Exploring the Antibacterial Potential of Artemisia judaica Compounds Targeting the Hydrolase/Antibiotic Protein in Klebsiella pneumoniae: In Vitro and In Silico Investigations
Abstract
1. Introduction
2. Results
2.1. Protein and Phytochemical Preparation
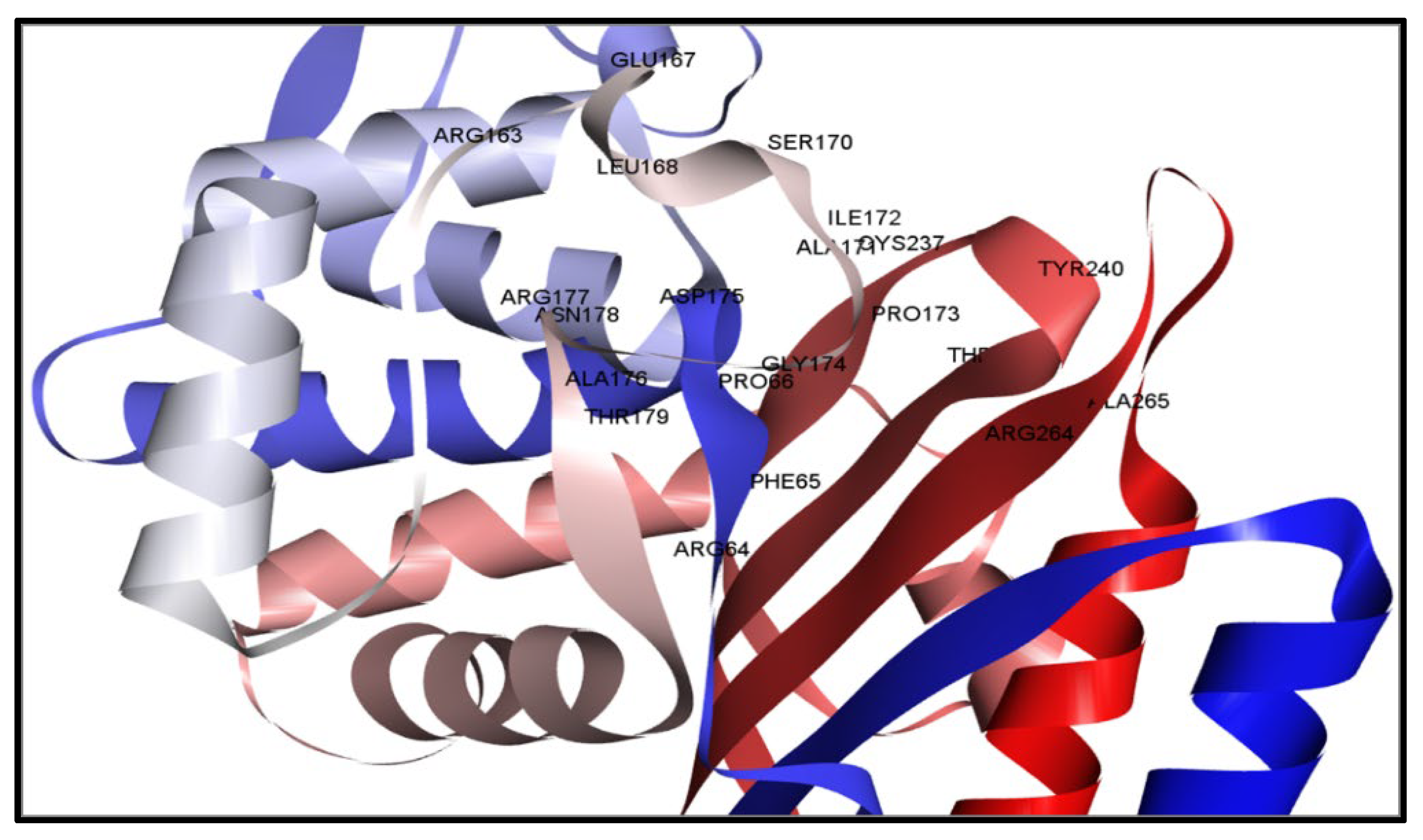
2.2. Active Position Detection and Generation of Receptor Grids
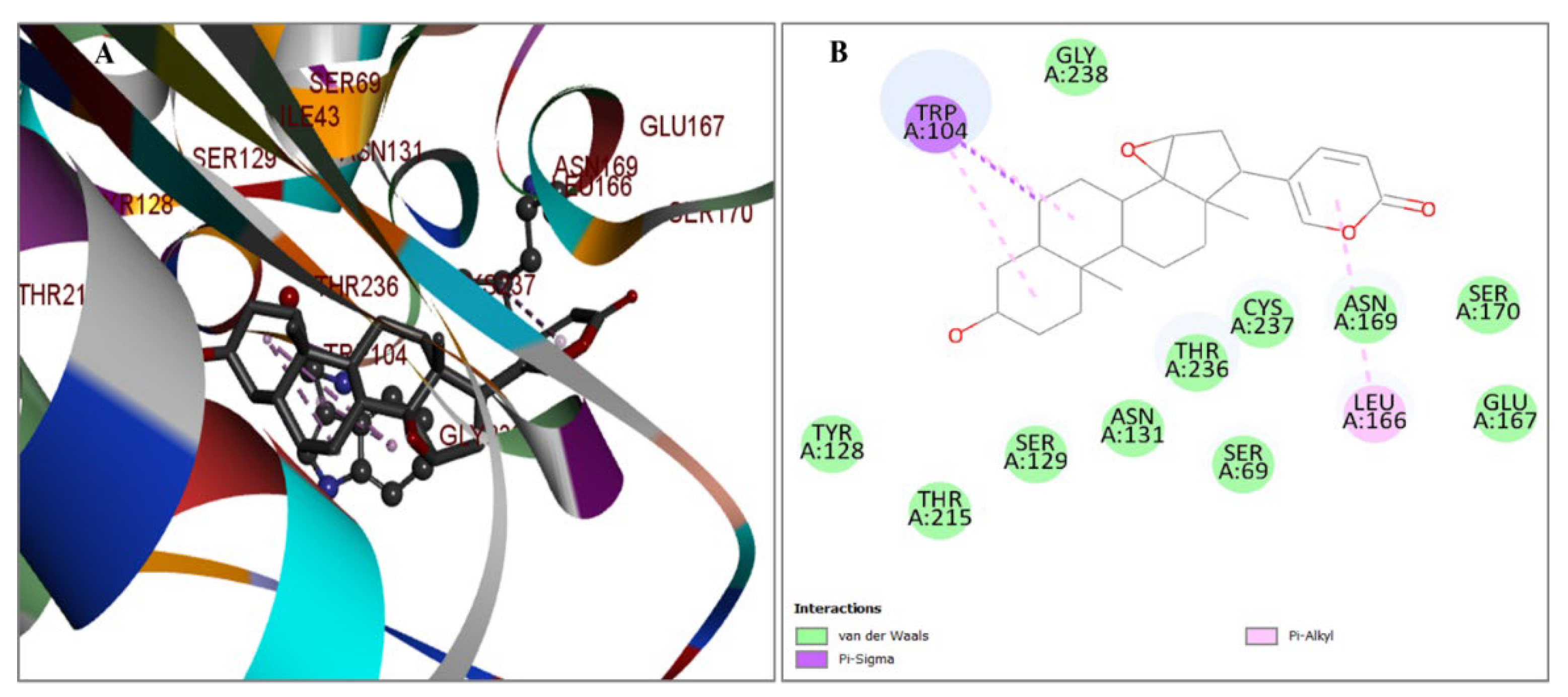
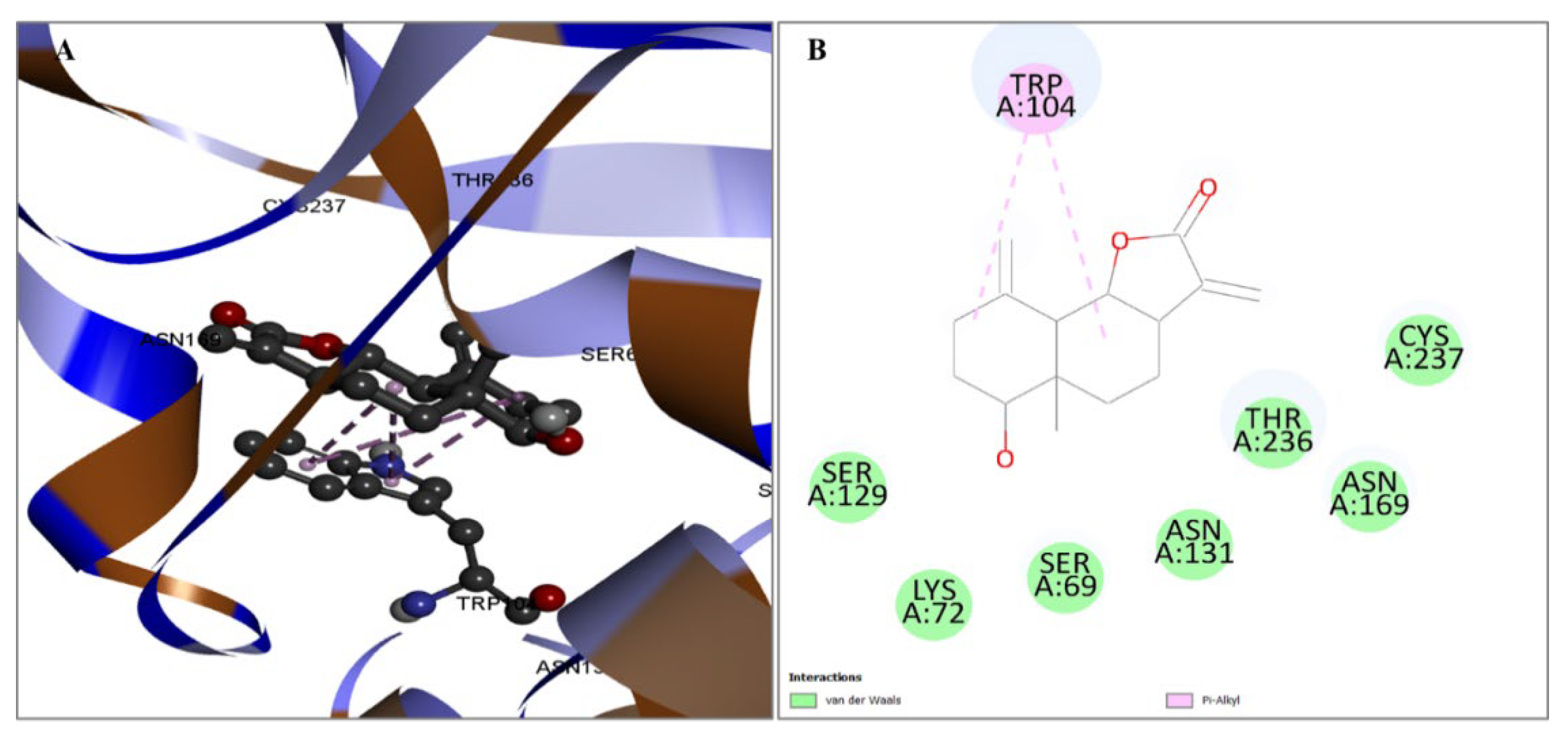
2.3. Evaluation of Protein and Ligand Interaction
2.4. ADME Calculation
2.5. Antibacterial Activity
2.5.1. MIC
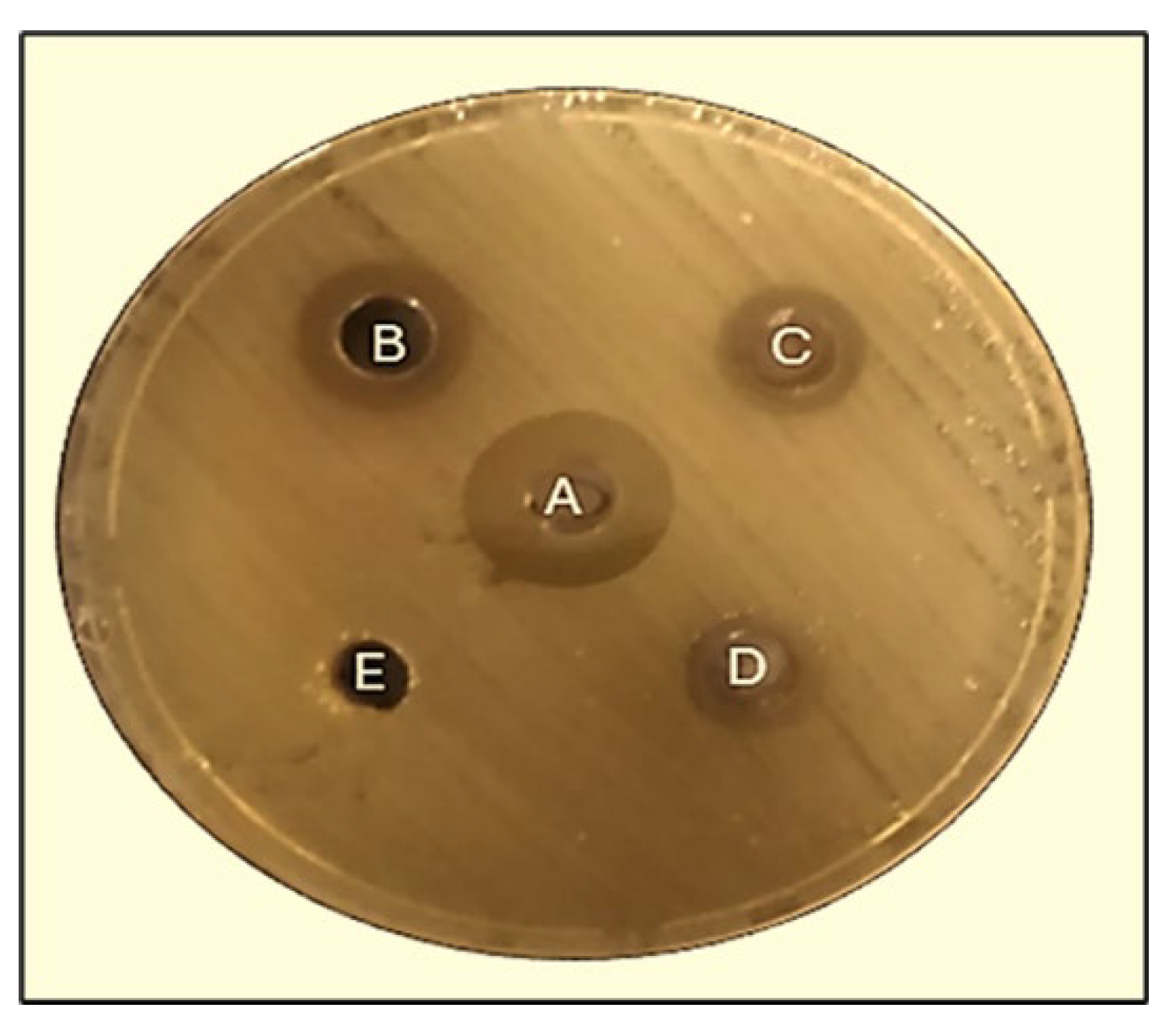
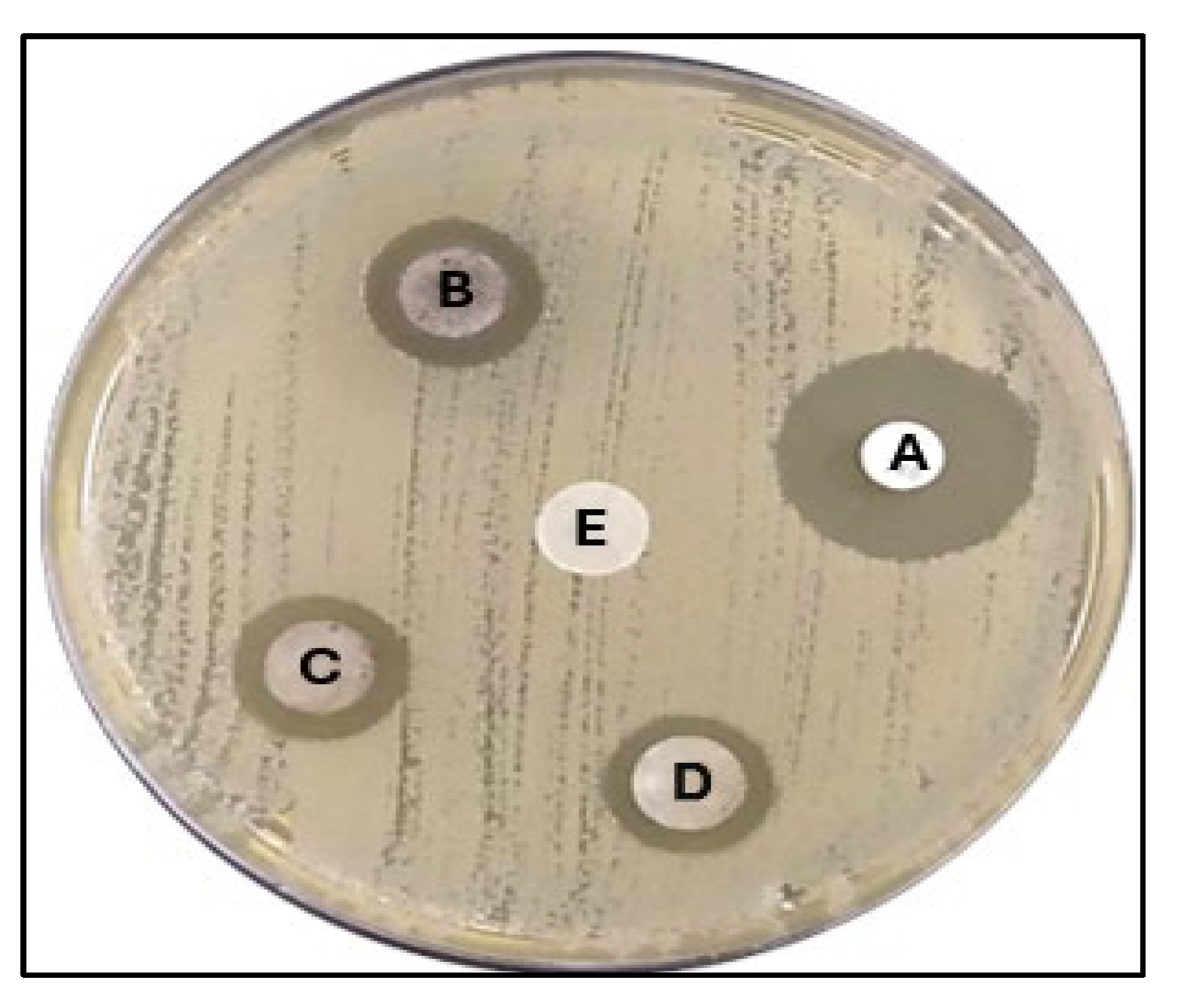
2.5.2. Well Diffusion Method
2.5.3. Disk Diffusion Method
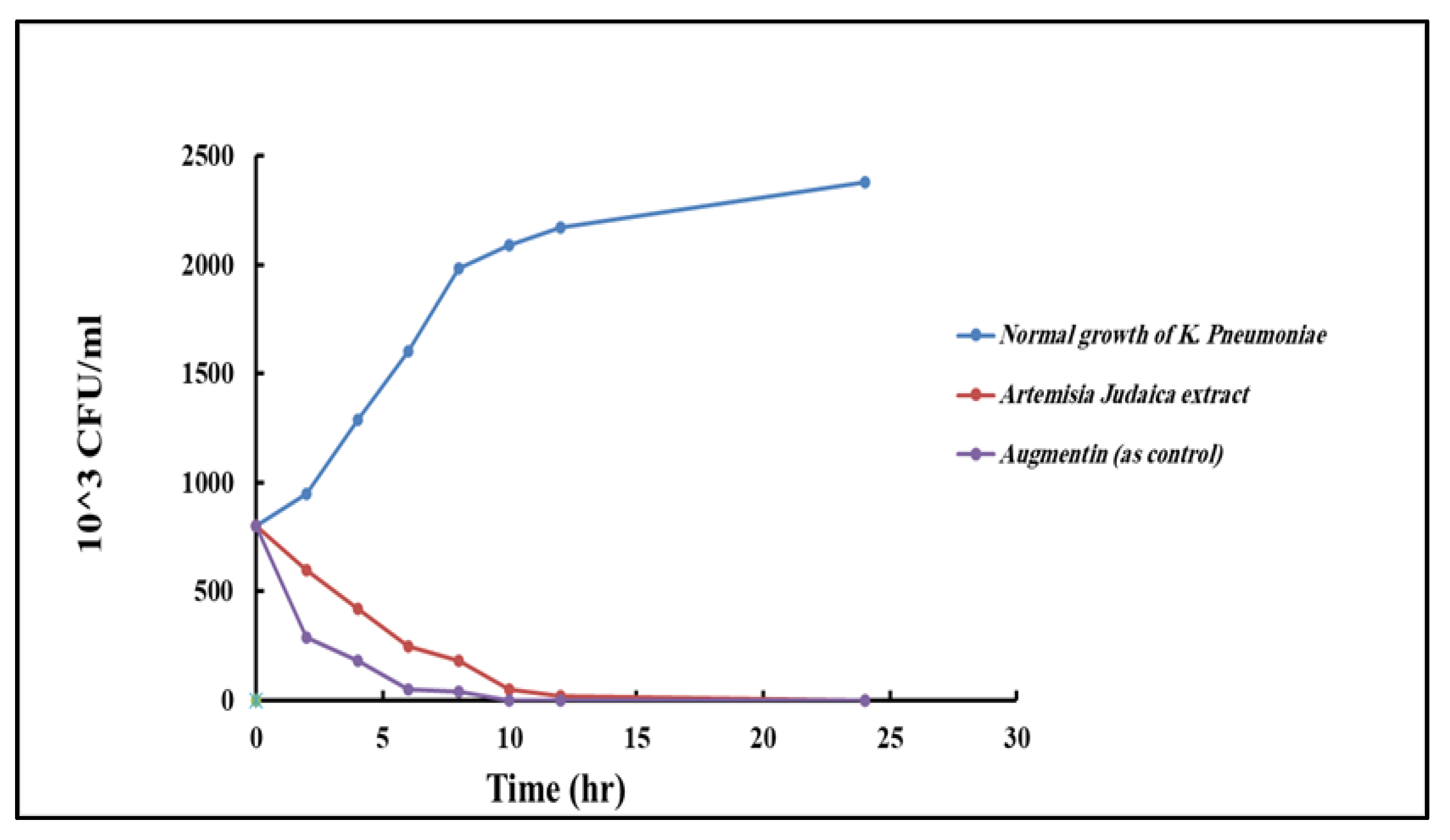
2.5.4. Time–Kill Curve
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.1.1. Preparation of Crude Extracts
4.1.2. GS/MS Analysis
4.2. Identifying and Preparing Proteins
4.3. Detection of Active Sites
4.4. Model Enhancement and Confirmation
4.5. Protein and Ligand Processing
4.6. Finding Protein Active Positions and Generating Receptor Grids
4.7. Simulation of Molecular Docking
4.8. ADME Analysis
4.9. Bacterial Strain
4.10. Antimicrobial Activity
4.10.1. Estimation of Minimum Inhibition Concentration (MIC)
4.10.2. Disk Diffusion Method
4.10.3. Well Diffusion Method
4.10.4. Time–Kill Curve
5. Conclusions
Supplementary Materials
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tumbarello, M.; Viale, P.; Viscoli, C.; Trecarichi, E.M.; Tumietto, F.; Marchese, A.; Spanu, T.; Ambretti, S.; Ginocchio, F.; Cristini, F. Predictors of mortality in bloodstream infections caused by Klebsiella pneumoniae carbapenemase–producing K. pneumoniae: Importance of combination therapy. Clin. Infect. Dis. 2012, 55, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Hobson, C.A.; Pierrat, G.; Tenaillon, O.; Bonacorsi, S.; Bercot, B.; Jaouen, E.; Jacquier, H.; Birgy, A. Klebsiella pneumoniae carbapenemase variants resistant to ceftazidime-avibactam: An evolutionary overview. Antimicrob. Agents Chemother. 2022, 66, e0044722. [Google Scholar] [CrossRef] [PubMed]
