Proteomic Network Analysis of Bronchoalveolar Lavage Fluid in Ex-Smokers to Discover Implicated Protein Targets and Novel Drug Treatments for Chronic Obstructive Pulmonary Disease
Abstract
:1. Introduction
2. Results
2.1. Study Population Characteristics
2.2. BALF Proteome Characteristics
2.3. Manually Curated Pathway Analysis
2.3.1. Functional Annotation of Differential Expressed Proteins and Transcription Factor Interactions
2.3.2. Bioinformatic Pathway Analysis of BALF Proteomic Data
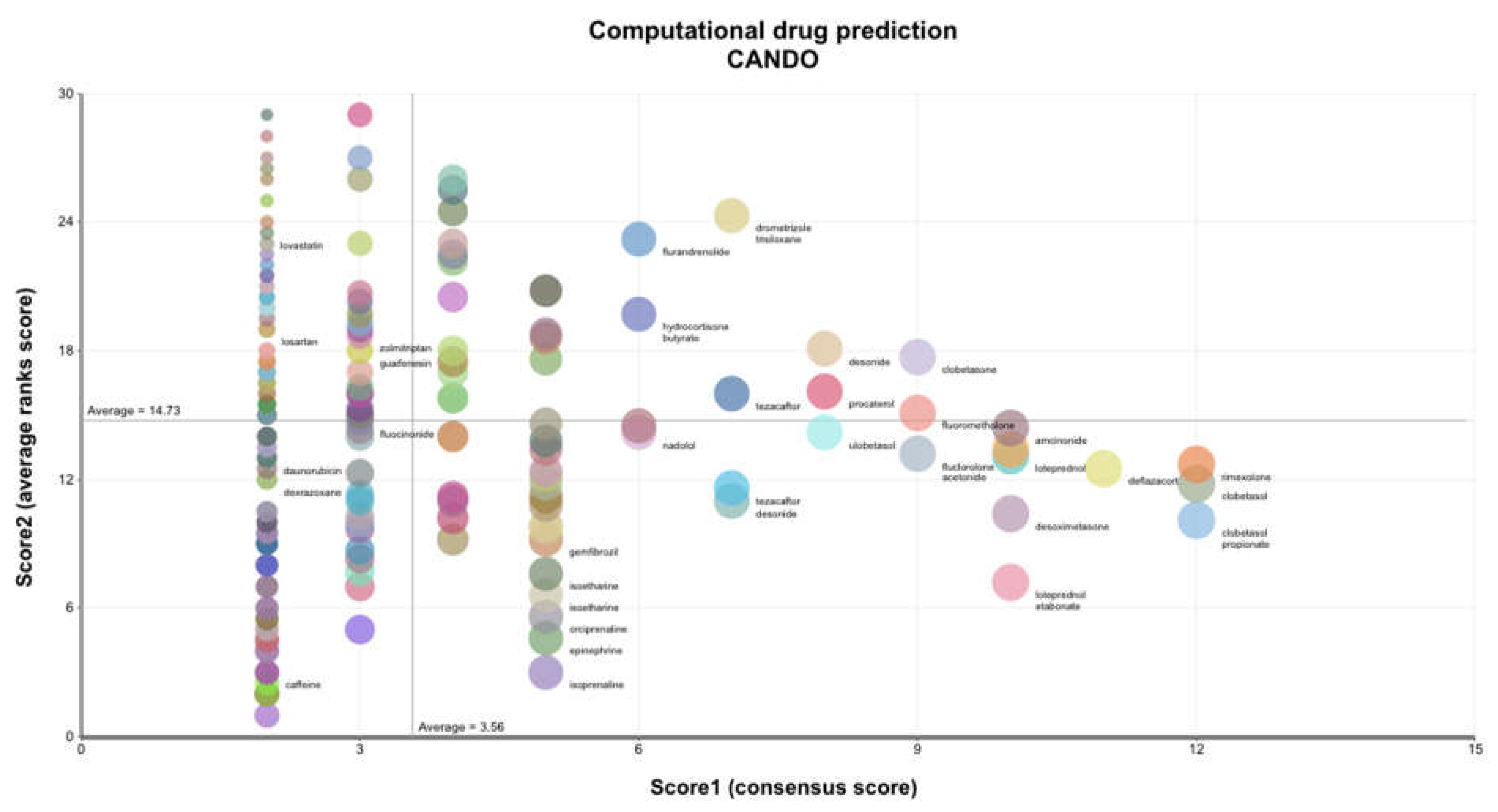
