From Gene Networks to Therapeutics: A Causal Inference and Deep Learning Approach for Drug Discovery
Abstract
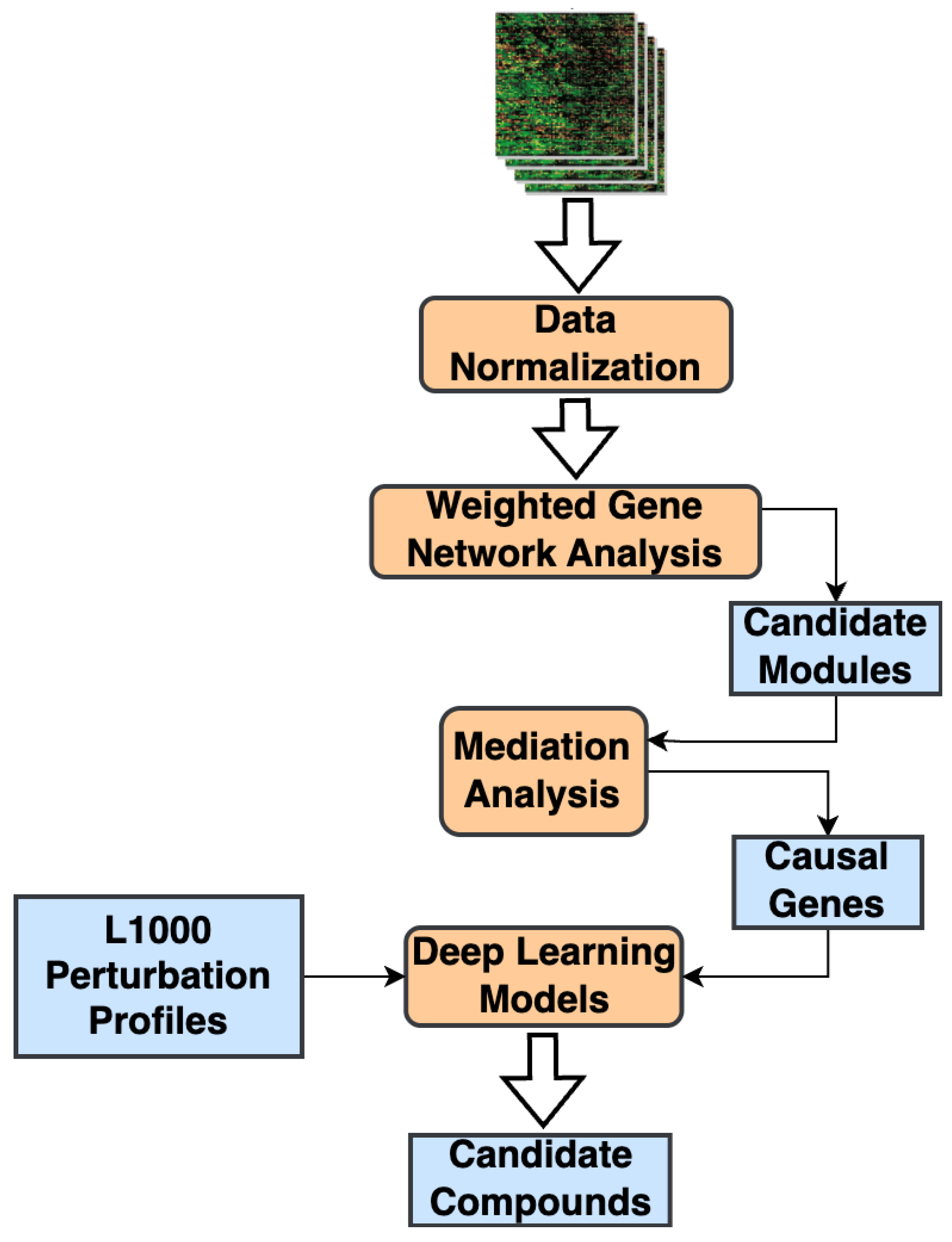
1. Introduction
2. Results
2.1. Transcriptomic Datasets
2.2. Correlated Gene Modules
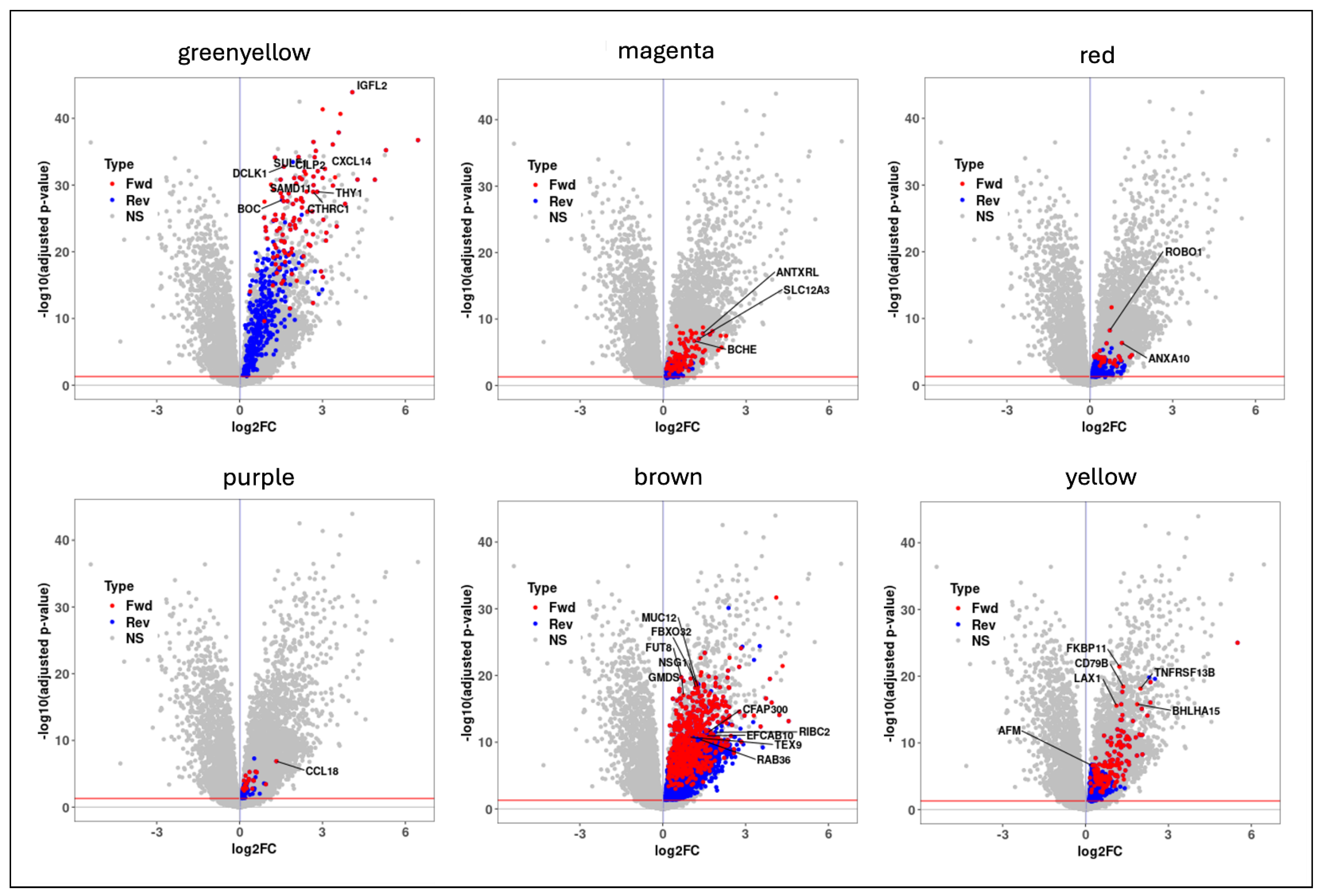
2.3. Candidate Causal Genes in IPF
2.4. Candidate Genes Enriched in Pro-Fibrotic Niches
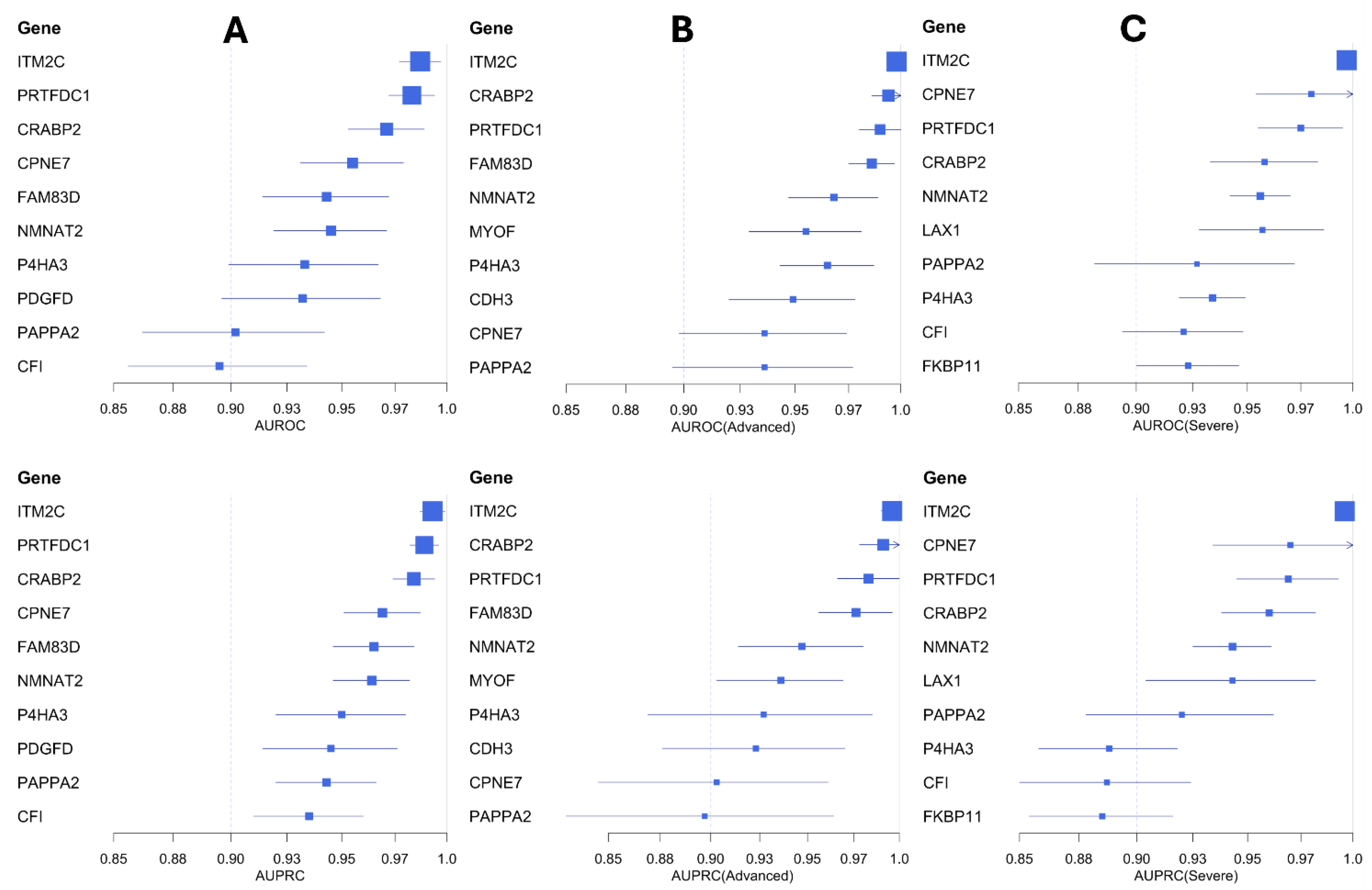
2.5. Mediator Genes Associated with IPF Severity
2.6. Biomarker Candidates in IPF
2.7. Small-Molecule Compounds Targeting the Causal Genes
2.8. Causal Candidate Genes and Small-Molecule Targets
3. Discussion
4. Materials and Methods
4.1. WGCNA and Candidate Correlated Modules
4.2. Mediation Analysis and Candidate Causal Genes
4.3. Machine Learning Models for Biomarker Analysis
4.4. DeepCE Model for Small-Molecule Screening
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sun, D.; Gao, W.; Hu, H.; Zhou, S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm. Sin. B 2022, 12, 3049–3062. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.H.; Qiu, Y.; Zeng, J.; Xie, L.; Zhang, P. A deep learning framework for high-throughput mechanism-driven phenotype compound screening and its application to COVID-19 drug repurposing. Nat. Mach. Intell. 2021, 3, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Ge, X.; Tan, H.; Xie, L.; Zhang, Y.; Hart, T.; Yang, X.; Bourne, P.E. Towards Structural Systems Pharmacology to Study Complex Diseases and Personalized Medicine. PLoS Comput. Biol. 2014, 10, e1003554. [Google Scholar] [CrossRef] [PubMed]
- Zhavoronkov, A.; Ivanenkov, Y.A.; Aliper, A.; Veselov, M.S.; Aladinskiy, V.A.; Aladinskaya, A.V.; Terentiev, V.A.; Polykovskiy, D.A.; Kuznetsov, M.D.; Asadulaev, A.; et al. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019, 37, 1038–1040. [Google Scholar] [CrossRef]
- Lounkine, E.; Keiser, M.J.; Whitebread, S.; Mikhailov, D.; Hamon, J.; Jenkins, J.L.; Lavan, P.; Weber, E.; Doak, A.K.; Côté, S.; et al. Large-scale prediction and testing of drug activity on side-effect targets. Nature 2012, 486, 361–367. [Google Scholar] [CrossRef]