- Hansen, G.T. Continuous evolution: Perspective on the epidemiology of carbapenemase resistance among Enterobacterales and other Gram-negative bacteria. Infect. Dis. Ther. 2021, 10, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Yigit, H.; Queenan, A.M.; Anderson, G.J.; Domenech-Sanchez, A.; Biddle, J.W.; Steward, C.D.; Alberti, S.; Bush, K.; Tenover, F.C. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob. Agents Chemother. 2001, 45, 1151–1161. [Google Scholar] [CrossRef] [PubMed]
- Nordmann, P.; Poirel, L. Epidemiology and diagnostics of carbapenem resistance in gram-negative bacteria. Clin. Infect. Dis. 2019, 69, S521–S528. [Google Scholar] [CrossRef] [PubMed]
- Tooke, C.L.; Hinchliffe, P.; Bragginton, E.C.; Colenso, C.K.; Hirvonen, V.H.; Takebayashi, Y.; Spencer, J. β-Lactamases and β-Lactamase Inhibitors in the 21st Century. J. Mol. Biol. 2019, 431, 3472–3500. [Google Scholar] [CrossRef] [PubMed]
- Zhanel, G.G.; Lawson, C.D.; Adam, H.; Schweizer, F.; Zelenitsky, S.; Lagacé-Wiens, P.R.; Denisuik, A.; Rubinstein, E.; Gin, A.S.; Hoban, D.J. Ceftazidime-avibactam: A novel cephalosporin/β-lactamase inhibitor combination. Drugs 2013, 73, 159–177. [Google Scholar] [CrossRef] [PubMed]
- Docquier, J.-D.; Mangani, S. An update on β-lactamase inhibitor discovery and development. Drug Resist. Updates 2018, 36, 13–29. [Google Scholar] [CrossRef]
- Jubair, N.; Rajagopal, M.; Chinnappan, S.; Abdullah, N.B.; Fatima, A. Review on the antibacterial mechanism of plant-derived compounds against multidrug-resistant bacteria (MDR). Evid.-Based Complement. Altern. Med. 2021, 2021, 3663315. [Google Scholar] [CrossRef]
