Pharmacokinetic Equations Applied to Obtain New Topological Models in the Search of Antibacterial Compounds
Abstract
1. Introduction
2. Results and Discussion
N = 76 λ = 0.2653522 F = 38.76
N = 76 λ = 0.3138095 F = 25.146
N = 14 r2 = 0.85378 r2cv = 0.74026 SEE = 6.306649 F = 19.46
N = 16 r2 = 0.85816 r2cv = 0.75868 SEE = 0.694465 F = 24.20
N = 12 r2 = 0.92043 r2cv = 0.70282 SEE = 65.42953 F = 30.85
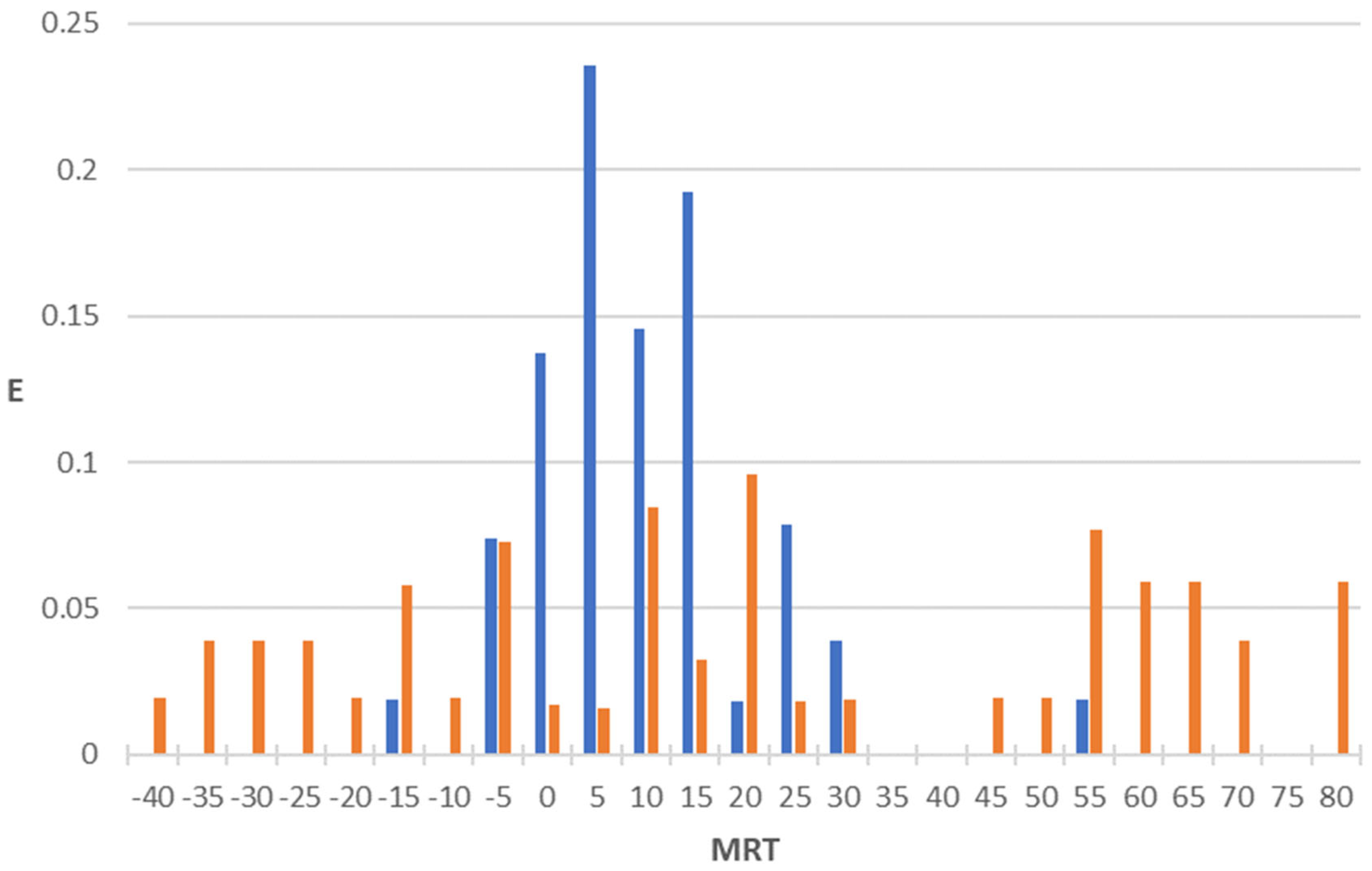
2.1. Antibacterial + Mean Residence Time Model (AB+MRT Model)
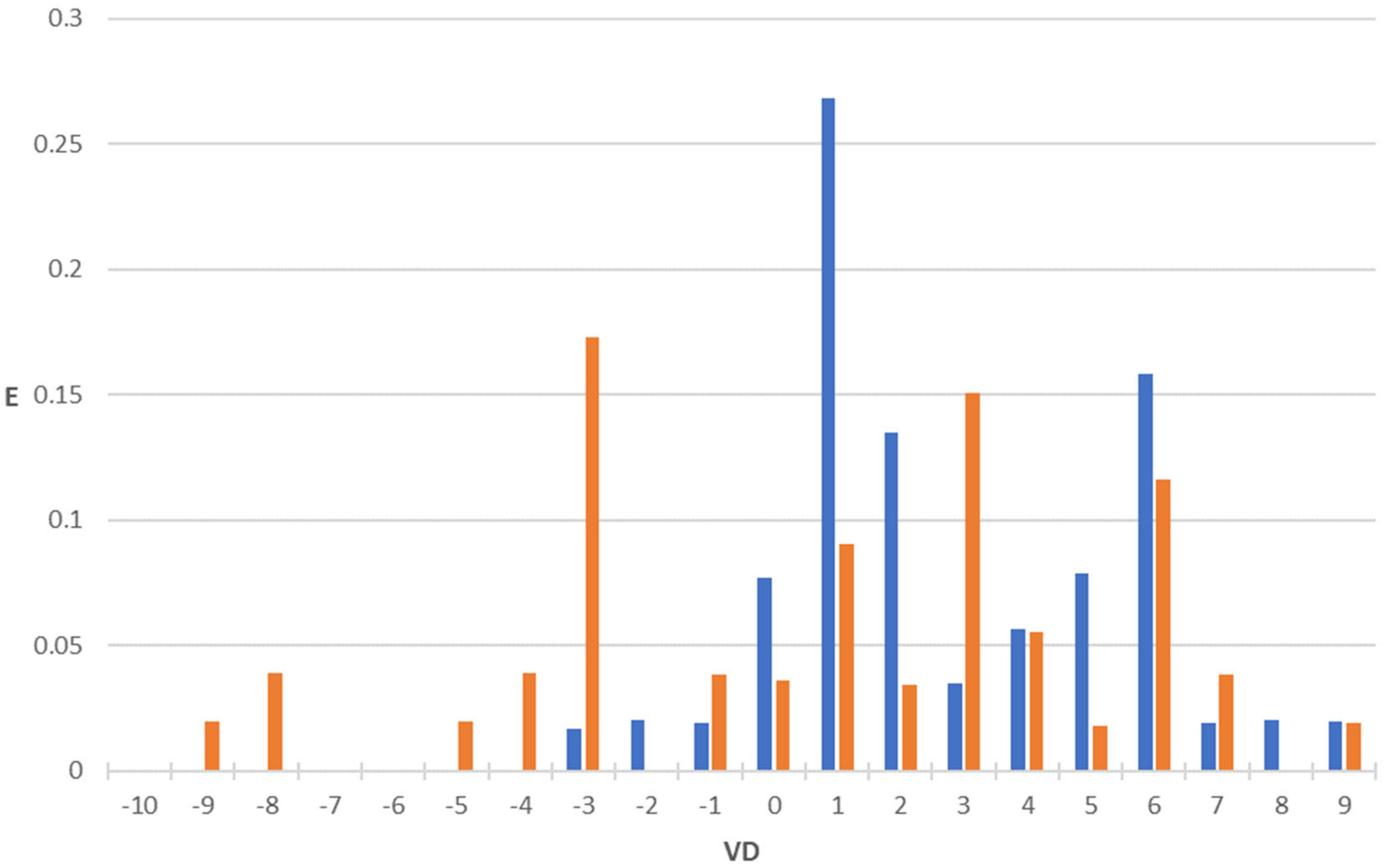
2.2. Antibacterial + Volume of Distribution Model (AB+VD Model)