- Ghandikota, S.K.; Jegga, A.G. Chapter Seven—Application of artificial intelligence and machine learning in drug repurposing. In Progress in Molecular Biology and Translational Science; Singh, V., Ed.; Academic Press: Cambridge, MA, USA, 2024; pp. 171–211. [Google Scholar]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Zhang, B.; Horvath, S. A General Framework for Weighted Gene Co-Expression Network Analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 17. [Google Scholar] [CrossRef]
- Liu, Y. CWGCNA: An R package to perform causal inference from the WGCNA framework. NAR Genom. Bioinform. 2024, 6, lqae042. [Google Scholar] [CrossRef]
- Porcu, E.; Sadler, M.C.; Lepik, K.; Auwerx, C.; Wood, A.R.; Weihs, A.; Sleiman, M.S.B.; Ribeiro, D.M.; Bandinelli, S.; Tanaka, T.; et al. Differentially expressed genes reflect disease-induced rather than disease-causing changes in the transcriptome. Nat. Commun. 2021, 12, 5647. [Google Scholar] [CrossRef]
- Kaur, A.; Mathai, S.K.; Schwartz, D.A. Genetics in Idiopathic Pulmonary Fibrosis Pathogenesis, Prognosis, and Treatment. Front. Med. 2017, 4, 154. [Google Scholar] [CrossRef]
- Maher, T.M. Interstitial Lung Disease: A Review. JAMA 2024, 331, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yella, J.K.; Ghandikota, S.; Cherukuri, T.C.; Ediga, H.H.; Madala, S.K.; Jegga, A.G. Pan-transcriptome-based candidate therapeutic discovery for idiopathic pulmonary fibrosis. Ther. Adv. Respir. Dis. 2020, 14, 1753466620971143. [Google Scholar] [CrossRef] [PubMed]
- Kolb, M.; Bonella, F.; Wollin, L. Therapeutic targets in idiopathic pulmonary fibrosis. Respir. Med. 2017, 131, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, H.; Cardwell, J.H.; Okamoto, T.; Walts, A.D.; Konigsberg, I.R.; Kurche, J.S.; Bang, T.J.; Schwarz, M.I.; Brown, K.K.; Kropski, J.A.; et al. Chronic Hypersensitivity Pneumonitis, an Interstitial Lung Disease with Distinct Molecular Signatures. Am. J. Respir. Crit. Care Med. 2020, 202, 1430–1444. [Google Scholar] [CrossRef]