- Hrytsyk, R.A.; Kutsyk, R.V.; Yurchyshyn, O.I.; Struk, O.A.; Kireev, I.V.; Grytsyk, A.R. The investigation of antimicrobial and antifungal activity of some Artemisia L. species. Pharmacia 2021, 68, 93–100. [Google Scholar] [CrossRef]
- Adewumi, O.A.; Singh, V.; Singh, G. Chemical composition, traditional uses and biological activities of artemisia species. J. Pharmacogn. Phytochem. 2020, 9, 1124–1140. [Google Scholar]
- Dhifi, W.; Bellili, S.; Jazi, S.; Bahloul, N.; Mnif, W. Essential oils’ chemical characterization and investigation of some biological activities: A critical review. Medicines 2016, 3, 25. [Google Scholar] [CrossRef]
- Salih, A.M.; Qahtan, A.A.; Al-Qurainy, F. Phytochemicals identification and bioactive compounds estimation of Artemisia Species grown in Saudia Arabia. Metabolites 2023, 13, 443. [Google Scholar] [CrossRef] [PubMed]
- Nigam, M.; Atanassova, M.; Mishra, A.P.; Pezzani, R.; Devkota, H.P.; Plygun, S.; Salehi, B.; Setzer, W.N.; Sharifi-Rad, J. Bioactive compounds and health benefits of Artemisia species. Nat. Prod. Commun. 2019, 14, 1934578X19850354. [Google Scholar]
- Petretto, G.L.; Chessa, M.; Piana, A.; Masia, M.D.; Foddai, M.; Mangano, G.; Culeddu, N.; Afifi, F.U.; Pintore, G. Chemical and biological study on the essential oil of Artemisia caerulescens L. ssp. densiflora (Viv.). Nat. Prod. Res. 2013, 27, 1709–1715. [Google Scholar] [CrossRef]
- Ekiert, H.; Klimek-Szczykutowicz, M.; Rzepiela, A.; Klin, P.; Szopa, A. Artemisia species with high biological values as a potential source of medicinal and cosmetic raw materials. Molecules 2022, 27, 6427. [Google Scholar] [CrossRef]
- Moharram, F.A.; Nagy, M.M.; El Dib, R.A.; El-Tantawy, M.M.; El Hossary, G.G.; El-Hosari, D.G. Pharmacological activity and flavonoids constituents of Artemisia judaica L. aerial parts. J. Ethnopharmacol. 2021, 270, 113777. [Google Scholar] [CrossRef] [PubMed]
