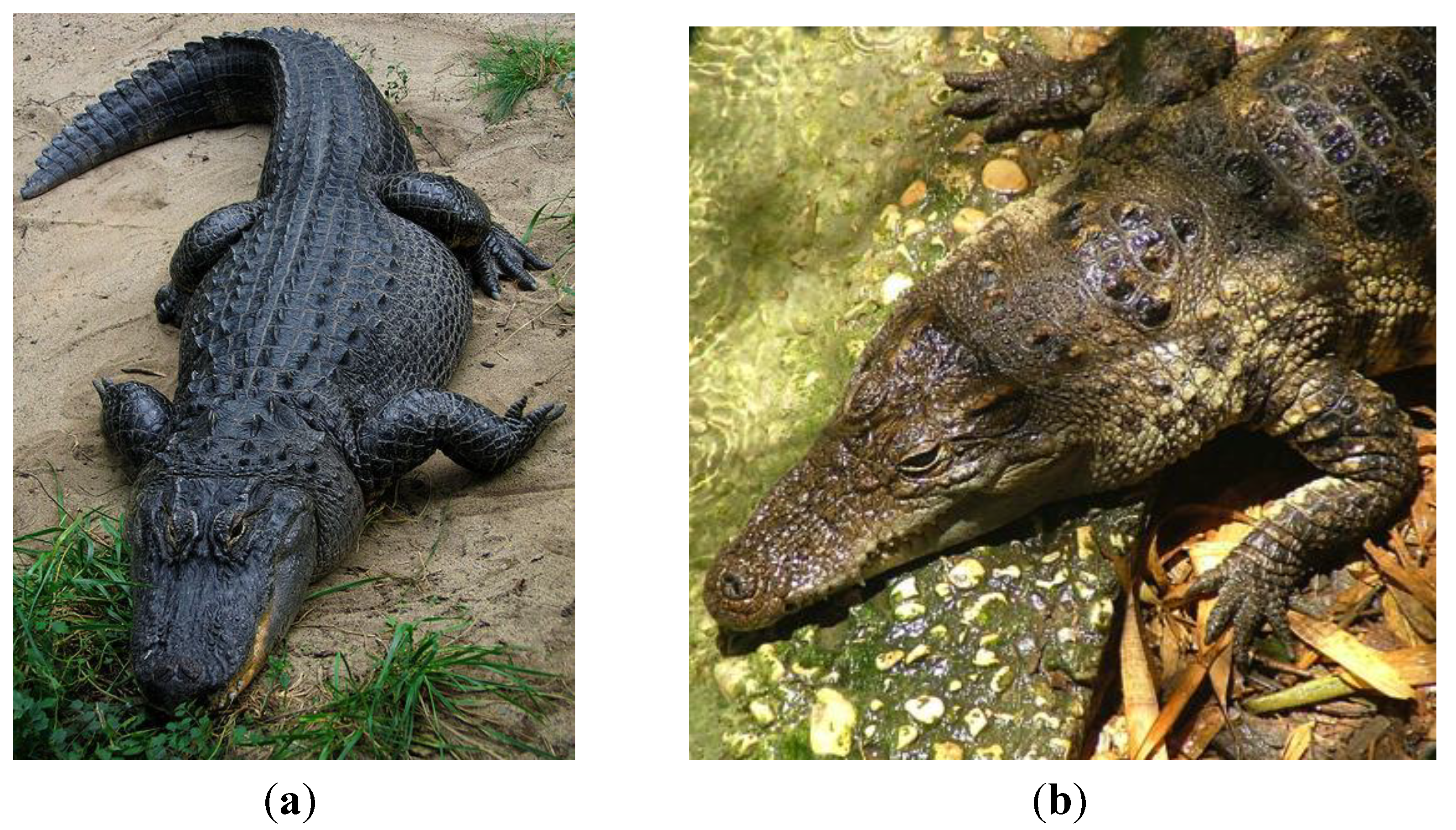
2.4. Crocodilia (Crocodilians)
3. Antimicrobial Peptides of Reptiles
3.1. Defensin Peptides in Reptiles
3.1.1. Three Sub-Classes of Defensins
3.1.2. Inducible Expression of β-Defensins in Wounded Lizards
| Organism | Peptide annotation | aa | Locus- Accession # |
|---|---|---|---|
| Alligator mississippiensis | Gallinacin-14-like | 58 | XP_006270781.1 |
| Alligator sinensis | Gallinacin-14-like | 58 | XP_006033878.1 |
| Anolis carolinensis | β-Defensin-like protein 5 | 62 | CBY85058.1 |
| Anolis carolinensis | β-Defensin-like protein 8 | 65 | CBY85059.1 |
| Anolis carolinensis | β-Defensin-like protein 9 | 66 | CBY85060.1 |
| Anolis carolinensis | β-Defensin-like protein 10 | 67 | CBY85061.1 |
| Anolis carolinensis | β-Defensin-like protein 15 | 63 | CCA62931.1 |
| Anolis carolinensis | β-Defensin-like protein 21 | 89 | CBY85062.1 |
| Anolis carolinensis | β-Defensin-like protein 22 | 95 | CBY85063.1 |
| Anolis carolinensis | β-Defensin-like protein 27 | 81 | CBY85064.1 |
| Anolis carolinensis | Gallinacin-10-like | 68 | XP_003225602.1 |
| Anolis carolinensis | Gallinacin-13-like | 60 | XP_003225598.1 |
| Anolis carolinensis | Hypothetical protein LOC100555370 | XP_003227809.1 | |
| Anolis carolinensis | Hypothetical protein LOC100555565 | XP_003227810.1 | |
| Anolis carolinensis | Hypothetical protein LOC100555756 | XP_003227811.1 | |
| Anolis carolinensis | Hypothetical protein LOC100562305 | XP_003225604.1 | |
| Anolis carolinensis | Hypothetical protein LOC100562502 | 65 | XP_003225605.1 |
| Anolis carolinensis | Hypothetical protein LOC100562898 | XP_003225607.1 | |
| Anolis carolinensis | Hypothetical protein LOC100563098 | XP_003225608.1 | |
| Chrysemys picta bellii | β-Defensin 1-like | 80 | XP_005308390.1 |
| Chrysemys picta bellii | Gallinacin-5-like | XP_005290738.1 | |
| Chrysemys picta bellii | Gallinacin-5-like, partial | XP_005314963.1 | |
| Chrysemys picta bellii | Gallinacin-14-like | 58 | XP_005308403.1 |
| Pelodiscus sinensis | Gallinacin-1 α-like | XP_006137072.1 | |
| Pelodiscus sinensis | Lingual antimicrobial peptide-like isoform X2 | XP_006127561.1 | |
| Bothrops neuwiedi | β-Defensin-like protein | AGF25392.1 | |
| Bothrops jararacussu | β-Defensin-like protein | AGF25388.1 | |
| Bothrops leucurus | β-Defensin-like protein | AGF25389.1 | |
| Bothrops matogrossensis | β-Defensin-like protein | AGF25391.1, AGF25390.1 | |
| Bothrops diporus | β-Defensin-like protein | AGF25384.1 | |
| Bothrops pauloensis | β-Defensin-like protein | AGF25393.1 | |
| Bothrops jararaca | β-Defensin-like protein | AGF25386.1, AGF25387.1 | |
| Bothrops atrox | β-Defensin-like protein | AGF25383.1 | |
| Bothrops erythromelas | β-defensin-like protein | AGF25385.1 |
3.1.3. Expression of β-Defensins in Reptile Eggs
3.2. Cathelicidin Peptides in Reptiles
3.3. Cathelicidin Peptides in Snakes
3.3.1. King Cobra (Ophiophagus (O.) Hannah)
| Protein name [Organism] | Accession | Sequence | |
|---|---|---|---|
| Cathelicidin-NA antimicrobial peptide [Naja atra] | B6S2X0.1 | 1 | MEGFFWKTLLVVGALTISGTSSFPHKPLTYEEAVDLAVSVYNSKSG EDSLYRLLEAVPALKWDALSESNQELNFSVKETV 80 |
| 81 | CQMAEERSLEECDFQEAGAVMGCTGYYFFGESPPVLVLTCKSVGNE- EEQKQEEGNEEEKEVEKEEKEEDQKDQPKR 156 | ||
| 157 | V  191 191 | ||
| Cathelicidin-BF antimicrobial peptide [Bungarus fasciatus] | B6D434.1 | 1 | MEGFFWKTLLVVGALAIAGTSSLPHKPLIYEEAVDLAVSIYNSKSGEDS LYRLLEAVSPPKWDPLSESNQELNFTMKETV 80 |
| 81 | CLVAEERSLEECDFQEDGAIMGCTGYYFFGESPPVLVLTCKPVGEE- EEQKQEEGNEEEKEVEKEEKEEDEKDQPRR 156 | ||
| 157 | V  191 191 | ||
| Cathelicidin-OH antimicrobial peptide [Ophiophagus hannah] | B6S2X2.1 | 1 | MEGFFWKTLLVVGALAIGGTSSLPHKPLTYEEAVDLAVSIYNSKSGEDSL YRLLEAVPPPEWDPLSESNQELNFTIKETV 80 |
| 81 | CLVAEERSLEECDFQEDGVVMGCTGYYFFGESPPVVVLTCKPVGEE- GEQKQEEGNEEEKEVEEEEQEEDEKDQPRR 156 | ||
| 157 | V  191 191 | ||
| cathelicidin-like peptide precursor [Bothrops atrox] | AGS36140.1 | 1 | MQGFFWKTWLVVALC--GTSSSLAHRPLSYGEALELALSIYNSKAG EESLFRLLEAVPQPEWDPLSEGSQQLNFTIKETV 78 |
| 79 | CQVEEERPLEECGFQEDGVVLECTGYYFFGETPPVVVLTCVPVGG V-EEEEEDE-EEQKAEVEKDEKEDEEKDRPKR 154 | ||
| 155 | VKRFKKFFKKLKNSVKKRVKKFFRKPRVIGVTFPF 189 | ||
| cathelicidin-related antimicrobial peptide isoform precursor [Pseudonaja textilis] | AGS36144.1 | 1 | MEGFFWKTWLVVAAFAIGGTSSLPHKPLTYEEAVDLAVSTYNGKSGEE SLYRLLEAVPPPKWDPLSESNQELNLTIKETV 80 |
| 81 | CLVAEERSLEECDFQDDGAVMGCTGYFFFGESPPVLVLTCEPLGED-EE QNQEEE-------EEEEKEEDEKDQPRR 149 | ||
| 150 | VKRFKKFFMKLKKSVKKRVMKFFKKPMVIGVTFPF 184 | ||
| cathelicidin-related antimicrobial peptide precursor [Pseudonaja textilis] | AGS36143.1 | 1 | MDGFFWKTWLVVAALAIGGTSSLPHKPLTYEEAVDLAVSTYNGKSGEES LYRLLEAVPPPKWDPLSESNQELNLTIKETV 80 |