- McDonough, J.E.; Ahangari, F.; Li, Q.; Jain, S.; Verleden, S.E.; Herazo-Maya, J.; Vukmirovic, M.; DeIuliis, G.; Tzouvelekis, A.; Tanabe, N.; et al. Transcriptional regulatory model of fibrosis progression in the human lung. JCI Insight 2019, 4, e131597. [Google Scholar] [CrossRef]
- Jaffar, J.; Wong, M.; Fishbein, G.A.; Alhamdoosh, M.; McMillan, L.; Gamell-Fulla, C.; Ng, M.; Wilson, N.; Symons, K.; Glaspole, I.; et al. Matrix metalloproteinase-7 is increased in lung bases but not apices in idiopathic pulmonary fibrosis. ERJ Open Res. 2022, 8, 00191–2022. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef]
- Subramanian, A.; Narayan, R.; Corsello, S.M.; Peck, D.D.; Natoli, T.E.; Lu, X.; Gould, J.; Davis, J.F.; Tubelli, A.A.; Asiedu, J.K.; et al. A Next Generation Connectivity Map: L1000 Platform and the First 1,000,000 Profiles. Cell 2017, 171, 1437–1452.e17. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Cannon, M.; Stevenson, J.; Stahl, K.; Basu, R.; Coffman, A.; Kiwala, S.; McMichael, J.F.; Kuzma, K.; Morrissey, D.; Cotto, K.; et al. DGIdb 5.0: Rebuilding the drug-gene interaction database for precision medicine and drug discovery platforms. Nucleic Acids Res. 2024, 52, D1227–D1235. [Google Scholar] [CrossRef]
- Corsello, S.M.; Bittker, J.A.; Liu, Z.; Gould, J.; McCarren, P.; Hirschman, J.E.; Johnston, S.E.; Vrcic, A.; Wong, B.; Khan, M.; et al. The Drug Repurposing Hub: A next-generation drug library and information resource. Nat. Med. 2017, 23, 405–408. [Google Scholar] [CrossRef] [PubMed]
- Buniello, A.; Suveges, D.; Cruz-Castillo, C.; Llinares, M.B.; Cornu, H.; Lopez, I.; Tsukanov, K.; Roldan-Romero, J.M.; Mehta, C.; Fumis, L.; et al. Open Targets Platform: Facilitating therapeutic hypotheses building in drug discovery. Nucleic Acids Res. 2025, 53, D1467–D1475. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.H.; Santacruz, D.; Jarosch, S.; Bleck, M.; Dalton, J.; McNabola, A.; Lempp, C.; Neubert, L.; Rath, B.; Kamp, J.C.; et al. Spatial transcriptomic characterization of pathologic niches in IPF. Sci. Adv. 2024, 10, eadl5473. [Google Scholar] [CrossRef] [PubMed]