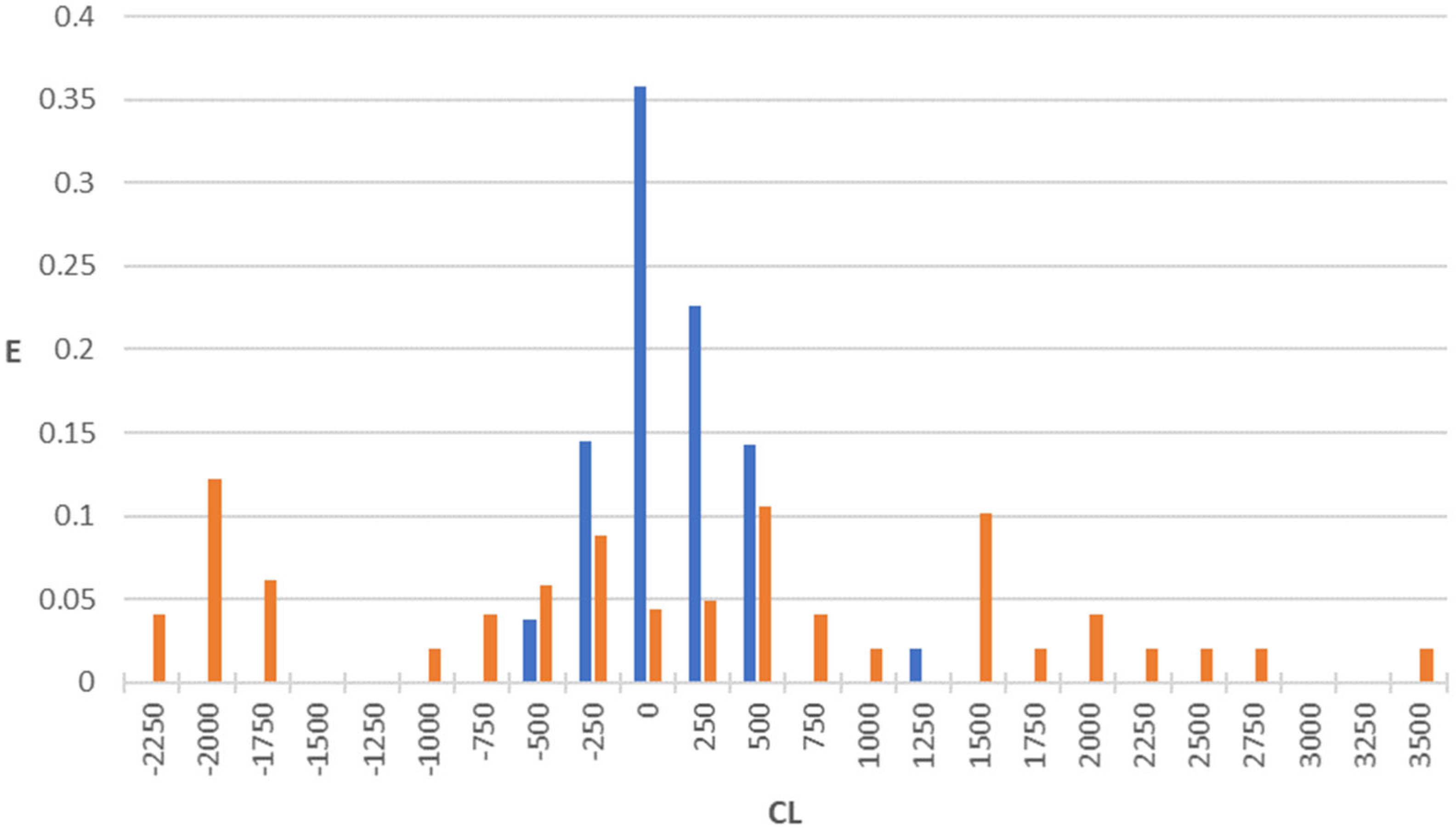
2.3. Antibacterial + Clearance Model (AB+CL Model)
2.4. Study of Drug-like Properties in Compounds Selected by Models
3. Materials and Methods
3.1. Compound Selection
3.2. Linear Discriminant Analysis and Multilinear Regression
3.3. Pharmacological Distribution Diagrams
3.4. Topological Models
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pagalilauan, G.L.; Limaye, A.P. Infections in transplant patients. Med. Clin. N. Am. 2013, 97, 581–600. [Google Scholar] [CrossRef] [PubMed]
- Pham, T.D.M.; Ziora, Z.M.; Blaskovich, M.A.T. Quinolone antibiotics. MedChemComm 2019, 10, 1719–1739. [Google Scholar] [CrossRef] [PubMed]
- Levy, S.B.; Marshall, B. Antibacterial resistance worldwide: Causes, challenges and responses. Nat. Med. 2004, 10, S122–S129. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.; Moreno-Morales, J.; Ballesté-Delpierre, C. Current landscape in the discovery of novel antibacterial agents. Clin. Microbiol. Infect. 2020, 26, 596–603. [Google Scholar] [CrossRef]
- World Health Organization. The WHO AWaRe (Access, Watch, Reserve) Antibiotic Book. 2022. Available online: https://apps.who.int/iris/bitstream/handle/10665/365237/9789240062382-eng.pdf (accessed on 20 April 2023).