| 81 | CLVAEERSLEECDFQDDGAVMGCTGYFFFGESPPVLVLTCEPLGED- EEQNQEEE-------EEEEKEEDEKDQPRR 149 | ||
| 150 | VKRFKKFFRKLKKSVKKRVKKFFKKPRVIGVTIPF 184 | ||
| cathelicidin-like peptide precursor [Bothrops lutzi] | AGS36141.1 | 1 | MQGFFWKTLLVVALC—GTSSSLAHRPLSYGEALELALSVYNSKAGE ESLFRLLEAVPQPEWDPLSEGSQQLNFTIKETV 78 |
| 79 | CQVEEERPLEECGFQEDGVVLECTGYYFFGETPPVVVLTCVPVGG V-EEEEEDE-EEQKAEVEKDEKEDEEKDRPKR 154 | ||
| 155 | VKRFKKFFKKLKNNVKKRVKKFFRKPRVIGVTIPF 189 | ||
| cathelicidin-like peptide precursor [Lachesis muta rhombeata] | AGS36142.1 | 1 | MQGFFWKTWLVLAVC--GTPASLAHRPLSYGEALELAVSVYNGKAGEAS LYRLLEAVPQPEWDPSSEGSQQLNFTLKETA 78 |
| 79 | CQVEEERSLEECGFQEDGVVLECTGYYFFGETPPVVVLSCVPVGGV eEEEEEEE-EEQKAEAENDEKEDEEKDQPKR 159 | ||
| 160 | 160 VKRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTFPF 194 | ||
| cathelicidin-like peptide precursor [Crotalus durissus terrificus] | AGS36138.1 | 1 | MQGFFWKTWLVLAVC—GTPASLAHRPLSYGEALELAVSVYNGKAG EASLYRLLEAVPQPEWDPSSEGSQQLNFTLKETA 78 |
| 79 | CQVEEERSLEECGFQEDGVVLECTGYYFFGETPPVVVLSCVPVGGV eEEEEEEE-EEQKAEAENDEKGDEEKDQPKR 159 | ||
| 160 | VKRFKKFFKKVKKSVKKRLKKIFKKPMVIGVTIPF 194 | ||
3.3.2. Chinese King Cobra (Naja (N.) Atra)
3.3.3. Banded Krait (B. fasciatus)
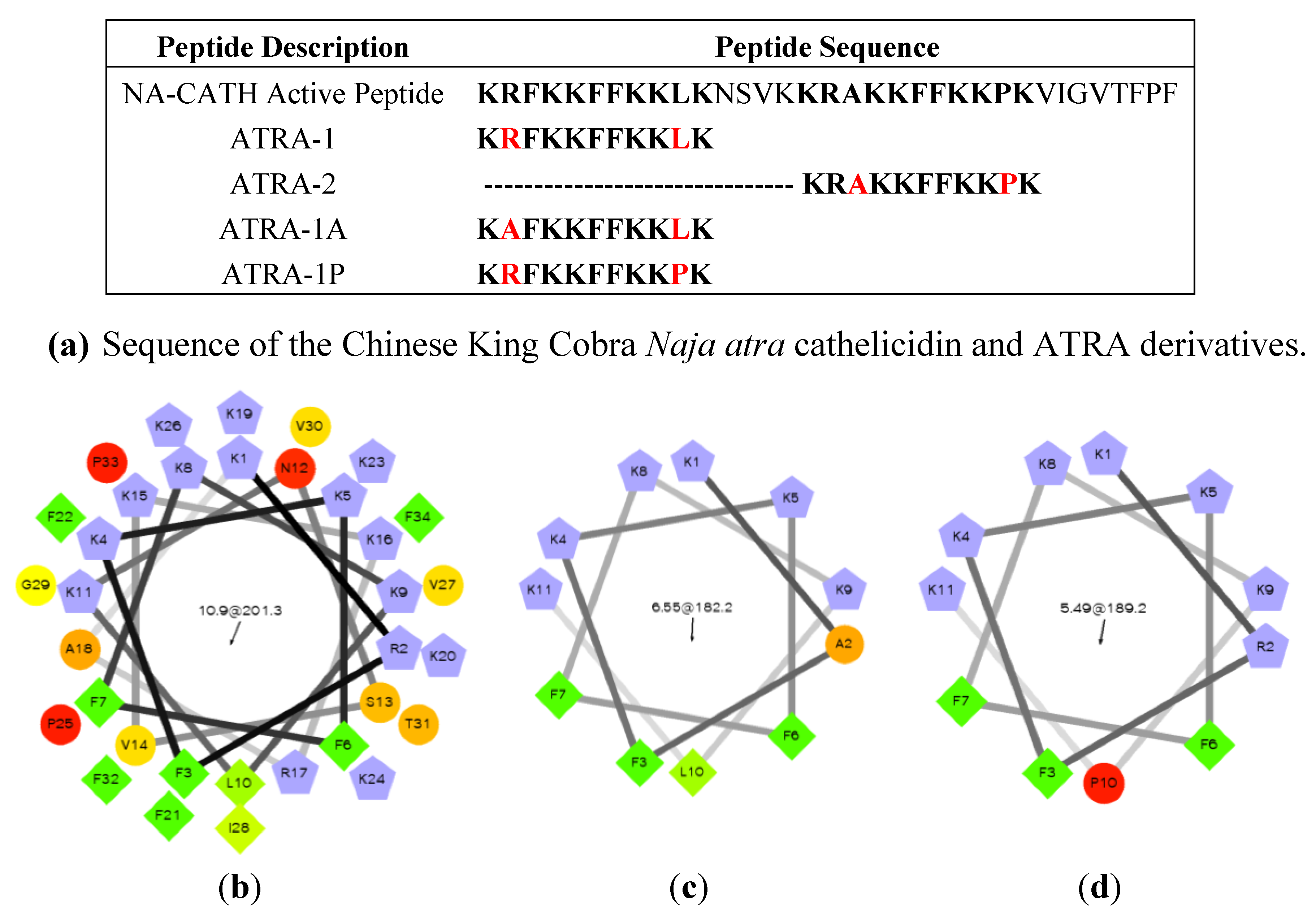
3.3.4. Predicted Cathelicidins in Other Snake Species
3.4. Cathelicidin Peptides in Lizards
3.5. Cathelicidin Peptides in Turtles
| Organism | Peptide annotation | Locus- Accession # |
|---|---|---|
| Chrysemys picta bellii | Cathelicidin-OH antimicrobial peptide-like | XM_005295113.1 |
| Chrysemys picta bellii | Cathelicidin-OH antimicrobial peptide-like | XP_005295170.1 |
| Chrysemys picta bellii | Cathelicidin-2-like | XP_005295171.1 |
| Chrysemys picta bellii | Uncharacterized LOC101951069 | XR_255838.1 |
| Chrysemys picta bellii | Uncharacterized LOC101951243 | XR_255839.1 |
| Pelodiscus sinensis | Cathelicidin-2-like | XM_006114422.1 |
| Pelodiscus sinensis | Cathelicidin-2-like | XM_006114419.1 |
| Pelodiscus sinensis | Cathelicidin-OH antimicrobial peptide-like transcript variant X1 | XM_006129620.1 |
| Pelodiscus sinensis | Cathelicidin-OH antimicrobial peptide-like transcript variant X2 | XM_006129621.1 |
| Pelodiscus sinensis | Cathelicidin-OH antimicrobial peptide-like | XM_006129625.1 |
| Pelodiscus sinensis | Cathelicidin-BF antimicrobial peptide-like | XP_006114480.1 |
| Pelodiscus sinensis | Cathelicidin-BF antimicrobial peptide-like | XM_006114418.1 |
| Pelodiscus sinensi | Cathelicidin-2-like | XP_006114484. |
| Pelodiscus sinensis | Cathelicidin-2-like | XP_006114481.1 |
| Pelodiscus sinensis | Cathelicidin-OH antimicrobial peptide-like isoform X1 | XP_006129682.1 |
| Pelodiscus sinensis | Cathelicidin-OH antimicrobial peptide-like isoform X2 | XP_006129683.1 |
| Pelodiscus sinensis | Cathelicidin-OH antimicrobial peptide-like | XP_006129687.1 |
| Chelonia mydas | Hypothetical protein UY3_13361 | EMP29519.1 |
| Chelonia mydas | Hypothetical protein UY3_13360 | EMP29518.1 |
3.6. Cathelicidin Peptides in Crocodilians
| Organism | Peptide annotation | Locus- Accession # |
|---|---|---|
| Alligator mississippiensis | Cathelicidin-2-like | XM_006262429.1 |
| Alligator mississippiensis | Cathelicidin-2-like | XP_006262491.1 |
| Alligator mississippiensis | Cathelicidin-OH antimicrobial peptide-like | XM_006262431.1 |
| Alligator mississippiensis | Cathelicidin-OH antimicrobial peptide-like | XM_006262430.1 |
| Alligator mississippiensis | Cathelicidin-OH antimicrobial peptide-like | XP_006262492.1 |
| Alligator mississippiensis | Cathelicidin-OH antimicrobial peptide-like | XP_006262493.1 |
| Alligator sinensis | Cathelicidin-2-like | XM_006026412.1 |
| Alligator sinensis | Cathelicidin-2-like | XP_006026474.1 |
| Alligator sinensis | Cathelicidin-2-like | XP_006026475.1 |
| Alligator sinensis | Cathelicidin-3-like | XM_006026409.1 |
| Alligator sinensis | Cathelicidin-3-like | XP_006026471.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XM_006037224.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XM_006026410.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XM_006026411.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XM_006037211.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XP_006037286.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XP_006026472.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XP_006037273.1 |
| Alligator sinensis | Cathelicidin-OH antimicrobial peptide-like | XP_006026487.1 |
4.1. Liver-Derived Peptides in Reptiles
4.1.1. Hepcidin (HAMP1)
4.1.2. LEAP-2, Liver Expressed Antimicrobial Peptide-2
| Species | Predicted LEAP-2 full sequence | Accession Number |
|---|---|---|
| Anolis carolinensis | MTPLKITAVILICSALLFQTQGASLYPPNSQLVRQR RMTPFWRGISRPIGASCRDNSECSTRLCRSKHCSLRTSQE | XP_003217432.1 |