- Ghandikota, S.; Sharma, M.; Ediga, H.H.; Madala, S.K.; Jegga, A.G. Consensus Gene Co-Expression Network Analysis Identifies Novel Genes Associated with Severity of Fibrotic Lung Disease. Int. J. Mol. Sci. 2022, 23, 5447. [Google Scholar] [CrossRef]
- Hanaka, T.; Kido, T.; Noguchi, S.; Yamada, S.; Noguchi, H.; Guo, X.; Nawata, A.; Wang, K.Y.; Oda, K.; Takaki, T.; et al. The overexpression of peroxiredoxin-4 affects the progression of idiopathic pulmonary fibrosis. BMC Pulm. Med. 2019, 19, 265. [Google Scholar] [CrossRef]
- Friedman, J.H.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Tay, J.K.; Narasimhan, B.; Hastie, T. Elastic Net Regularization Paths for All Generalized Linear Models. J. Stat. Softw. 2023, 106, 1–31. [Google Scholar] [CrossRef]
- Zhang, T.; Hou, Z.; Ding, Z.; Wang, P.; Pan, X.; Li, X. Single Cell RNA-Seq Identifies Cell Subpopulations Contributing to Idiopathic Pulmonary Fibrosis in Humans. J. Cell. Mol. Med. 2025, 29, e70402. [Google Scholar] [CrossRef]
- Prele, C.M.; Miles, T.; Pearce, D.R.; O’Donoghue, R.J.; Grainge, C.; Barrett, L.; Birnie, K.; Lucas, A.D.; Baltic, S.; Ernst, M.; et al. Plasma cell but not CD20-mediated B-cell depletion protects from bleomycin-induced lung fibrosis. Eur. Respir. J. 2022, 60, 2101469. [Google Scholar] [CrossRef]
- Cui, H.; Xie, N.; Jiang, D.; Banerjee, S.; Ge, J.; Sanders, Y.Y.; Liu, G. Inhibition of Glutaminase 1 Attenuates Experimental Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 2019, 61, 492–500. [Google Scholar] [CrossRef]
- Ge, J.; Cui, H.; Xie, N.; Banerjee, S.; Guo, S.; Dubey, S.; Barnes, S.; Liu, G. Glutaminolysis Promotes Collagen Translation and Stability via alpha-Ketoglutarate-mediated mTOR Activation and Proline Hydroxylation. Am. J. Respir. Cell Mol. Biol. 2018, 58, 378–390. [Google Scholar] [CrossRef]
- He, A.R.; Cohen, R.B.; Denlinger, C.S.; Sama, A.; Birnbaum, A.; Hwang, J.; Sato, T.; Lewis, N.; Mynderse, M.; Niland, M.; et al. First-in-Human Phase I Study of Merestinib, an Oral Multikinase Inhibitor, in Patients with Advanced Cancer. Oncologist 2019, 24, e930–e942. [Google Scholar] [CrossRef]