- Ncibi, S.S.; Aymen, M.M.; Oraiby, M.E.; Almalki, J.A.; Khadashi, H.A.; Mobarki, A.A. Antioxidant, Antibacterial and Cytotoxic Activities of Artemisia judaica, Ruta graveolens and Suaeda monoica from Saudi Arabia. Biomed. Pharmacol. J. 2023, 16, 1451–1462. [Google Scholar] [CrossRef]
- Wang, K.; Zhou, M.; Gong, X.; Zhou, Y.; Chen, J.; Ma, J.; Zhang, P. Starch–protein interaction effects on lipid metabolism and gut microbes in host. Front. Nutr. 2022, 9, 1018026. [Google Scholar] [CrossRef]
- Al-Wahaibi, L.H.N.; Mahmood, A.; Khan, M.; Alkhathlan, H.Z. Comparative study on the essential oils of Artemisia judaica and A. herba-alba from Saudi Arabia. Arab. J. Chem. 2020, 13, 2053–2065. [Google Scholar] [CrossRef]
- Malik, M.A.; Albeladi, S.S.; Al-Maaqar, S.M.; Alshehri, A.A.; Al-Thabaiti, S.A.; Khan, I.; Kamli, M.R. Biosynthesis of Novel Ag-Cu Bimetallic Nanoparticles from Leaf Extract of Salvia officinalis and Their Antibacterial Activity. Life 2023, 13, 653. [Google Scholar] [CrossRef] [PubMed]
- Qaeed, M.A.; Hendi, A.; Obaid, A.S.; Thahe, A.A.; Osman, A.M.; Ismail, A.; Mindil, A.; Eid, A.A.; Aqlan, F.; Osman, N.M. The effect of different aqueous solutions ratios of Ocimum basilicum utilized in AgNPs synthesis on the inhibition of bacterial growth. Sci. Rep. 2023, 13, 5866. [Google Scholar] [CrossRef] [PubMed]
- Qaeed, M.A.; Hendi, A.; Thahe, A.A.; Al-Maaqar, S.M.; Osman, A.M.; Ismail, A.; Mindil, A.; Eid, A.A.; Aqlan, F.; Al-Nahari, E. Effect of Different Ratios of Mentha spicata Aqueous Solution Based on a Biosolvent on the Synthesis of AgNPs for Inhibiting Bacteria. J. Nanomater. 2023, 2023, 3599501. [Google Scholar] [CrossRef]