| Alligator sinensis | MHWLKVIAVMLLFALHLFQIHCASLHQPNSQPKRQRRM TPFWRGVSSLRPIGASCRDDIECVTMLCRKSHCSLRTSRE | XP_006023615.1 |
| Alligator mississippiensis | MHWLKVIAVMLLFALHLFQIHCASLHQPNSQPKRQRRM TPFWRGVSSLRPIGASCKDDGECITMRCRKSHCSLRTSRE | XP_006263463.1 |
| Pelodiscus sinensis | MQCLKVIALLLFCAALLTQTHCASLHHSSSQLTRQRRMTP FWRGISLRPIGALCRHDNECISMLCRKNRCSLRISCE | XP_006128591.1 |
| Chrysemys picta belli | MQYLKVIAVLLLCAALLSQIHSASLHRPSSHLTRQRRMTPF WRGISLRPIGAICRDDSECVSRLCRKNHCSIRISRA | XP_005302895.1 |
4.2. Lysozyme in Reptiles
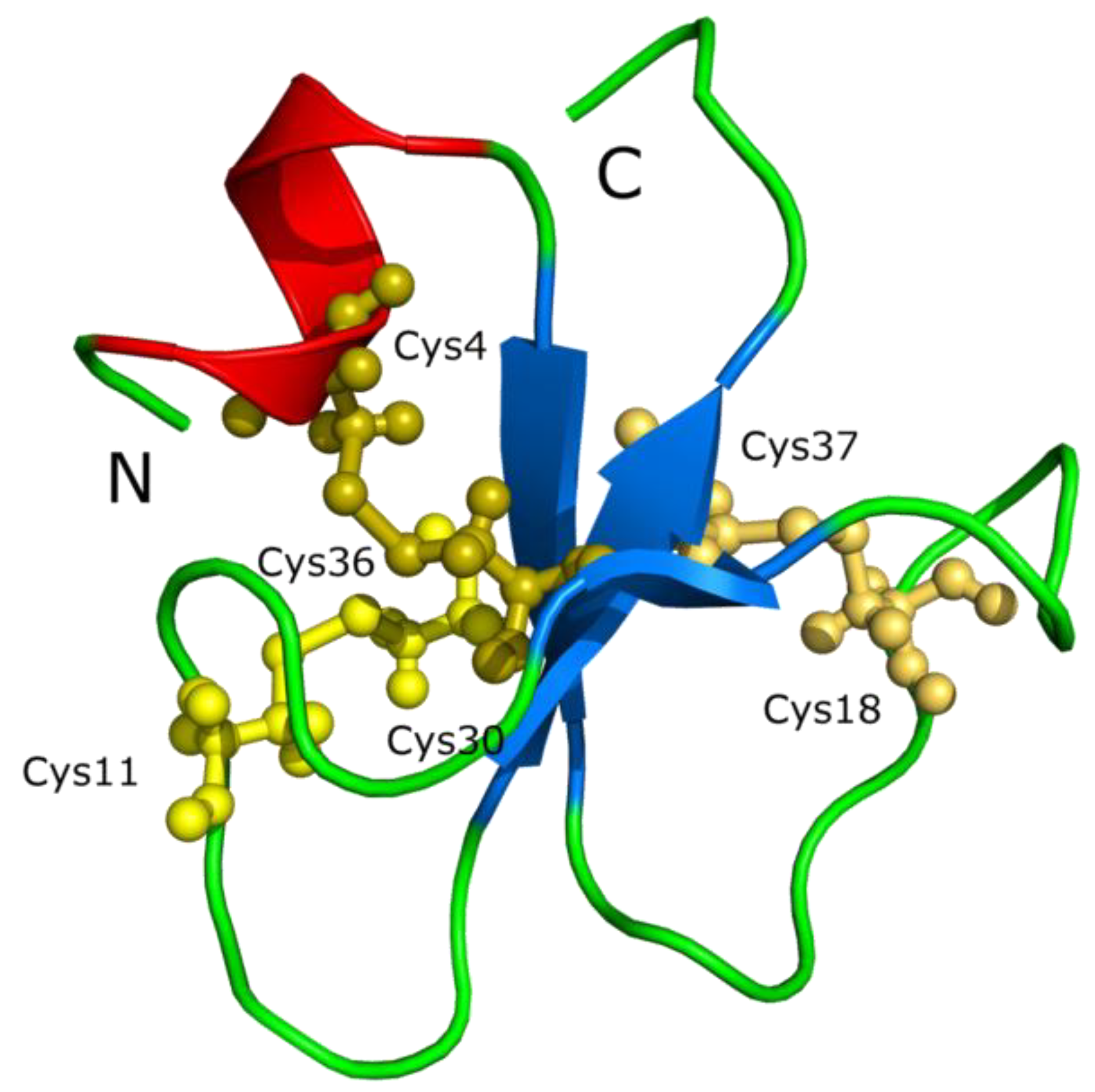
4.3. Crotamine Peptides in Reptiles
| Reptile name | Enzyme Name | Accession Number |
|---|---|---|
| Softshell turtle lysozyme C(SSTL) | Lysozyme C (1,4-β-N-acetylmuramidase C) | Q7LZQ1.3 |
| Asiatic softshell turtle lysozyme C (ASTL) | Lysozyme C | P85345.1 |
| Pelodiscus sinensis | Lysozyme | ADR51676.1 |
| Pelodiscus sinensis | PREDICTED: lysozyme g-like isoform X2 | XP_006113603.1 |
| Pelodiscus sinensis | PREDICTED: lysozyme g-like isoform X1 | XP_006113602.1 |
| Pelodiscus sinensis | PREDICTED: lysozyme g-like | XP_006113601.1 |
| Chelonia mydas | Lysozyme C | EMP38935.1 |
| Chelonia mydas | Lysozyme G | EMP27176.1 |
| Chrysemys picta bellii | PREDICTED: lysozyme C-like | XP_005314893.1 |
| Chrysemys picta bellii | PREDICTED: lysozyme C-like | XP_005312037.1 |
| Chrysemys picta bellii | PREDICTED: lysozyme g-like protein 2 | XP_005283410.1 |
| Chrysemys picta bellii | PREDICTED: lysozyme G-like | XP_005283294.1 |
| Ophiophagus hannah | Lysozyme C, partial | ETE58503.1 |
| XP_003225844.1, | ||
| Anolis carolinensis | Lysozyme C, milk isozyme-like | XP_003216710.1, |
| XP_003216704.1 | ||
| Anolis carolinensis | Lysozyme C II-like | XP_003224512.1 |
| Anolis carolinensis | Lysozyme g-like | XP_003227178.1 |
| Alligator sinensis | Lysozyme C-like | XP_006027022.1, XP_006027021.1 |
| Alligator sinensis | Lysozyme G-like | XP_006026406.1, XP_006026397.1, XP_006026395.1, XP_006026396.1 |
| Reptile | AA | Name | Accession numbers |
|---|---|---|---|
| Uromastyx aegyptia | 67 | crotamine-Uro-1 | AGI97143.1 |
| Crotalus durissus terrificus | 65 | crotamine | AAF34911.1, AAF34910.1, AAC02995.1, AAC06241.1 |
| Crotalus durissus | 34 | crotamine | ACA63453.1, ACA63452.1, ACA63451.1, ACA63450.1, ACA63449.1, ACA63448.1, ACA63447.1, ACA63446.1 |
| Crotalus oreganus helleri | 65 | crotamine 7 | AEU60015.1 |
| Crotalus oreganus helleri | 65 | crotamine 6 | AEU60014.1 |
| Crotalus oreganus helleri | 65 | crotamine 5 | AEU60013.1 |
| Crotalus oreganus helleri | 70 | crotamine 4 | AEU60012.1 |
| Crotalus oreganus helleri | 70 | crotamine 3 | AEU60011.1 |
| Crotalus oreganus helleri | 83 | crotamine 2 | AEU60010.1 |
| Crotalus oreganus helleri | 70 | crotamine 1 | AEU60009.1 |
| Pogona barbata | 102 | CLP-POGL1 | AAZ75614.1 |
| Pogona barbata | 67 | CLP-POGL2 | AAZ75615.1 |
| Pogona barbata | 61 | CLP-POGU3 | AAZ75613.1 |
| Pogona barbata | 98 | CLP-POGU2 | AAZ75612.1 |
| Pogona barbata | 76 | CLP-POGU1 | AAZ75611.1 |
| Varanus tristis | 83 | crotamine-Var-5 | AGI97148.1 |
| Varanus glauerti | 83 | crotamine-Var-4 | AGI97147.1 |
| Varanus glauerti | 83 | crotamine-Var-3 | AGI97146.1 |
| Varanus glauerti | 83 | crotamine-Var-2 | AGI97145.1 |
| Varanus glauerti | 83 | crotamine-Var-1 | AGI97144.1 |
4.4. Other Peptides in Reptiles
4.4.1. Leucrocin
4.4.2. Omwaprin
4.4.3. Hemocidin
4.4.4. Other Peptides
5. Conclusions
Acknowledgments
Conflicts of Interest
References
- Wang, G. Antimicrobial Peptide Database. Available online: http://aps.unmc.edu/AP (accessed on 9 May 2014).
- Wang, G.; Li, X.; Wang, Z. APD2: The updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res. 2009, 37, D933–D937. [Google Scholar] [CrossRef]
- Zhao, H.; Gan, T.X.; Liu, X.D.; Jin, Y.; Lee, W.H.; Shen, J.H.; Zhang, Y. Identification and characterization of novel reptile cathelicidins from elapid snakes. Peptides 2008, 29, 1685–1691. [Google Scholar] [CrossRef]
- Li, S.A.; Lee, W.H.; Zhang, Y. Efficacy of OH-CATH30 and its analogs against drug-resistant bacteria in vitro and in mouse models. Antimicrob. Agents Chemother. 2012, 56, 3309–3317. [Google Scholar] [CrossRef]
- Zhang, B.Y.; Li, S.M.; Gao, Z.H.; Shen, J.H. Protective effects of snake venom antimicrobial peptide OH-CATH on E. coli induced rabbit urinary tract infection models. Dong Wu Xue Yan Jiu 2013, 34, 27–32. [Google Scholar]
- Zhang, Y.; Zhao, H.; Yu, G.Y.; Liu, X.D.; Shen, J.H.; Lee, W.H.; Zhang, Y. Structure-function relationship of king cobra cathelicidin. Peptides 2010, 31, 1488–1493. [Google Scholar] [CrossRef]