- Crestani, B.; Marchand-Adam, S.; Quesnel, C.; Plantier, L.; Borensztajn, K.; Marchal, J.; Mailleux, A.; Soler, P.; Dehoux, M. Hepatocyte growth factor and lung fibrosis. Proc. Am. Thorac. Soc. 2012, 9, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Stella, G.M.; Gentile, A.; Balderacchi, A.; Meloni, F.; Milan, M.; Benvenuti, S. Ockham’s razor for the MET-driven invasive growth linking idiopathic pulmonary fibrosis and cancer. J. Transl. Med. 2016, 14, 256. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stella, G.M.; D’Agnano, V.; Piloni, D.; Saracino, L.; Lettieri, S.; Mariani, F.; Lancia, A.; Bortolotto, C.; Rinaldi, P.; Falanga, F.; et al. The oncogenic landscape of the idiopathic pulmonary fibrosis: A narrative review. Transl. Lung Cancer Res. 2022, 11, 472–496. [Google Scholar] [CrossRef] [PubMed]
- El Awdan, S.A.; Abdel Rahman, R.F.; Ibrahim, H.M.; Hegazy, R.R.; El Marasy, S.A.; Badawi, M.; Arbid, M.S. Regression of fibrosis by cilostazol in a rat model of thioacetamide-induced liver fibrosis: Up regulation of hepatic cAMP, and modulation of inflammatory, oxidative stress and apoptotic biomarkers. PLoS ONE 2019, 14, e0216301. [Google Scholar] [CrossRef]
- Hada, Y.; Uchida, H.A.; Umebayashi, R.; Yoshida, M.; Wada, J. Cilostazol Attenuates AngII-Induced Cardiac Fibrosis in apoE Deficient Mice. Int. J. Mol. Sci. 2022, 23, 9065. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, X.; Hecker, L.; Kurundkar, D.; Kurundkar, A.; Liu, H.; Jin, T.-H.; Desai, L.; Bernard, K.; Thannickal, V.J. Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J. Clin. Investig. 2013, 123, 1096–1108. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem 2025 update. Nucleic Acids Res. 2025, 53, D1516–D1525. [Google Scholar] [CrossRef]