- Alós, J.I. Antibiotic resistance: A global crisis. Enferm. Infecc. Microbiol. Clin. 2015, 33, 692–699. [Google Scholar] [CrossRef]
- Pushpakom, S.; Iorio, F.; Eyers, P.A.; Escott, K.J.; Hopper, S.; Wells, A.; Doig, A.; Guilliams, T.; Latimer, J.; McNamee, C.; et al. Drug repurposing: Progress, challenges and recommendations. Nat. Rev. Drug Discov. 2019, 18, 41–58. [Google Scholar] [CrossRef]
- Pinzi, L.; Bisi, N.; Rastelli, G. How drug repurposing can advance drug discovery: Challenges and opportunities. Front. Drug Discov. 2024, 4, 1460100. [Google Scholar] [CrossRef]
- Suay-Garcia, B.; Bueso-Bordils, J.I.; Falco, A.; Perez-Gracia, M.T.; Anton-Fos, G.; Aleman-Lopez, P. Quantitative structure-activity relationship methods in the discovery and development of antibacterials. WIREs Comput. Mol. Sci. 2020, 10, e1472. [Google Scholar] [CrossRef]
- Hopkins, A.L. Network pharmacology: The next paradigm in drug discovery. Nat. Chem. Biol. 2008, 4, 682–690. [Google Scholar] [CrossRef]
- Suay-García, B.; Bueso-Bordils, J.I.; Falcó, A.; Antón-Fos, G.M.; Alemán-López, P.A. Virtual combinatorial chemistry and pharmacological screening: A short guide to drug design. Int. J. Mol. Sci. 2022, 23, 1620. [Google Scholar] [CrossRef]
- Suay-Garcia, B.; Falcó, A.; Bueso-Bordils, J.I.; Anton-Fos, G.M.; Pérez-Gracia, M.T.; Alemán-López, P.A. Tree-based QSAR model for drug repurposing in the discovery of new antibacterial compounds against Escherichia coli. Pharm. 2020, 13, 431. [Google Scholar] [CrossRef] [PubMed]
- Bueso-Bordils, J.I.; Alemán-López, P.A.; Martín-Algarra, R.; Duart, M.J.; Falcó, A.; Antón-Fos, G.M. Molecular topology for the search of new anti-MRSA compounds. Int. J. Mol. Sci. 2021, 22, 5823. [Google Scholar] [CrossRef] [PubMed]
- Muratov, E.N.; Bajorath, J.; Sheridan, R.P.; Tetko, I.V.; Filimonov, D.; Poroikov, V.; Oprea, T.I.; Baskin, I.I.; Varnek, A.; Roitberg, A.; et al. QSAR without borders. Chem. Soc. Rev. 2020, 49, 3525–3564. [Google Scholar] [CrossRef]
- Farha, M.A.; Brown, E.D. Drug repurposing for antimicrobial discovery. Nat. Microbiol. 2019, 4, 565–577. [Google Scholar] [CrossRef]
- Hanser, T.; Barber, C.; Marchaland, J.F.; Werner, S. Applicability domain: Towards a more formal definition. SAR QSAR Environ. Res. 2016, 27, 865–881. [Google Scholar] [CrossRef]
- Organization for Economic Cooperation and Development. OECD Principles for the Validation, for Regulatory Purposes, of (Quantitative) Structure–Activity Relationship Models. Available online: https://www.oecd.org/chemicalsafety/risk-assessment/37849783.pdf (accessed on 26 May 2025).