- Wang, Y.; Hong, J.; Liu, X.; Yang, H.; Liu, R.; Wu, J.; Wang, A.; Lin, D.; Lai, R. Snake cathelicidin from Bungarus fasciatus is a potent peptide antibiotics. PloS One 2008, 3, e3217. [Google Scholar]
- Chen, W.; Yang, B.; Zhou, H.; Sun, L.; Dou, J.; Qian, H.; Huang, W.; Mei, Y.; Han, J. Structure-activity relationships of a snake cathelicidin-related peptide, BF-15. Peptides 2011, 32, 2497–2503. [Google Scholar] [CrossRef]
- Wang, H.; Ke, M.; Tian, Y.; Wang, J.; Li, B.; Wang, Y.; Dou, J.; Zhou, C. BF-30 selectively inhibits melanoma cell proliferation via cytoplasmic membrane permeabilization and DNA-binding in vitro and in B16F10-bearing mice. Eur. J. Pharmacol. 2013, 707, 1–10. [Google Scholar]
- Zhou, H.; Dou, J.; Wang, J.; Chen, L.; Wang, H.; Zhou, W.; Li, Y.; Zhou, C. The antibacterial activity of BF-30 in vitro and in infected burned rats is through interference with cytoplasmic membrane integrity. Peptides 2011, 32, 1131–1138. [Google Scholar] [CrossRef]
- Nair, D.G.; Fry, B.G.; Alewood, P.; Kumar, P.P.; Kini, R.M. Antimicrobial activity of omwaprin, a new member of the waprin family of snake venom proteins. Biochem. J. 2007, 402, 93–104. [Google Scholar] [CrossRef]
- Banigan, J.R.; Mandal, K.; Sawaya, M.R.; Thammavongsa, V.; Hendrickx, A.P.; Schneewind, O.; Yeates, T.O.; Kent, S.B. Determination of the X-ray structure of the snake venom protein omwaprin by total chemical synthesis and racemic protein crystallography. Protein Sci. 2010, 19, 1840–1849. [Google Scholar] [CrossRef]
- Schmidt, J.J.; Weinstein, S.A.; Smith, L.A. Molecular properties and structure-function relationships of lethal peptides from venom of Wagler’s pit viper, Trimeresurus wagleri. Toxicon 1992, 30, 1027–1036. [Google Scholar] [CrossRef]
- Stegemann, C.; Kolobov, A., Jr.; Leonova, Y.F.; Knappe, D.; Shamova, O.; Ovchinnikova, T.V.; Kokryakov, V.N.; Hoffmann, R. Isolation, purification and de novo sequencing of TBD-1, the first beta-defensin from leukocytes of reptiles. Proteomics 2009, 9, 1364–1373. [Google Scholar] [CrossRef]
- Lakshminarayanan, R.; Vivekanandan, S.; Samy, R.P.; Banerjee, Y.; Chi-Jin, E.O.; Teo, K.W.; Jois, S.D.; Kini, R.M.; Valiyaveettil, S. Structure, self-assembly, and dual role of a beta-defensin-like peptide from the Chinese soft-shelled turtle eggshell matrix. J. Am. Chem. Soc. 2008, 130, 4660–4668. [Google Scholar] [CrossRef]
- Lakshminarayanan, R.; Chi-Jin, E.O.; Loh, X.J.; Kini, R.M.; Valiyaveettil, S. Purification and characterization of a vaterite-inducing peptide, pelovaterin, from the eggshells of Pelodiscus sinensis (Chinese soft-shelled turtle). Biomacromolecules 2005, 6, 1429–1437. [Google Scholar] [CrossRef]
- Thammasirirak, S.; Ponkham, P.; Preecharram, S.; Khanchanuan, R.; Phonyothee, P.; Daduang, S.; Srisomsap, C.; Araki, T.; Svasti, J. Purification, characterization and comparison of reptile lysozymes. Comp. biochem. Phys. Toxicol. Pharmacol. 2006, 143, 209–217. [Google Scholar]
- Chattopadhyay, S.; Sinha, N.K.; Banerjee, S.; Roy, D.; Chattopadhyay, D.; Roy, S. Small cationic protein from a marine turtle has beta-defensin-like fold and antibacterial and antiviral activity. Proteins 2006, 64, 524–531. [Google Scholar] [CrossRef]
- Ponkham, P.; Daduang, S.; Kitimasak, W.; Krittanai, C.; Chokchaichamnankit, D.; Srisomsap, C.; Svasti, J.; Kawamura, S.; Araki, T.; Thammasirirak, S. Complete amino acid sequence of three reptile lysozymes. Comp. Biochem. Phys. Toxicol. Pharmacol. 2010, 151, 75–83. [Google Scholar] [CrossRef]
- Yount, N.Y.; Kupferwasser, D.; Spisni, A.; Dutz, S.M.; Ramjan, Z.H.; Sharma, S.; Waring, A.J.; Yeaman, M.R. Selective reciprocity in antimicrobial activity versus cytotoxicity of hBD-2 and crotamine. Proc. Natl. Acad. Sci. USA 2009, 106, 14972–14977. [Google Scholar] [CrossRef]
- Coronado, M.A.; Georgieva, D.; Buck, F.; Gabdoulkhakov, A.H.; Ullah, A.; Spencer, P.J.; Arni, R.K.; Betzel, C. Purification, crystallization and preliminary X-ray diffraction analysis of crotamine, a myotoxic polypeptide from the Brazilian snake Crotalus durissus terrificus. Acta Crystallogr. F 2012, 68, 1052–1054. [Google Scholar] [CrossRef]
- Modesto, S.P.; Anderson, J.S. The phylogenetic definition of reptilia. Syst. Biol. 2004, 53, 815–821. [Google Scholar] [CrossRef]
- Alfoldi, J.; Di Palma, F.; Grabherr, M.; Williams, C.; Kong, L.; Mauceli, E.; Russell, P.; Lowe, C.B.; Glor, R.E.; Jaffe, J.D.; et al. The genome of the green anole lizard and a comparative analysis with birds and mammals. Nature 2011, 477, 587–591. [Google Scholar] [CrossRef]
- Cladogram by Benchill, licensed under the Creative Commons Attribution 3.0 Unported license. Available online: http://en.wikipedia.org/wiki/File:Tuatara_cladogram.svg (accessed on 9 May 2014).