- Travaglini, K.J.; Nabhan, A.N.; Penland, L.; Sinha, R.; Gillich, A.; Sit, R.V.; Chang, S.; Conley, S.D.; Mori, Y.; Seita, J.; et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature 2020, 587, 619–625. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Bravo, À.; Queralt-Rosinach, N.; Gutiérrez-Sacristán, A.; Deu-Pons, J.; Centeno, E.; García-García, J.; Sanz, F.; Furlong, L.I. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017, 45, D833–D839. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37 (Suppl. 2), W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Langfelder, P.; Zhang, B.; Horvath, S. Defining clusters from A hierarchical cluster tree: The Dynamic Tree Cut package for R. Bioinformatics 2008, 24, 719–720. [Google Scholar] [CrossRef]
- VanderWeele, T.J. Mediation Analysis: A Practitioner’s Guide. Annu. Rev. Public Health 2016, 37, 17–32. [Google Scholar] [CrossRef]
- Weininger, D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988, 28, 31–36. [Google Scholar] [CrossRef]
- Grover, A.; Leskovec, J. node2vec: Scalable feature learning for networks. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 855–864. [Google Scholar]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]


| GEO Accession ID | # IPF | # Controls | Reference |
|---|---|---|---|
| GSE150910 | 103 | 103 | [15] |
| GSE124685 | 49 (19 = mild (IPF1); 16 = moderate (IPF2); 14 = advanced (IPF3)) | 35 | [16] |
| GSE213001 | 61 (22 = Advanced, 27 = Severe) | 40 | [17] |
| Gene | AUPRC | AUROC | |
|---|---|---|---|
| IPF vs. Controls | ITM2C | 0.006 | 0.01 |
| PRTFDC1 | 0.007 | 0.011 | |
| CRABP2 | 0.01 | 0.018 | |
| CPNE7 | 0.018 | 0.024 | |
| FAM83D | 0.018 | 0.029 | |
| NMNAT2 | 0.006 | 0.026 | |
| P4HA3 | 0.03 | 0.034 | |
| PDGFD | 0.031 | 0.036 | |
| PAPPA2 | 0.023 | 0.04 | |
| Severe IPF vs. Controls | ITM2C | 0.004 | 0.003 |
| CPNE7 | 0.036 | 0.026 | |
| PRTFDC1 | 0.024 | 0.02 | |
| CRABP2 | 0.022 | 0.025 | |
| NMNAT2 | 0.018 | 0.014 | |
| LAX1 | 0.039 | 0.029 | |
| PAPPA2 | 0.042 | 0.045 | |
| Advanced IPF vs. Controls | ITM2C | 0.006 | 0.004 |
| CRABP2 | 0.013 | 0.008 | |
| PRTFDC1 | 0.017 | 0.01 | |
| FAM83D | 0.02 | 0.011 | |
| NMNAT2 | 0.033 | 0.021 | |
| MYOF | 0.033 | 0.026 | |
| P4HA3 | 0.058 | 0.022 | |
| CDH3 | 0.047 | 0.029 | |
| CPNE7 | 0.058 | 0.038 |