- Bueso-Bordils, J.I.; Aleman, P.A.; Zamora, L.L.; Martin-Algarra, R.; Duart, M.J.; Antón-Fos, G.M. Topological model for the search of new antibacterial drugs. 158 theoretical candidates. Curr. Comput. Aided Drug Des. 2015, 11, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Bueso-Bordils, J.I.; Aleman-López, P.A.; Costa-Piles, S.; Duart, M.J.; Lahuerta-Zamora, L.; Martin-Algarra, R.; Anton-Fos, G.M. Obtaining microbiological and pharmacokinetic highly predictive equations. Curr. Top Med. Chem. 2018, 18, 908–916. [Google Scholar] [CrossRef]
- Bueso-Bordils, J.I.; Antón-Fos, G.M.; Falcó, A.; Duart, M.J.; Martín-Algarra, R.; Alemán-López, P.A. New pharmacokinetic and microbiological prediction equations to be used as models for the search of antibacterial drugs. Pharmaceuticals 2022, 15, 122. [Google Scholar] [CrossRef]
- Allen, D.M. The relationship between variable selection and data augmentation and a method for prediction. Technometrics 1974, 16, 125–127. [Google Scholar] [CrossRef]
- Levison, M.E.; Levison, J.H. Pharmacokinetics and pharmacodynamics of antibacterial agents. Infect. Dis. Clin. N. Am. 2009, 23, 791–815. [Google Scholar] [CrossRef]
- Adjei, M.D.; Deck, J.; Heinze, T.M.; Freeman, J.P.; Williams, A.J.; Sutherland, J.B. Identification of metabolites produced from N-phenylpiperazine by Mycobacterium spp. J. Ind. Microbiol. Biotechnol. 2007, 34, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xu, C.; Zhao, X.; Domagala, J.; Drlica, K. Fluoroquinolone action against mycobacteria: Effects of C-8 substituents on growth, survival, and resistance. Antimicrob. Agents Chemother. 1998, 42, 2978–2984. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scoper, S.V. Review of third-and fourth-generation fluoroquinolones in ophthalmology: In-vitro and in-vivo efficacy. Adv. Ther. 2008, 25, 979–994. [Google Scholar] [CrossRef]
- Bolon, M.K. The newer fluoroquinolones. Infect. Dis. Clin. N. Am. 2009, 23, 1027–1051. [Google Scholar] [CrossRef]
- Rusu, A.; Lungu, I.A.; Moldovan, O.L.; Tanase, C.; Hancu, G. Structural characterization of the millennial antibacterial (fluoro) quinolones-shaping the fifth generation. Pharmaceutics 2021, 13, 1289. [Google Scholar] [CrossRef]
- Cojutti, P.G.; Ramos-Martin, V.; Schiavon, I.; Rossi, P.; Baraldo, M.; Hope, W.; Pea, F. Population pharmacokinetics and pharmacodynamics of levofloxacin in acutely hospitalized older patients with various degrees of renal function. Antimicrob. Agents Chemother. 2017, 61, e02134-16. [Google Scholar] [CrossRef]
- Ball, P. Moxifloxacin (Avelox): An 8-methoxyquinolone antibacterial with enhanced potency. Int. J. Clin. Pract. 2000, 54, 329–332. [Google Scholar] [CrossRef]
- Brighty, K.E.; Gootz, T.D. The chemistry and biological profile of trovafloxacin. J. Antimicrob. Chemother. 1997, 39, 1–14. [Google Scholar] [CrossRef]
- Gai, X.; Shen, N.; He, B.; Zhou, Q.; Bo, S.; Li, X.; Zhai, S.; Yin, A.; Lu, W. Population pharmacokinetics of ciprofloxacin in Chinese elderly patients with lower respiratory tract infection. Zhonghua Yi Xue Za Zhi 2015, 95, 1581–1585. [Google Scholar]