- Ernst, C.H.; Lovich, J.E. Turtles of the United States and Canada, 2nd ed.; Johns Hopkins University Press: Baltimore, MD, USA, 2009; pp. 185–259. [Google Scholar]
- Photo from Oregon Department of Fish & Wildlife, licensed under the Creative Commons Attribution-Share Alike 2.0 Generic license. Photo by Gary M. Stolz/U. S. Fish and Wildlife Service in the Public domain. Available online: http://en.wikipedia.org/wiki/FileA4_Western_painted_turtle.jpg (accessed on 9 May 2014).
- Underside of a Western Painted Turtle. Photo by Matt Young, l.u.t.C.C.A.-S.A.G.l. Available online: http://en.wikipedia.org/wiki/File:B4_Western_painted_turtle_underside.jpg (accessed on 9 May 2014).
- Wang, Z.; Pascual-Anaya, J.; Zadissa, A.; Li, W.; Niimura, Y.; Huang, Z.; Li, C.; White, S.; Xiong, Z.; Fang, D.; et al. The draft genomes of soft-shell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Genet. 2013, 45, 701–706. [Google Scholar] [CrossRef]
- Jiang, J.J.; Xia, E.H.; Gao, C.W.; Gao, L.Z. The complete mitochondrial genome of western painted turtle, Chrysemys picta bellii (Chrysemys, Emydidae). Mitochondrial DNA 2014. [Google Scholar] [CrossRef]
- Shaffer, H.B.; Minx, P.; Warren, D.E.; Shedlock, A.M.; Thomson, R.C.; Valenzuela, N.; Abramyan, J.; Amemiya, C.T.; Badenhorst, D.; Biggar, K.K.; et al. The western painted turtle genome, a model for the evolution of extreme physiological adaptations in a slowly evolving lineage. Genome Biol. 2013, 14, R28. [Google Scholar] [CrossRef]
- Gilbert, S.F.; Corfe, I. Turtle origins: Picking up speed. Dev. Cell 2013, 25, 326–328. [Google Scholar] [CrossRef]
- Zardoya, R.; Meyer, A. Complete mitochondrial genome suggests diapsid affinities of turtles. Proc. Natl. Acad. Sci. USA 1998, 95, 14226–14231. [Google Scholar] [CrossRef]
- Iwabe, N.; Hara, Y.; Kumazawa, Y.; Shibamoto, K.; Saito, Y.; Miyata, T.; Katoh, K. Sister group relationship of turtles to the bird-crocodilian clade revealed by nuclear DNA-coded proteins. Mol. Biol. Evol. 2005, 22, 810–813. [Google Scholar] [CrossRef]
- Roos, J.; Aggarwal, R.K.; Janke, A. Extended mitogenomic phylogenetic analyses yield new insight into crocodylian evolution and their survival of the Cretaceous-Tertiary boundary. Mol. Phylogenet. Evol. 2007, 45, 663–673. [Google Scholar] [CrossRef]
- Katsu, Y.; Matsubara, K.; Kohno, S.; Matsuda, Y.; Toriba, M.; Oka, K.; Guillette, L.J., Jr.; Ohta, Y.; Iguchi, T. Molecular cloning, characterization, and chromosome mapping of reptilian estrogen receptors. Endocrinology 2010, 151, 5710–5720. [Google Scholar]
- Lyson, T.R.; Sperling, E.A.; Heimberg, A.M.; Gauthier, J.A.; King, B.L.; Peterson, K.J. MicroRNAs support a turtle + lizard clade. Biol. Lett. 2012, 8, 104–107. [Google Scholar] [CrossRef]
- Jones, M.E.; Cree, A. Tuatara. Curr. Biol. 2012, 22, R986–R987. [Google Scholar] [CrossRef]
- Hay, J.M.; Sarre, S.D.; Lambert, D.M.; Allendorf, F.W.; Daugherty, C.H. Genetic diversity and taxonomy: A reassessment of species designation in tuatara (Sphenodon: Reptilia). Conserv. Genet. 2010, 11, 1063–1081. [Google Scholar]
- Photo by PhillipC, licensed under the Creative Commons Attribution 2.0 Generic license. Available online: http://en.wikipedia.org/wiki/File:Sphenodon_punctatus_in_Waikanae,_New_Zealand.jpg (accessed on 9 May 2014).
- Ramstad, K.M.; Nelson, N.J.; Paine, G.; Beech, D.; Paul, A.; Paul, P.; Allendorf, F.W.; Daugherty, C.H. Species and cultural conservation in New Zealand: maori traditional ecological knowledge of tuatara. Conserv. Biol. 2007, 21, 455–464. [Google Scholar] [CrossRef]
- Miller, H.C.; Biggs, P.J.; Voelckel, C.; Nelson, N.J. De novo sequence assembly and characterisation of a partial transcriptome for an evolutionarily distinct reptile, the tuatara (Sphenodon punctatus). BMC Genomics 2012, 13, 439. [Google Scholar] [CrossRef]
- Vonk, F.J.; Casewell, N.R.; Henkel, C.V.; Heimberg, A.M.; Jansen, H.J.; McCleary, R.J.; Kerkkamp, H.M.; Vos, R.A.; Guerreiro, I.; Calvete, J.J.; et al. The king cobra genome reveals dynamic gene evolution and adaptation in the snake venom system. Proc. Natl. Acad. Sci. US A 2013, 110, 20651–20656. [Google Scholar] [CrossRef]
- Pyron, R.A.; Burbrink, F.T.; Guiher, T.J. Claims of potential expansion throughout the U.S. by invasive python species are contradicted by ecological niche models. PloS One 2008, 3, e2931. [Google Scholar] [CrossRef]
- Photo by Greg Hume, licensed under the Creative Commons Attribution 2.0 Generic license. Available online: http://en.wikipedia.org/wiki/File:King_Cobra_25.jpg (accessed on 9 May 2014).
- Found inside a water catchment on Lantau. 2011, Photo by: Thomas Brown. This file is licensed under the Creative Commons Attribution 2.0 Generic license. Available online: http://en.wikipedia.org/wiki/File:Naja_atra_juvenile.jpg (accessed on 9 May 2014).
- Photo by AshLin. Photo of Banded Krait captured in Binnaguri, N.B., India on 19 Sep 2006. This file is licensed under the Creative Commons Attribution-Share Alike 2.5 Generic license. Available online: http://en.wikipedia.org/wiki/File:AB_054_Banded_Krait.JPG (accessed on 9 May 2014).
- Photo by PiccoloNamek at en.wikipedia, licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license. Available online: http://en.wikipedia.org/wiki/File:Anolis_carolinensis.jpg (accessed on 9 May 2014).
- Aird, S.D.; Watanabe, Y.; Villar-Briones, A.; Roy, M.C.; Terada, K.; Mikheyev, A.S. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics 2013, 14, 790. [Google Scholar] [CrossRef]
- Dalla Valle, L.; Benato, F.; Maistro, S.; Quinzani, S.; Alibardi, L. Bioinformatic and molecular characterization of beta-defensins-like peptides isolated from the green lizard Anolis carolinensis. Dev. Comp. Immunol. 2012, 36, 222–229. [Google Scholar] [CrossRef]
- Erickson, G.M.; Gignac, P.M.; Steppan, S.J.; Lappin, A.K.; Vliet, K.A.; Brueggen, J.D.; Inouye, B.D.; Kledzik, D.; Webb, G.J. Insights into the ecology and evolutionary success of crocodilians revealed through bite-force and tooth-pressure experimentation. PloS One 2012, 7, e31781. [Google Scholar] [CrossRef]
- Shirley, M.H.; Vliet, K.A.; Carr, A.N.; Austin, J.D. Rigorous approaches to species delimitation have significant implications for African crocodilian systematics and conservation. Proc. Biol. Sci. 2014, 281, 20132483. [Google Scholar]
- An American Alligator (One of two) in captivity at the Columbus Zoo, Powell, Ohio. This file is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license. Available online: http://en.wikipedia.org/wiki/File:American_Alligator.jpg (accessed on 9 May 2014).