- Qanash, H.; Bazaid, A.S.; Aldarhami, A.; Alharbi, B.; Almashjary, M.N.; Hazzazi, M.S.; Felemban, H.R.; Abdelghany, T.M. Phytochemical characterization and efficacy of Artemisia judaica extract loaded chitosan nanoparticles as inhibitors of cancer proliferation and microbial growth. Polymers 2023, 15, 391. [Google Scholar] [CrossRef] [PubMed]
- Allam, H.; Benamar, H.; Ben Mansour, R.; Ksouri, R.; Bennaceur, M. Phenolic composition, antioxidant, and antibacterial activities of Artemisia judaica subsp. sahariensis. J. Herbs Spices Med. Plants 2019, 25, 347–362. [Google Scholar] [CrossRef]
- Brogi, S.; Ramalho, T.C.; Kuca, K.; Medina-Franco, J.L.; Valko, M. In silico methods for drug design and discovery. Front. Chem. 2020, 8, 612. [Google Scholar] [CrossRef] [PubMed]
- Alsenani, T.; Viviani, S.; Papp-Wallace, K.; Bonomo, R.; van den Akker, F. Exploring avibactam and relebactam inhibition of Klebsiella pneumoniae carbapenemase D179N variant: Role of the Ω loop-held deacylation water. Antimicrob. Agents Chemother. 2023, 67, e0035023. [Google Scholar] [CrossRef]
- Li, B.; Wang, W.; Zhao, L.; Li, M.; Yan, D.; Li, X.; Zhang, J.; Gao, Q.; Feng, Y.; Zheng, J. Aggregation-induced emission-based macrophage-like nanoparticles for targeted photothermal therapy and virus transmission blockage in Monkeypox. Adv. Mater. 2024, 36, 2305378. [Google Scholar] [CrossRef]
- Joshi, G.; Sindhu, J.; Thakur, S.; Rana, A.; Sharma, G.; Poduri, R. Recent efforts for drug identification from phytochemicals against SARS-CoV-2: Exploration of the chemical space to identify druggable leads. Food Chem. Toxicol. 2021, 152, 112160. [Google Scholar] [CrossRef]
- Egieyeh, S.A.; Oselusi, S.O.; Christoffels, A. Cheminformatic Characterization of Natural Antimicrob al Products for the Development of New Lead Compounds. 2021. Available online: https://repository.uwc.ac.za/handle/10566/6585 (accessed on 13 May 2024).