- Arjona, A. Nemonoxacin quinolone antibiotic. Drug Future 2009, 34, 196–203. [Google Scholar]
- Butranova, O.I.; Ushkalova, E.A.; Zyryanov, S.K.; Chenkurov, M.S.; Baybulatova, E.A. Pharmacokinetics of antibacterial agents in the elderly: The body of evidence. Biomedicines 2023, 11, 1633. [Google Scholar] [CrossRef]
- Póvoa, P.; Moniz, P.; Pereira, J.G.; Coelho, L. Optimizing antimicrobial drug dosing in critically ill patients. Microorganisms 2021, 9, 1401. [Google Scholar] [CrossRef]
- Domagala, J.M. Structure-activity and structure-side-effect relationships for the quinolone antibacterials. J. Antimicrob. Chemother. 1994, 33, 685–706. [Google Scholar] [CrossRef]
- Gootz, T.D.; Brighty, K.E. Fluoroquinolone antibacterials: SAR, mechanism of action, resistance, and clinical aspects. Med. Res. Rev. 1996, 16, 433–486. [Google Scholar] [CrossRef]
- Di, L.; Kerns, E.H. Drug-like Properties Concepts, Structure Design and Methods from ADME to Toxicity Optimization, 2nd ed.; Academic Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Mauri, A.; Bertola, M. Alvascience: A new software suite for the QSAR workflow applied to the blood-brain barrier permeability. Int. J. Mol. Sci. 2022, 23, 12882. [Google Scholar] [CrossRef]
- Veber, D.F.; Johnson, S.R.; Cheng, H.Y.; Smith, B.R.; Ward, K.W.; Kopple, K.D. Molecular properties that influence the oral bioavailability of drug candidates. J. Med. Chem. 2002, 45, 2615–2623. [Google Scholar] [CrossRef]
- Viswanadhan, V.N.; Ghose, A.K.; Revankar, G.R.; Robins, R.K. Atomic physicochemical parameters for three dimensional structure directed quantitative structure-activity relationships. 4. Additional parameters for hydrophobic and dispersive interactions and their application for an automated superposition of certain naturally occurring nucleoside antibiotics. J. Chem. Inf. Comput. Sci. 1989, 29, 163–172. [Google Scholar]
- Ertl, P.; Rohde, B.; Selzer, P. Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J. Med. Chem. 2000, 43, 3714–3717. [Google Scholar] [CrossRef]
- Oprea, T.I.; Gottfries, J.; Sherbukhin, V.; Svensson, P.; Kühler, T.C. Chemical information management in drug discovery: Optimizing the computational and combinatorial chemistry interfaces. J. Mol. Graph. Model. 2000, 18, 512–524. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- O’Neil, M.J. The Merck Index, 13th ed.; Merck & Co. Inc.: Rahway, NJ, USA, 2001. [Google Scholar]
- DESMOL Software, version 13; Unidad de Investigación de Diseño de Fármacos y Conectividad Molecular, Facultad de Farmacia, Universitat de València: Valencia, Spain, 2000.
- Hall, L.H. MOLCONN-Z Software, version 3.0; Eastern Nazarene College: Quincy, MA, USA, 1995.
- Roy, K.; Kar, S.; Das, R.N. A Primer on QSAR/QSPR Modeling: Fundamental Concepts, 1st ed.; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Dixon, W.J. BMDP Statistical Software, version 7.0; University of California: Berkeley, CA, USA, 1990.