- A Siamese Crocodile (Crocodylus siamensis) at the Jerusalem Biblical Zoo. This file is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license. Available online: http://commons.wikimedia.org/wiki/File:Siamese_Crocodile-Biblical_Zoo.JPG (accessed on 9 May 2014).
- Lance, V.A. Alligator physiology and life history: The importance of temperature. Exp. Gerontol. 2003, 38, 801–805. [Google Scholar] [CrossRef]
- Brown, D.R.; Schumacher, I.M.; Nogueira, M.F.; Richey, L.J.; Zacher, L.A.; Schoeb, T.R.; Vliet, K.A.; Bennett, R.A.; Jacobson, E.R.; Brown, M.B. Detection of antibodies to a pathogenic mycoplasma in American alligators (Alligator mississippiensis), broad-nosed Caimans (Caiman latirostris), and Siamese crocodiles (Crocodylus siamensis). J. Clin. Microbiol. 2001, 39, 285–292. [Google Scholar] [CrossRef]
- Meganathan, P.R.; Dubey, B.; Batzer, M.A.; Ray, D.A.; Haque, I. Molecular phylogenetic analyses of genus Crocodylus (Eusuchia, Crocodylia, Crocodylidae) and the taxonomic position of Crocodylus porosus. Mol. Phylogenet. Evol. 2010, 57, 393–402. [Google Scholar] [CrossRef]
- Kommanee, J.; Preecharram, S.; Daduang, S.; Temsiripong, Y.; Dhiravisit, A.; Yamada, Y.; Thammasirirak, S. Antibacterial activity of plasma from crocodile (Crocodylus siamensis) against pathogenic bacteria. Ann. Clin. Microbiol. Antimicrob. 2012, 11, 22. [Google Scholar] [CrossRef]
- Theansungnoen, T.; Yaraksa, N.; Daduang, S.; Dhiravisit, A.; Thammasirirak, S. Purification and Characterization of Antioxidant Peptides from Leukocyte Extract of Crocodylus siamensis. Protein J. 2014, 33, 24–31. [Google Scholar]
- Srihongthong, S.; Pakdeesuwan, A.; Daduang, S.; Araki, T.; Dhiravisit, A.; Thammasirirak, S. Complete amino acid sequence of globin chains and biological activity of fragmented crocodile hemoglobin (Crocodylus siamensis). Protein J. 2012, 31, 466–476. [Google Scholar]
- Jandaruang, J.; Siritapetawee, J.; Thumanu, K.; Songsiriritthigul, C.; Krittanai, C.; Daduang, S.; Dhiravisit, A.; Thammasirirak, S. The effects of temperature and pH on secondary structure and antioxidant activity of Crocodylus siamensis hemoglobin. Protein J. 2012, 31, 43–50. [Google Scholar]
- Johnston, M.A.; Porter, D.E.; Scott, G.I.; Rhodes, W.E.; Webster, L.F. Isolation of faecal coliform bacteria from the American alligator (Alligator mississippiensis). J. Appl. Microbiol. 2010, 108, 965–973. [Google Scholar] [CrossRef]
- Merchant, M.E.; Mills, K.; Leger, N.; Jerkins, E.; Vliet, K.A.; McDaniel, N. Comparisons of innate immune activity of all known living crocodylian species. Com. Biochem. Physiol. Part B 2006, 143, 133–137. [Google Scholar]
- Merchant, M.E.; Leger, N.; Jerkins, E.; Mills, K.; Pallansch, M.B.; Paulman, R.L.; Ptak, R.G. Broad spectrum antimicrobial activity of leukocyte extracts from the American alligator (Alligator mississippiensis). Vet. Immunol. Immunopathol. 2006, 110, 221–228. [Google Scholar] [CrossRef]
- Merchant, M.E.; Roche, C.; Elsey, R.M.; Prudhomme, J. Antibacterial properties of serum from the American alligator (Alligator mississippiensis). Comp. biochem. Physiol. Part B 2003, 136, 505–513. [Google Scholar] [CrossRef]
- Preecharram, S.; Jearranaiprepame, P.; Daduang, S.; Temsiripong, Y.; Somdee, T.; Fukamizo, T.; Svasti, J.; Araki, T.; Thammasirirak, S. Isolation and characterisation of crocosin, an antibacterial compound from crocodile (Crocodylus siamensis) plasma. Nihon chikusan Gakkaiho 2010, 81, 393–401. [Google Scholar]
- Pata, S.; Yaraksa, N.; Daduang, S.; Temsiripong, Y.; Svasti, J.; Araki, T.; Thammasirirak, S. Characterization of the novel antibacterial peptide Leucrocin from crocodile (Crocodylus siamensis) white blood cell extracts. Dev. Comp. Immunol. 2011, 35, 545–553. [Google Scholar]
- Pata, S.; Daduang, S.; Svasti, J.; Thammasirirak, S. Isolation of Lysozyme like protein from crocodile leukocyte extract (Crocodylus siamensis). KMITL Sci. Technol. J. 2007, 7, 70–85. [Google Scholar]
- Castoe, T.A.; Pollock, D.D. Chinese alligator genome illustrates molecular adaptations. Cell res. 2013, 23, 1254–1255. [Google Scholar] [CrossRef]
- St John, J.A.; Braun, E.L.; Isberg, S.R.; Miles, L.G.; Chong, A.Y.; Gongora, J.; Dalzell, P.; Moran, C.; Bed'hom, B.; Abzhanov, A.; et al. Sequencing three crocodilian genomes to illuminate the evolution of archosaurs and amniotes. Genome Biol. 2012, 13, 415. [Google Scholar] [CrossRef]
- Wan, Q.H.; Pan, S.K.; Hu, L.; Zhu, Y.; Xu, P.W.; Xia, J.Q.; Chen, H.; He, G.Y.; He, J.; Ni, X.W.; et al. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 2013, 23, 1091–1105. [Google Scholar] [CrossRef]
- Chromek, M.; Arvidsson, I.; Karpman, D. The antimicrobial peptide cathelicidin protects mice from Escherichia coli O157:H7-mediated disease. PloS One 2012, 7, e46476. [Google Scholar] [CrossRef]
- Oguro-Okano, M.; Honda, M.; Yamazaki, K.; Okano, K. Mutations in the melanocortin 1 receptor, beta-defensin103 and agouti signaling protein genes, and their association with coat color phenotypes in Akita-inu dogs. J. Vet. Med. Sci. 2011, 73, 853–858. [Google Scholar] [CrossRef]
- Schmutz, S.M.; Berryere, T.G. Genes affecting coat colour and pattern in domestic dogs: A review. Anim. Genet. 2007, 38, 539–549. [Google Scholar] [CrossRef]
- Zasloff, M. Magainins, a class of antimicrobial peptides from Xenopus skin: isolation, characterization of two active forms, and partial cDNA sequence of a precursor. Proc. Natl. Acad. Sci. USA 1987, 84, 5449–5453. [Google Scholar] [CrossRef]
- Wilson, C.L.; Schmidt, A.P.; Pirila, E.; Valore, E.V.; Ferri, N.; Sorsa, T.; Ganz, T.; Parks, W.C. Differential Processing of {alpha}- and {beta}-Defensin Precursors by Matrix Metalloproteinase-7 (MMP-7). J. Biol. Chem. 2009, 284, 8301–8311. [Google Scholar]
- Zhao, L.; Lu, W. Defensins in innate immunity. Curr. Opin. Hematol. 2014, 21, 37–42. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Lu, W. alpha-Defensins in human innate immunity. Immunol. Rev. 2012, 245, 84–112. [Google Scholar] [CrossRef]
- Garcia, J.R.; Jaumann, F.; Schulz, S.; Krause, A.; Rodriguez-Jimenez, J.; Forssmann, U.; Adermann, K.; Kluver, E.; Vogelmeier, C.; Becker, D.; et al. Identification of a novel, multifunctional beta-defensin (human beta-defensin 3) with specific antimicrobial activity. Its interaction with plasma membranes of Xenopus oocytes and the induction of macrophage chemoattraction. Cell Tissue Res. 2001, 306, 257–264. [Google Scholar] [CrossRef]
- Kaplinsky, N.J.; Gilbert, S.F.; Cebra-Thomas, J.; Lillevali, K.; Saare, M.; Chang, E.Y.; Edelman, H.E.; Frick, M.A.; Guan, Y.; Hammond, R.M.; et al. The Embryonic Transcriptome of the Red-Eared Slider Turtle. PloS one 2013, 8, e66357. [Google Scholar]
- Lorenzo, A. Immunolocalization of a beta-defensin (Tu-BD-1) in the skin and subdermal granulocytes of turtles indicate the presence of an antimicrobial skin barrier. Ann. Anat. 2013, 195, 554–561. [Google Scholar] [CrossRef]
- Benato, F.; Dalla Valle, L.; Skobo, T.; Alibardi, L. Biomolecular identification of beta-defensin-like peptides from the skin of the soft-shelled turtle Apalone spinifera. J. Exp. Zool. Part B 2013, 320, 210–217. [Google Scholar] [CrossRef]
- Alibardi, L. Ultrastructural immunolocalization of beta-defensin-27 in granulocytes of the dermis and wound epidermis of lizard suggests they contribute to the anti-microbial skin barrier. Anat. Cell Biol. 2013, 46, 246–253. [Google Scholar] [CrossRef]
- Alibardi, L. Histochemical, Biochemical and Cell Biological aspects of tail regeneration in lizard, an amniote model for studies on tissue regeneration. Prog. Histochem. Cytochem. 2014, 48, 143–244. [Google Scholar] [CrossRef]
- Alibardi, L. Granulocytes of reptilian sauropsids contain beta-defensin-like peptides: a comparative ultrastructural survey. J. Morphol. 2013, 274, 877–886. [Google Scholar] [CrossRef]
- Alibardi, L.; Celeghin, A.; Dalla Valle, L. Wounding in lizards results in the release of beta-defensins at the wound site and formation of an antimicrobial barrier. Dev. Comp. Immunol. 2012, 36, 557–565. [Google Scholar] [CrossRef]
- Abdel Mageed, A.M.; Isobe, N.; Yoshimura, Y. Immunolocalization of avian beta-defensins in the hen oviduct and their changes in the uterus during eggshell formation. Reproduction 2009, 138, 971–978. [Google Scholar] [CrossRef]
- Gong, D.; Wilson, P.W.; Bain, M.M.; McDade, K.; Kalina, J.; Herve-Grepinet, V.; Nys, Y.; Dunn, I.C. Gallin; an antimicrobial peptide member of a new avian defensin family, the ovodefensins, has been subject to recent gene duplication. BMC Immunol. 2010, 11, 12. [Google Scholar]
- Herve, V.; Meudal, H.; Labas, V.; Rehault Godbert, S.; Gautron, J.; Berges, M.; Guyot, N.; Delmas, A.F.; Nys, Y.; Landon, C. 3D NMR structure of hen egg gallin (chicken ovo-defensin) reveals a new variation of the beta-defensin fold. J. Biol. Chem. 2014.