- El-Amier, Y.A.; Al Borki, A.E.-N.S.; Elagami, S.A. Potential of wild plant Artemisia judaica L. as sustainable source of antioxidant and antimicrobial compounds. J. Exp. Sci. 2019, 10, 4–8. [Google Scholar] [CrossRef]
- Mohammed, S.; Dekabo, A.; Hailu, T. Phytochemical analysis and anti-microbial activities of Artemisia spp. and rapid isolation methods of artemisinin. AMB Express 2022, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Younes, K.M.; Bin, M.K.; Unissa, R.; Almarshdi, A.A.; Alharbi, F.M.; Alenzi, S.S.; Albsher, B.N.; Abouzied, A.S. In-Vitro Evaluation of Anti-Microbial and Cytotoxic Activity of Artemisia judaica Leaves and Stem Extracts via Induction of Caspase Dependent Apoptosis. Indian J. Pharm. Educ. Res 2022, 56, s52–s57. [Google Scholar] [CrossRef]
- Rouhi, S.; Ramazanzadeh, R.; Mohammadi, S.; Abodollahi, A.; Shakib, P.; Mohammadi, B.; Ahmadi, A. Antibacterial effects of Artemisa aucheri leaf and Spirulina Blue-Green algae aqueous and alcoholic extracts on the multidrug-resistant Klebsiella pneumoniae isolated from the patients with pneumonia. Sci. J. Kurd. Univ. Med. Sci. 2020, 25, 124–139. [Google Scholar] [CrossRef]
- Gheith, I.; El-Mahmoudy, A. Assessment of the antimicrobial potential of the hydro-methanolic extract of Sidr (Ziziphus spina-christi) plant against selected pathogens in vitro. Life Sci. J. 2018, 15, 27–34. [Google Scholar]
- Abubakar, A.R.; Haque, M. Preparation of medicinal plants: Basic extraction and fractionation procedures for experimental purposes. J. Pharm. Bioallied Sci. 2020, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Makhawi, A.M.; Mustafa, M.I.; Uagoub, H.A. Phytochemical Screening and Antimicrobial Activity of Ziziphus spina-christi Stem Barks. bioRxiv 2020. bioRxiv:2020.02.24.963157. [Google Scholar]
- Kızıl, G.; Kızıl, M.; Yavuz, M.; Emen, S.; Hakimoğlu, F. Antioxidant Activities of Ethanol Extracts of Hypericum triquetrifolium. and Hypericum scabroides. Pharm. Biol. 2008, 46, 231–242. [Google Scholar] [CrossRef]
- Abdul Qadir, M.; Shahzadi, S.K.; Bashir, A.; Munir, A.; Shahzad, S. Evaluation of phenolic compounds and antioxidant and antimicrobial activities of some common herbs. Int. J. Anal. Chem. 2017, 2017, 3475738. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Kareem, M.S.; Rabbih, M.A.E.F.; Selim, E.T.M.; Elsherbiny, E.A.E.-m.; El-Khateeb, A.Y. Application of GC/EIMS in combination with semi-empirical calculations for identification and investigation of some volatile components in basil essential oil. Int. J. Anal. Mass Spectrom. Chromatogr. 2016, 4, 14–25. [Google Scholar] [CrossRef]
- Billones, J.B.; Carrillo, M.C.O.; Organo, V.G.; Macalino, S.J.Y.; Sy, J.B.A.; Emnacen, I.A.; Clavio, N.A.B.; Concepcion, G.P. Toward antituberculosis drugs: In silico screening of synthetic compounds against Mycobacterium tuberculosis L,D-transpeptidase 2. Drug Des. Dev. Ther. 2016, 10, 1147–1157. [Google Scholar] [CrossRef]
- Audat, S.A.; Al-Shar’i, N.A.; Al-Oudat, B.A.; Bryant-Friedrich, A.; Bedi, M.F.; Zayed, A.L.; Al-Balas, Q.A. Identification of human leukotriene A4 hydrolase inhibitors using structure-based pharmacophore modeling and molecular docking. Molecules 2020, 25, 2871. [Google Scholar] [CrossRef] [PubMed]