- CLSI. CLSI Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Second Informational Supplement; CLSI Document M100-S22; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2012; Volume 32. [Google Scholar]
- Hawkins, D.M. The problem of overfitting. J. Chem. Inf. Comput. Sci. 2004, 44, 1–12. [Google Scholar] [CrossRef]
- Duart, M.J.; García-Domenech, R.; Gálvez, J.; Alemán, P.A.; Martín-Algarra, R.V.; Antón-Fos, G.M. Application of a mathematical topological pattern of antihistaminic activity for the selection of new drug candidates and pharmacology assays. J. Med. Chem. 2006, 49, 3667–3673. [Google Scholar] [CrossRef]
- Balasubramaniyan, D.; Chidambaramb, N. On some neighbourhood degree-based topological indices with QSPR analysis of asthma drugs. Eur. Phys. J. Plus 2023, 138, 823. [Google Scholar] [CrossRef]
- García-García, Á.; Julián-Ortiz, J.V.; Gálvez, J.; Font, D.; Ayats, C.; Guna Serrano, M.D.R.; Muñoz-Collado, C.; Borrás, R.; Villalgordo, J.M. Similarity-Based Virtual Screening to Find Antituberculosis Agents Based on Novel Scaffolds: Design, Syntheses and Pharmacological Assays. Int. J. Mol. Sci. 2022, 23, 15057. [Google Scholar] [CrossRef]
- Gálvez, J.; García-Domenech, R.; de Gregorio Alapont, C.; de Julián-Ortiz, J.V.; Popa, L. Pharmacological distribution diagrams: A tool for de novo drug design. J. Mol. Graph. 1996, 14, 272–276. [Google Scholar] [CrossRef]
- Kier, L.B.; Hall, L.H. General definition of valence delta-values for molecular connectivity. J. Pharm. Sci. 1983, 72, 1170–1173. [Google Scholar] [CrossRef] [PubMed]
- Gálvez, J.; García-Domenech, R.; Salabert, M.T.; Soler, R. Charge indexes. New topological descriptors. J. Chem. Inf. Comput. Sci. 1994, 34, 520–525. [Google Scholar] [CrossRef]
- Kier, L.B.; Hall, L.H. The E-state as an extended free valence. J. Chem. Inf. Comput. Sci. 1997, 37, 548–552. [Google Scholar] [CrossRef]
- Basak, S.C.; Mills, D. Quantitative structure-property relationships (QSPRs) for the estimation of vapor pressure: A hierarchical approach using mathematical structural descriptors. J. Chem. Inf. Comput. Sci. 2001, 41, 692–701. [Google Scholar] [CrossRef]
- Shannon, C.E.; Weaver, W. The Mathematical Theory of Communication; University of Illinois Press: Champaign, IL, USA, 1949. [Google Scholar]
- Moliner, R.; Garcia, F.; Galvez, J.; Garcia-Domenech, R.; Serrano, C. Nuevos índices topológicos en conectividad molecular. Su aplicación a algunas propiedades fisicoquímicas de un grupo de hidrocarburos alifáticos. An. Real Acad. Farm. 1991, 57, 287–298. [Google Scholar]
- Cummins, D.J.; Andrews, C.W.; Bentley, J.A.; Cory, M. Molecular diversity in chemical databases: Comparison of medicinal chemistry knowledge bases and databases of commercially available compounds. J. Chem. Inf. Comput. Sci. 1996, 36, 750–763. [Google Scholar] [CrossRef] [PubMed]
- Bonchev, D.; Trinajstic, N. On topological characterization of molecular branching. Int. J. Quantum Chem. 1978, 14, 293–303. [Google Scholar] [CrossRef]
- Kier, L.B. A shape index from molecular graphs. Quant. Struct.-Act. Relat. 1985, 4, 109–116. [Google Scholar] [CrossRef]
- Kier, L.B. Distinguishing atom differences in a molecular graph shape index. Quant. Struct.-Act. Relat. 1986, 5, 7–12. [Google Scholar] [CrossRef]
- Kier, L.B. An index of molecular flexibility from Kappa shape attributes. Quant. Struct.-Act. Relat. 1989, 8, 221–224. [Google Scholar] [CrossRef]
- Suay-García, B.; Alemán-López, P.; Bueso-Bordils, J.I.; Falcó, A.; Pérez-Gracia, M.T.; Antón-Fos, G.M. Opological index Nclass as a factor determining the antibacterial activity of quinolones against Escherichia coli. Future Med. Chem. 2019, 11, 2255–2262. [Google Scholar] [CrossRef] [PubMed]