| Drug | A375 | HA1E | HELA | HT29 | MCF7 | PC3 | YAPC |
|---|---|---|---|---|---|---|---|
| 1,4-Bis((3,4-dimethoxyphenyl)sulfonyl)-1,4-diazepane | −0.25 (0.051) | −0.27 (0.036) | −0.34 (0.006) | −0.28 (0.027) | −0.33 (0.009) | −0.25 (0.053) | −0.28 (0.03) |
| CB-839 (Telaglenastat) | −0.29 (0.023) | 0.04 (0.76) | −0.29 (0.02) | −0.26 (0.044) | −0.25 (0.049) | −0.28 (0.03) | −0.18 (0.16) |
| [2-(4-Amino-1,2,5-oxadiazol-3-yl)-1-ethylimidazo[4,5-c]pyridin-7-yl]-[(3S)-3-aminopyrrolidin-1-yl]methanone | −0.31 (0.014) | −0.18 (0.17) | −0.26 (0.041) | −0.32 (0.014) | −0.31 (0.015) | −0.29 (0.02) | −0.19 (0.13) |
| Aminofurazanyl-azabenzimidazole 6n | −0.35 (0.005) | −0.27 (0.03) | −0.3 (0.02) | −0.23 (0.07) | −0.29 (0.023) | −0.25 (0.053) | −0.19 (0.15) |
| [2-(4-Amino-furazan-3-yl)-1-ethyl-1H-imidazo[4,5-c]pyridin-7-ylmethyl]-piperidin-4-yl-amine | −0.28 (0.025) | −0.1 (0.42) | −0.25 (0.052) | −0.3 (0.02) | −0.28 (0.026) | −0.3 (0.02) | −0.09 (0.51) |
| Pentamidine | −0.31 (0.015) | −0.16 (0.24) | −0.21 (0.098) | −0.21 (0.11) | −0.26 (0.046) | −0.26 (0.044) | −0.27 (0.034) |
| RHC-80267 | −0.22 (0.089) | 0.08 (0.52) | −0.18 (0.16) | −0.35 (0.006) | −0.29 (0.023) | −0.23 (0.075) | −0.26 (0.039) |
| RK-682 | −0.27 (0.033) | 0.12 (0.35) | 0.019 (0.88) | 0.22 (0.097) | −0.18 (0.17) | −0.22 (0.091) | −0.29 (0.025) |
| Merestinib | −0.08 (0.54) | −0.09 (0.48) | −0.33 (0.008) | −0.06 (0.64) | −0.28 (0.03) | −0.07 (0.57) | −0.05 (0.73) |
| LY-255283 | −0.25 (0.053) | 0.17 (0.2) | −0.18 (0.16) | −0.26 (0.047) | −0.2 (0.12) | −0.28 (0.028) | −0.07 (0.58) |
| Cilostazol | −0.31 (0.015) | 0.018 (0.89) | −0.15 (0.25) | −0.16 (0.22) | −0.11 (0.41) | −0.06 (0.67) | −0.28 (0.033) |
| M2-PK-activator | −0.19 (0.15) | −0.26 (0.04) | −0.21 (0.11) | 0.25 (0.05) | −0.22 (0.089) | −0.23 (0.07) | −0.29 (0.02) |
| NNC-711 | −0.11 (0.4) | −0.03 (0.79) | −0.2 (0.11) | −0.3 (0.018) | −0.27 (0.038) | 0.1 (0.44) | 0.009 (0.95) |
| CID 11973736 | −0.23 (0.077) | −0.2 (0.12) | −0.33 (0.009) | −0.24 (0.06) | −0.32 (0.014) | −0.18 (0.16) | −0.07 (0.6) |