- Herve-Grepinet, V.; Rehault-Godbert, S.; Labas, V.; Magallon, T.; Derache, C.; Lavergne, M.; Gautron, J.; Lalmanach, A.C.; Nys, Y. Purification and characterization of avian beta-defensin 11, an antimicrobial peptide of the hen egg. Antimicrob. Agents Chemother. 2010, 54, 4401–4409. [Google Scholar] [CrossRef]
- Mine, Y.; Oberle, C.; Kassaify, Z. Eggshell matrix proteins as defense mechanism of avian eggs. J. Agric. Food Chem. 2003, 51, 249–253. [Google Scholar] [CrossRef]
- Kosciuczuk, E.M.; Lisowski, P.; Jarczak, J.; Strzalkowska, N.; Jozwik, A.; Horbanczuk, J.; Krzyzewski, J.; Zwierzchowski, L.; Bagnicka, E. Cathelicidins: family of antimicrobial peptides. A review. Mol. Biol. Reports 2012, 39, 10957–10970. [Google Scholar] [CrossRef]
- Lehrer, R.I.; Ganz, T. Cathelicidins: A family of endogenous antimicrobial peptides. Curr. Opin. Hematol. 2002, 9, 18–22. [Google Scholar] [CrossRef]
- Cole, A.M.; Shi, J.; Ceccarelli, A.; Kim, Y.H.; Park, A.; Ganz, T. Inhibition of neutrophil elastase prevents cathelicidin activation and impairs clearance of bacteria from wounds. Blood 2001, 97, 297–304. [Google Scholar] [CrossRef]
- Tongaonkar, P.; Golji, A.E.; Tran, P.; Ouellette, A.J.; Selsted, M.E. High fidelity processing and activation of the human alpha-defensin HNP1 precursor by neutrophil elastase and proteinase 3. PloS One 2012, 7, e32469. [Google Scholar]
- Nizet, V.; Gallo, R.L. Cathelicidins and innate defense against invasive bacterial infection. Scand. J. Infect. Dis. 2003, 35, 670–676. [Google Scholar] [CrossRef]
- Zanetti, M.; Gennaro, R.; Scocchi, M.; Skerlavaj, B. Structure and biology of cathelicidins. Adv. Exp. Med. Biol. 2000, 479, 203–218. [Google Scholar]
- van Dijk, A.; Molhoek, E.M.; Bikker, F.J.; Yu, P.L.; Veldhuizen, E.J.; Haagsman, H.P. Avian cathelicidins: Paradigms for the development of anti-infectives. Vet. Microbiol. 2011, 153, 27–36. [Google Scholar] [CrossRef]
- Van Dijk, A.; Veldhuizen, E.J.; van Asten, A.J.; Haagsman, H.P. CMAP27, a novel chicken cathelicidin-like antimicrobial protein. Vet. Immunol. Immunopathol. 2005, 106, 321–327. [Google Scholar] [CrossRef]
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006, 281, 2858–2867. [Google Scholar]
- Van Dijk, A.; Tersteeg-Zijderveld, M.H.; Tjeerdsma-van Bokhoven, J.L.; Jansman, A.J.; Veldhuizen, E.J.; Haagsman, H.P. Chicken heterophils are recruited to the site of Salmonella infection and release antibacterial mature Cathelicidin-2 upon stimulation with LPS. Mol. Immunol. 2009, 46, 1517–1526. [Google Scholar] [CrossRef]
- Sorensen, O.E.; Borregaard, N. Cathelicidins--nature's attempt at combinatorial chemistry. Comb. Chem. High Throughput Screen. 2005, 8, 273–280. [Google Scholar] [CrossRef]
- De Latour, F.A.; Amer, L.S.; Papanstasiou, E.A.; Bishop, B.M.; van Hoek, M.L. Antimicrobial activity of the Naja atra cathelicidin and related small peptides. Biochem. Biophys. Res. Commun. 2010, 396, 825–830. [Google Scholar] [CrossRef]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Susceptibility of Pseudomonas aeruginosa Biofilm to Alpha-Helical Peptides: d-enantiomer of LL-37. Front. Microbiol. 2011, 2, 128. [Google Scholar]
- Amer, L.S.; Bishop, B.M.; van Hoek, M.L. Antimicrobial and antibiofilm activity of cathelicidins and short, synthetic peptides against Francisella. Biochem. Biophys. Res. Commun. 2010, 396, 246–251. [Google Scholar] [CrossRef]
- Dean, S.N.; Bishop, B.M.; van Hoek, M.L. Natural and synthetic cathelicidin peptides with anti-microbial and anti-biofilm activity against Staphylococcus aureus. BMC Microbiol. 2011, 11, 114. [Google Scholar] [CrossRef]
- Juba, M.; Porter, D.; Dean, S.; Gillmor, S.; Bishop, B. Characterization and performance of short cationic antimicrobial peptide isomers. Biopolymers 2013, 100, 387–401. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Z.; Chen, L.; Guang, H.; Li, Z.; Yang, H.; Li, J.; You, D.; Yu, H.; Lai, R. Cathelicidin-BF, a snake cathelicidin-derived antimicrobial peptide, could be an excellent therapeutic agent for acne vulgaris. PloS One 2011, 6, e22120. [Google Scholar]
- Hao, Q.; Wang, H.; Wang, J.; Dou, J.; Zhang, M.; Zhou, W.; Zhou, C. Effective antimicrobial activity of Cbf-K16 and Cbf-A7 A13 against NDM-1-carrying Escherichia coli by DNA binding after penetrating the cytoplasmic membrane in vitro. J. Pept. Sci. 2013, 19, 173–180. [Google Scholar] [CrossRef]
- Wang, J.; Li, B.; Li, Y.; Dou, J.; Hao, Q.; Tian, Y.; Wang, H.; Zhou, C. BF-30 effectively inhibits ciprofloxacin-resistant bacteria in vitro and in a rat model of vaginosis. Arch. Pharm. Res. 2013. [Google Scholar] [CrossRef]
- Dalla Valle, L.; Benato, F.; Paccanaro, M.C.; Alibardi, L. Bioinformatic and molecular characterization of cathelicidin-like peptides isolated from the green lizard Anolis carolinensis (Reptilia: Lepidosauria: Iguanidae). Ital. J. Zool. 2013, 80, 177–186. [Google Scholar] [CrossRef]
- Alibardi, L. Ultrastructural immunolocalization of chatelicidin-like peptides in granulocytes of normal and regenerating lizard tissues. Acta Histochem. 2013.