- Wiener, H. Structural determination of paraffin boiling points. J. Am. Chem. Soc. 1947, 69, 17–20. [Google Scholar] [CrossRef]
- Wiener, H. Relation of the physical properties of the isomeric alkanes to molecular structure. Surface tension, specific dispersion, and critical solution temperature in aniline. J. Phys. Chem. 1948, 52, 1082–1089. [Google Scholar] [CrossRef]
- Randic, M.; Guo, X.; Oxley, T.; Krishnapriyan, H.; Naylor, L. Wiener matrix invariants. J. Chem. Inf. Comput. Sci. 1994, 34, 361–367. [Google Scholar] [CrossRef]
- Niederfellner, J.; Lenoir, D.; Matuschek, G.; Rehfeldt, F.; Utschick, H.; Brügemann, R. Description of vapor pressures of polycyclic aromatic compounds by graph theoretical indices. Quant. Struct.-Act. Relat. 1997, 16, 38–48. [Google Scholar] [CrossRef]
- Platt, J.R. Influence of neighbor bonds on additive bond properties in paraffins. J. Chem. Phys. 1947, 15, 419–420. [Google Scholar] [CrossRef]
- Harras, M.F.; Sabour, R.; Farghaly, T.A.; Ibrahim, M.H. Drug repurposing approach in developing new furosemide analogs as antimicrobial candidates and anti-PBP: Design, synthesis, and molecular docking. Bioorg. Chem. 2023, 137, 106585. [Google Scholar] [CrossRef] [PubMed]
- Wien, R.; Harrison, J.; Freeman, W.A. Diamidines as antibacterial compounds. Brit. J. Pharmacol. 1948, 3, 211–218. [Google Scholar] [CrossRef]
- Szatlóczky, E. Cause, diagnosis, and chemotherapy of lactose intolerance. Br. Med. J. 1982, 284, 1405. [Google Scholar] [CrossRef]
- Wasi, N.; Singh, H.B. In vitro evaluation of bacteriostatic activity of metal complexes of amodiaquine and primaquine. Inorg. Chim. Acta 1988, 151, 287–289. [Google Scholar] [CrossRef]
- Bichowsky-Slomnitzki, L. The effect of aromatic diamidines on bacterial growth. J. Bacteriol. 1948, 55, 27–31. [Google Scholar] [CrossRef][Green Version]
- Piotrowski, W.; Sas-Piotrowska, B.; Slizak, W. Response of Rhizobacterium leguminosarum (strain B-73B) to some fungicides and antibiotics. Prog. Plant Prot. 1999, 39, 845–847. [Google Scholar]
- Kamath, A.; Ojima, I. Advances in the chemistry of β-lactam and its medicinal applications. Tetrahedron 2012, 68, 10640–10664. [Google Scholar] [CrossRef]
| DL Filter a | IM Dataset b | AB Model c | AB+MRT Model d | AB+VD Model e | AB+CL Model f |
|---|---|---|---|---|---|
| Ro5 ≥ 0.75 | 75.8 | 91.6 | 100 | 100 | 100 |
| 20 ≤ nAT ≤ 70 | 77.5 | 91.6 | 100 | 97.4 | 100 |
| 40 < AMR < 130 | 82.6 | 93.4 | 100 | 94.6 | 100 |
| TPSA < 140 | 78.9 | 82.8 | 89.9 | 97.4 | 94.9 |
| 1 ≤ RBN ≤ 9 | 81.7 | 85.3 | 100 | 92.1 | 94.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bueso-Bordils, J.I.; Antón-Fos, G.M.; Martín-Algarra, R.; Alemán-López, P.A. Pharmacokinetic Equations Applied to Obtain New Topological Models in the Search of Antibacterial Compounds. Pharmaceuticals 2025, 18, 865. https://doi.org/10.3390/ph18060865
Bueso-Bordils JI, Antón-Fos GM, Martín-Algarra R, Alemán-López PA. Pharmacokinetic Equations Applied to Obtain New Topological Models in the Search of Antibacterial Compounds. Pharmaceuticals. 2025; 18(6):865. https://doi.org/10.3390/ph18060865
Chicago/Turabian StyleBueso-Bordils, Jose I., Gerardo M. Antón-Fos, Rafael Martín-Algarra, and Pedro A. Alemán-López. 2025. "Pharmacokinetic Equations Applied to Obtain New Topological Models in the Search of Antibacterial Compounds" Pharmaceuticals 18, no. 6: 865. https://doi.org/10.3390/ph18060865
APA StyleBueso-Bordils, J. I., Antón-Fos, G. M., Martín-Algarra, R., & Alemán-López, P. A. (2025). Pharmacokinetic Equations Applied to Obtain New Topological Models in the Search of Antibacterial Compounds. Pharmaceuticals, 18(6), 865. https://doi.org/10.3390/ph18060865