| Azeloyl diethyl salicylate | −0.15 (0.26) | 0.1 (0.45) | −0.34 (0.008) | −0.32 (0.011) | −0.16 (0.22) | −0.19 (0.13) | 0.02 (0.88) |
| Cetrimonium | −0.31 (0.015) | −0.04 (0.78) | −0.15 (0.26) | 0.23 (0.07) | −0.2 (0.13) | −0.28 (0.027) | −0.24 (0.06) |
| Decamethonium | −0.26 (0.043) | −0.04 (0.75) | −0.06 (0.65) | −0.26 (0.047) | −0.23 (0.072) | −0.23 (0.07) | −0.24 (0.056) |
| Gemcadiol | −0.29 (0.021) | −0.004 (0.98) | −0.06 (0.63) | −0.25 (0.049) | −0.14 (0.29) | −0.25 (0.056) | 0.24 (0.066) |
| Clofilium | −0.28 (0.026) | 0.23 (0.079) | −0.19 (0.14) | −0.25 (0.051) | −0.14 (0.29) | −0.27 (0.03) | −0.06 (0.64) |
| 1-[[4,5-Bis(4-methoxyphenyl)-2-thiazolyl]carbonyl]-4-methylpiperazine | −0.13 (0.33) | −0.15 (0.24) | −0.29 (0.025) | −0.17 (0.19) | −0.28 (0.029) | −0.17 (0.19) | 0.04 (0.78) |
| Zindotrine | −0.03 (0.82) | 0.17 (0.18) | 0.05 (0.68) | −0.28 (0.03) | −0.19 (0.15) | 0.04 (0.74) | −0.31 (0.015) |
| Eliglustat | −0.04 (0.78) | −0.01 (0.94) | −0.26 (0.04) | −0.27 (0.036) | −0.1 (0.44) | −0.16 (0.22) | −0.01 (0.93) |
| 3-(Azepan-1-ylsulfonyl)-N-(3-bromophenyl)benzamide | −0.02 (0.87) | −0.25 (0.054) | −0.26 (0.04) | −0.08 (0.54) | −0.11 (0.43) | −0.08 (0.53) | −0.25 (0.047) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghandikota, S.; Jegga, A.G. From Gene Networks to Therapeutics: A Causal Inference and Deep Learning Approach for Drug Discovery. Pharmaceuticals 2025, 18, 1304. https://doi.org/10.3390/ph18091304
Ghandikota S, Jegga AG. From Gene Networks to Therapeutics: A Causal Inference and Deep Learning Approach for Drug Discovery. Pharmaceuticals. 2025; 18(9):1304. https://doi.org/10.3390/ph18091304
Chicago/Turabian StyleGhandikota, Sudhir, and Anil G. Jegga. 2025. "From Gene Networks to Therapeutics: A Causal Inference and Deep Learning Approach for Drug Discovery" Pharmaceuticals 18, no. 9: 1304. https://doi.org/10.3390/ph18091304
APA StyleGhandikota, S., & Jegga, A. G. (2025). From Gene Networks to Therapeutics: A Causal Inference and Deep Learning Approach for Drug Discovery. Pharmaceuticals, 18(9), 1304. https://doi.org/10.3390/ph18091304