- Park, C.H.; Valore, E.V.; Waring, A.J.; Ganz, T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001, 276, 7806–7810. [Google Scholar]
- Rodriguez, R.; Jung, C.L.; Gabayan, V.; Deng, J.C.; Ganz, T.; Nemeth, E.; Bulut, Y. Hepcidin induction by pathogens and pathogen-derived molecules is strongly dependent on interleukin-6. Infect. Immun. 2014, 82, 745–752. [Google Scholar] [CrossRef]
- Hao, J.; Li, Y.W.; Xie, M.Q.; Li, A.X. Molecular cloning, recombinant expression and antibacterial activity analysis of hepcidin from Simensis crocodile (Crocodylus siamensis). Comp. Biochem. Phys. Part B 2012, 163, 309–315. [Google Scholar] [CrossRef]
- Hilton, K.B.; Lambert, L.A. Molecular evolution and characterization of hepcidin gene products in vertebrates. Gene 2008, 415, 40–48. [Google Scholar]
- Sang, Y.; Ramanathan, B.; Minton, J.E.; Ross, C.R.; Blecha, F. Porcine liver-expressed antimicrobial peptides, hepcidin and LEAP-2: cloning and induction by bacterial infection. Dev. Comp. Immunol. 2006, 30, 357–366. [Google Scholar] [CrossRef]
- Hocquellet, A.; Odaert, B.; Cabanne, C.; Noubhani, A.; Dieryck, W.; Joucla, G.; le Senechal, C.; Milenkov, M.; Chaignepain, S.; Schmitter, J.M.; et al. Structure-activity relationship of human liver-expressed antimicrobial peptide 2. Peptides 2010, 31, 58–66. [Google Scholar] [CrossRef]
- Henriques, S.T.; Tan, C.C.; Craik, D.J.; Clark, R.J. Structural and functional analysis of human liver-expressed antimicrobial peptide 2. Chembiochem 2010, 11, 2148–2157. [Google Scholar] [CrossRef]
- Krause, A.; Sillard, R.; Kleemeier, B.; Kluver, E.; Maronde, E.; Conejo-Garcia, J.R.; Forssmann, W.G.; Schulz-Knappe, P.; Nehls, M.C.; Wattler, F.; et al. Isolation and biochemical characterization of LEAP-2, a novel blood peptide expressed in the liver. Protein Sci. 2003, 12, 143–152. [Google Scholar] [CrossRef]
- Araki, T.; Yamamoto, T.; Torikata, T. Reptile lysozyme: the complete amino acid sequence of soft-shelled turtle lysozyme and its activity. Biosci. Biotechnol. Biochem. 1998, 62, 316–324. [Google Scholar] [CrossRef]
- Chijiiwa, Y.; Kawamura, S.; Torikata, T.; Araki, T. Amino acid sequence and activity of green turtle (Chelonia mydas) lysozyme. Protein J. 2006, 25, 336–344. [Google Scholar] [CrossRef]
- Prajanban, B.O.; Shawsuan, L.; Daduang, S.; Kommanee, J.; Roytrakul, S.; Dhiravisit, A.; Thammasirirak, S. Identification of five reptile egg whites protein using MALDI-TOF mass spectrometry and LC/MS-MS analysis. J. Proteomics 2012, 75, 1940–1959. [Google Scholar] [CrossRef]
- Radis-Baptista, G.; Kerkis, I. Crotamine, a small basic polypeptide myotoxin from rattlesnake venom with cell-penetrating properties. Curr. Pharm. Design 2011, 17, 4351–4361. [Google Scholar] [CrossRef]
- Kerkis, I.; Silva Fde, S.; Pereira, A.; Kerkis, A.; Radis-Baptista, G. Biological versatility of crotamine—A cationic peptide from the venom of a South American rattlesnake. Expert Opin. Investig. Drugs 2010, 19, 1515–1525. [Google Scholar] [CrossRef]
- Structure of crotamine, a Na+ channel affecting toxin from Crotalus durissus terrificus venom (PDB 1H5O). This image is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license. Available online: http://en.wikipedia.org/wiki/File:Crotamin_1H5O.png (accessed on 9 May 2014).
- Radis-Baptista, G.; Kubo, T.; Oguiura, N.; Prieto da Silva, A.R.; Hayashi, M.A.; Oliveira, E.B.; Yamane, T. Identification of crotasin, a crotamine-related gene of Crotalus durissus terrificus. Toxicon 2004, 43, 751–759. [Google Scholar] [CrossRef]
- Yamane, E.S.; Bizerra, F.C.; Oliveira, E.B.; Moreira, J.T.; Rajabi, M.; Nunes, G.L.; de Souza, A.O.; da Silva, I.D.; Yamane, T.; Karpel, R.L.; et al. Unraveling the antifungal activity of a South American rattlesnake toxin crotamine. Biochimie 2013, 95, 231–240. [Google Scholar]
- Yaraksa, N.; Anunthawan, T.; Theansungnoen, T.; Daduang, S.; Araki, T.; Dhiravisit, A.; Thammasirirak, S. Design and synthesis of cationic antibacterial peptide based on Leucrocin I sequence, antibacterial peptide from crocodile (Crocodylus siamensis) white blood cell extracts. J. Antibiot. 2013, 67, 205–212. [Google Scholar]
- Mak, P. Hemocidins in a functional and structural context of human antimicrobial peptides. Front. Biosci. 2008, 13, 6859–6871. [Google Scholar]
- Sheshadri, P.; Abraham, J. Antimicrobial properties of hemoglobin. Immunopharm. Immunotoxicol. 2012, 34, 896–900. [Google Scholar] [CrossRef]
- Parish, C.A.; Jiang, H.; Tokiwa, Y.; Berova, N.; Nakanishi, K.; McCabe, D.; Zuckerman, W.; Xia, M.M.; Gabay, J.E. Broad-spectrum antimicrobial activity of hemoglobin. Bioorg. Med. Chem. 2001, 9, 377–382. [Google Scholar]
- Anwised, P.; Kabbua, T.; Temsiripong, T.; Dhiravisit, A.; Jitrapakdee, S.; Araki, T.; Yoneda, K.; Thammasirirak, S. Molecular cloning and expression of alpha-globin and beta-globin genes from crocodile (Crocodylus siamensis). Protein J. 2013, 32, 172–182. [Google Scholar]
- Santamaria, C.; Larios, S.; Quiros, S.; Pizarro-Cerda, J.; Gorvel, J.P.; Lomonte, B.; Moreno, E. Bactericidal and antiendotoxic properties of short cationic peptides derived from a snake venom Lys49 phospholipase A2. Antimicrob. Agents Chemother. 2005, 49, 1340–1345. [Google Scholar] [CrossRef]
- Santamaria, C.; Larios, S.; Angulo, Y.; Pizarro-Cerda, J.; Gorvel, J.P.; Moreno, E.; Lomonte, B. Antimicrobial activity of myotoxic phospholipases A2 from crotalid snake venoms and synthetic peptide variants derived from their C-terminal region. Toxicon 2005, 45, 807–815. [Google Scholar] [CrossRef]
- Yu, H.Y.; Huang, K.C.; Yip, B.S.; Tu, C.H.; Chen, H.L.; Cheng, H.T.; Cheng, J.W. Rational design of tryptophan-rich antimicrobial peptides with enhanced antimicrobial activities and specificities. Chembiochem 2010, 11, 2273–2282. [Google Scholar] [CrossRef]
- Chu, H.L.; Yu, H.Y.; Yip, B.S.; Chih, Y.H.; Liang, C.W.; Cheng, H.T.; Cheng, J.W. Boosting salt resistance of short antimicrobial peptides. Antimicrob. Agents Chemother. 2013, 57, 4050–4052. [Google Scholar] [CrossRef]
- Deslouches, B.; Steckbeck, J.D.; Craigo, J.K.; Doi, Y.; Mietzner, T.A.; Montelaro, R.C. Rational design of engineered cationic antimicrobial peptides consisting exclusively of arginine and tryptophan, and their activity against multidrug-resistant pathogens. Antimicrob. Agents Chemother. 2013, 57, 2511–2521. [Google Scholar] [CrossRef]
- Gomes, V.M.; Carvalho, A.O.; Da Cunha, M.; Keller, M.N.; Bloch, C., Jr.; Deolindo, P.; Alves, E.W. Purification and characterization of a novel peptide with antifungal activity from Bothrops jararaca venom. Toxicon 2005, 45, 817–827. [Google Scholar] [CrossRef]
- Xie, J.P.; Yue, J.; Xiong, Y.L.; Wang, W.Y.; Yu, S.Q.; Wang, H.H. In vitro activities of small peptides from snake venom against clinical isolates of drug-resistant Mycobacterium tuberculosis. Int. J. Antimicrob. Agents 2003, 22, 172–174. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Van Hoek, M.L. Antimicrobial Peptides in Reptiles. Pharmaceuticals 2014, 7, 723-753. https://doi.org/10.3390/ph7060723
Van Hoek ML. Antimicrobial Peptides in Reptiles. Pharmaceuticals. 2014; 7(6):723-753. https://doi.org/10.3390/ph7060723
Chicago/Turabian StyleVan Hoek, Monique L. 2014. "Antimicrobial Peptides in Reptiles" Pharmaceuticals 7, no. 6: 723-753. https://doi.org/10.3390/ph7060723
APA StyleVan Hoek, M. L. (2014). Antimicrobial Peptides in Reptiles. Pharmaceuticals, 7(6), 723-753. https://doi.org/10.3390/ph7060723