- Heo, L.; Park, H.; Seok, C. GalaxyRefine: Protein structure refinement driven by side-chain repacking. Nucleic Acids Res. 2013, 41, W384–W388. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Wu, F.; Hu, J.; Wang, W.; Zhang, J.; Guo, G. Structural investigation of the interaction mechanism between chlorogenic acid and AMPA receptor via in silico approaches. Molecules 2022, 27, 3394. [Google Scholar] [CrossRef] [PubMed]
- Sumon, M.A.A.; Asseri, A.H.; Molla, M.H.R.; Aljahdali, M.O.; Hasan, M.R.; Rahman, M.A.; Hasan, M.T.; Sumon, T.A.; Gabr, M.H.; Islam, M.S. Identification of natural antiviral drug candidates against Tilapia Lake Virus: Computational drug design approaches. PLoS ONE 2023, 18, e0287944. [Google Scholar] [CrossRef] [PubMed]
- Molla, M.H.R.; Aljahdali, M.O.; Sumon, M.A.A.; Asseri, A.H.; Altayb, H.N.; Islam, M.S.; Alsaiari, A.A.; Opo, F.D.M.; Jahan, N.; Ahammad, F. Integrative ligand-based pharmacophore modeling, virtual screening, and molecular docking simulation approaches identified potential lead compounds against pancreatic cancer by targeting FAK1. Pharmaceuticals 2023, 16, 120. [Google Scholar] [CrossRef] [PubMed]
- Mun, C.S.; Hui, L.Y.; Sing, L.C.; Karunakaran, R.; Ravichandran, V. Multi-targeted molecular docking, pharmacokinetics, and drug-likeness evaluation of coumarin based compounds targeting proteins involved in development of COVID-19. Saudi J. Biol. Sci. 2022, 29, 103458. [Google Scholar] [CrossRef] [PubMed]
- Aljahdali, M.O.; Molla, M.H.R.; Ahammad, F. Compounds identified from marine mangrove plant (Avicennia alba) as potential antiviral drug candidates against WDSV, an in-silico approach. Mar. Drugs 2021, 19, 253. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhou, Y.; Li, L.; Shen, X.; Chen, G.; Wang, X.; Liang, X.; Tan, M.; Huang, Z. Computational approaches in preclinical studies on drug discovery and development. Front. Chem. 2020, 8, 726. [Google Scholar] [CrossRef] [PubMed]
- Tibbitts, J.; Canter, D.; Graff, R.; Smith, A.; Khawli, L.A. Key factors influencing ADME properties of therapeutic proteins: A need for ADME characterization in drug discovery and development. MAbs 2016, 8, 229–245. [Google Scholar] [CrossRef]
- Shou, W.Z. Current status and future directions of high-throughput ADME screening in drug discovery. J. Pharm. Anal. 2020, 10, 201–208. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed]
- Al-Maaqar, S.M.; Al-Johny, B.O.; Al-Sharif, F.D.; Al-Shaeri, M.; AL-Kenani, N.; Hussain, M.B. Sensitivity of multidrug-resistant pathogenic bacteria to ethanolic extract of Ziziphus spina-christi L. (Sidr) leaves. Biosci. Res. 2022, 19, 1467–1475. [Google Scholar]
- Monica, C. District Laboratory Practice in Tropical Countries; Cambridge University Press: Cambridge, UK, 2006. [Google Scholar]

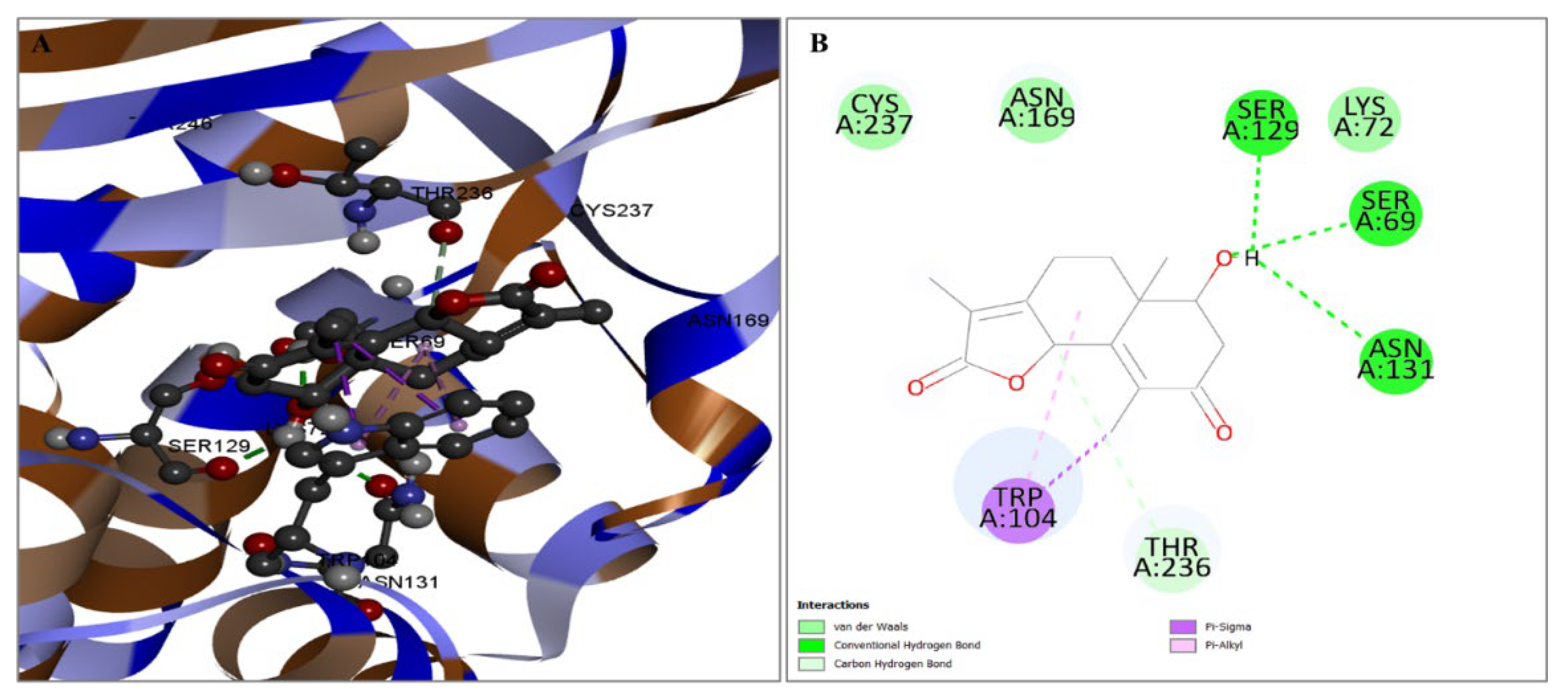
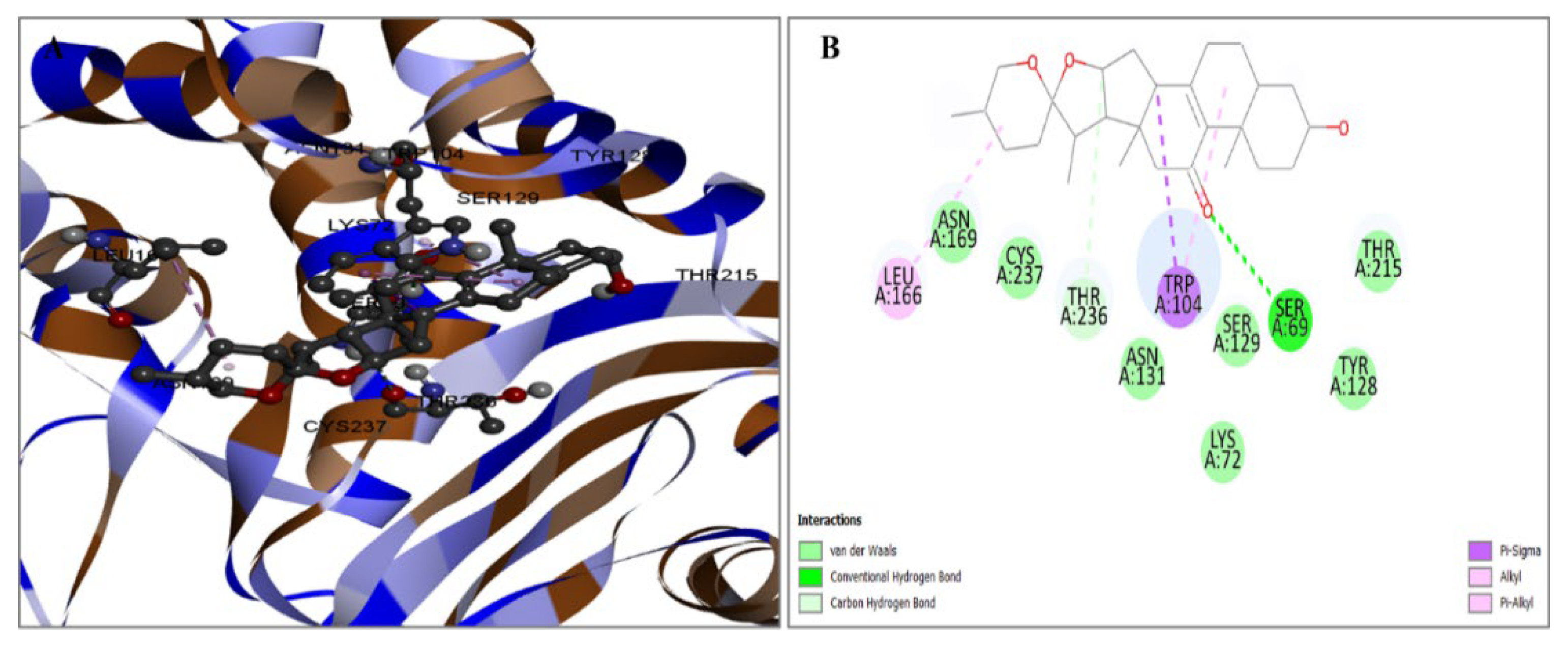

| PubChem CID | Chemical Name | Formula | Binding Affinity (Kcal/mol) |
|---|---|---|---|
| 6917974 | Resibufogenin | C24H32O4 | −9.0 |
| 159099 | Naphtho(1,2-b)furan-2,8(3H,4H)-dione, 3a,5,5a,6,7,9b-hexahydro-6-hydroxy-5a,9-dimethyl-3-methylene-,(3aS,5aR,6R,9bS) | C15H18O4 | −7.8 |
| 628694 | 16-hydroxy-5′,7,9,13-tetramethylspiro[5-oxapentacyclo[10.8.0.02,9.04,8.013,18]icos-1(12)-ene-6,2′-oxane]-11-one | C27H40O4 | −7.7 |
| 482788 | Reynosin | C15H20O3 | −7.5 |
| PubChem CID | Residue | Distance | Category | Type |
|---|---|---|---|---|
| CID6917974 | TRP104 | 3.69752 | Hydrophobic | Pi-Sigma |
| TRP104 | 4.65974 | Hydrophobic | Pi-Alkyl | |
| TRP104 | 4.07224 | Hydrophobic | Pi-Alkyl | |
| TRP104 | 5.46292 | Hydrophobic | Pi-Alkyl | |
| LEU166 | 5.42357 | Hydrophobic | Pi-Alkyl | |
| CID159099 | SER69 | 2.35134 | Hydrogen Bond | Conv-H-Bond |
| SER129 | 2.06128 | Hydrogen Bond | Conv-H-Bond | |
| ASN131 | 2.92787 | Hydrogen Bond | Conv-H-Bond | |
| THR236 | 3.51664 | Hydrogen Bond | Carbon Hydrogen Bond | |
| TRP104 | 3.50246 | Hydrophobic | Pi-Sigma | |
| TRP104 | 3.85502 | Hydrophobic | Pi-Sigma | |
| TRP104 | 5.23469 | Hydrophobic | Pi-Alkyl | |
| TRP104 | 4.48891 | Hydrophobic | Pi-Alkyl | |
| CID628694 | SER69 | 3.01252 | Hydrogen Bond | Conv-H-Bond |
| THR236 | 3.68071 | Hydrogen Bond | Carbon Hydrogen Bond | |
| TRP104 | 3.41301 | Hydrophobic | Pi-Sigma | |
| LEU166 | 5.36347 | Hydrophobic | Alkyl | |
| TRP104 | 4.06557 | Hydrophobic | Pi-Alkyl | |
| TRP104 | 5.18272 | Hydrophobic | Pi-Alkyl | |
| CID482788 | TRP104 | 5.13792 | Hydrophobic | Pi-Alkyl |
| TRP104 | 4.22344 | Hydrophobic | Pi-Alkyl | |
| TRP104 | 4.44737 | Hydrophobic | Pi-Alkyl | |
| TRP104 | 5.06342 | Hydrophobic | Pi-Alkyl |
| Properties | CID6917974 | CID159099 | CID628694 | CID482788 | |
|---|---|---|---|---|---|
| Phytochemical properties | MW (g/M) | 384.51 | 262.30 | 428.60 | 248.32 |
| Heavy atoms | 28 | 19 | 31 | 18 | |
| Aromatic heavy atoms | 6 | 0 | 0 | 0 | |
| Rotatable bond | 1 | 0 | 0 | 0 | |
| Hydrogen bond acceptors | 4 | 4 | 4 | 3 | |
| Hydrogen-bond donor | 1 | 1 | 1 | 1 | |
| Lipophilicity | Log Po/w | 3.88 | 1.64 | 4.30 | 2.34 |
| Water solubility | Log S (ESOL) | −4.67 (moderately soluble) | −1.98 (soluble) | −5.14 (moderately soluble) | −2.66 (soluble) |
| Pharmacokinetics | GI absorption | High | High | High | High |
| Drug-likeness | Lipinski | Yes (0 violations) | Yes (0 violations) | Yes (0 violations) | Yes (0 violations) |
| Medicinal chemistry synthesis accessibility | 5.98 | 4.41 | 6.70 | 4.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alshammari, F.A. Exploring the Antibacterial Potential of Artemisia judaica Compounds Targeting the Hydrolase/Antibiotic Protein in Klebsiella pneumoniae: In Vitro and In Silico Investigations. Pharmaceuticals 2024, 17, 667. https://doi.org/10.3390/ph17060667
Alshammari FA. Exploring the Antibacterial Potential of Artemisia judaica Compounds Targeting the Hydrolase/Antibiotic Protein in Klebsiella pneumoniae: In Vitro and In Silico Investigations. Pharmaceuticals. 2024; 17(6):667. https://doi.org/10.3390/ph17060667
Chicago/Turabian StyleAlshammari, Fahdah Ayed. 2024. "Exploring the Antibacterial Potential of Artemisia judaica Compounds Targeting the Hydrolase/Antibiotic Protein in Klebsiella pneumoniae: In Vitro and In Silico Investigations" Pharmaceuticals 17, no. 6: 667. https://doi.org/10.3390/ph17060667
APA StyleAlshammari, F. A. (2024). Exploring the Antibacterial Potential of Artemisia judaica Compounds Targeting the Hydrolase/Antibiotic Protein in Klebsiella pneumoniae: In Vitro and In Silico Investigations. Pharmaceuticals, 17(6), 667. https://doi.org/10.3390/ph17060667