2.4. Computational Drug Prediction
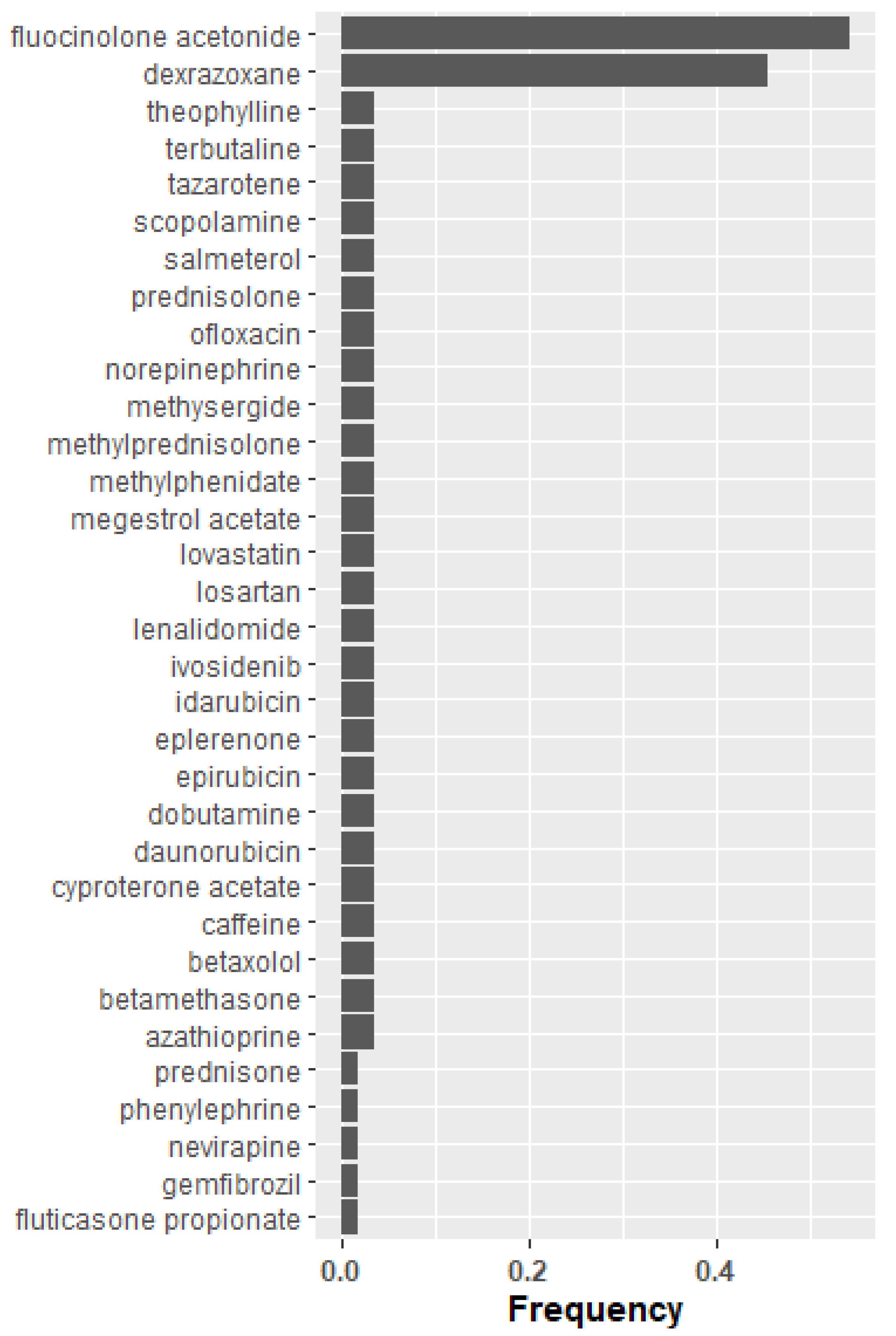
2.5. Candidate Key Mediators of COPD Pathology Based on Literature Derived Drug Enrichment
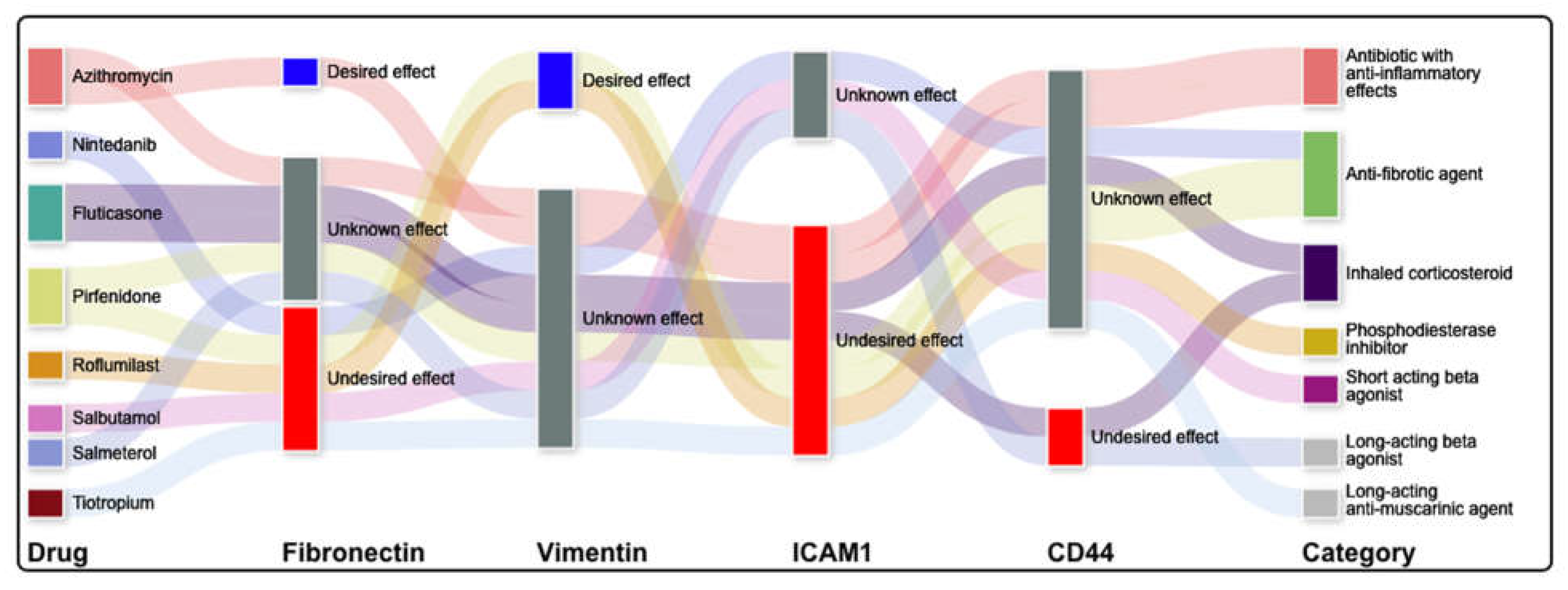
2.5.1. Literature Informed Protein-Protein and Protein-Drug Interaction Network
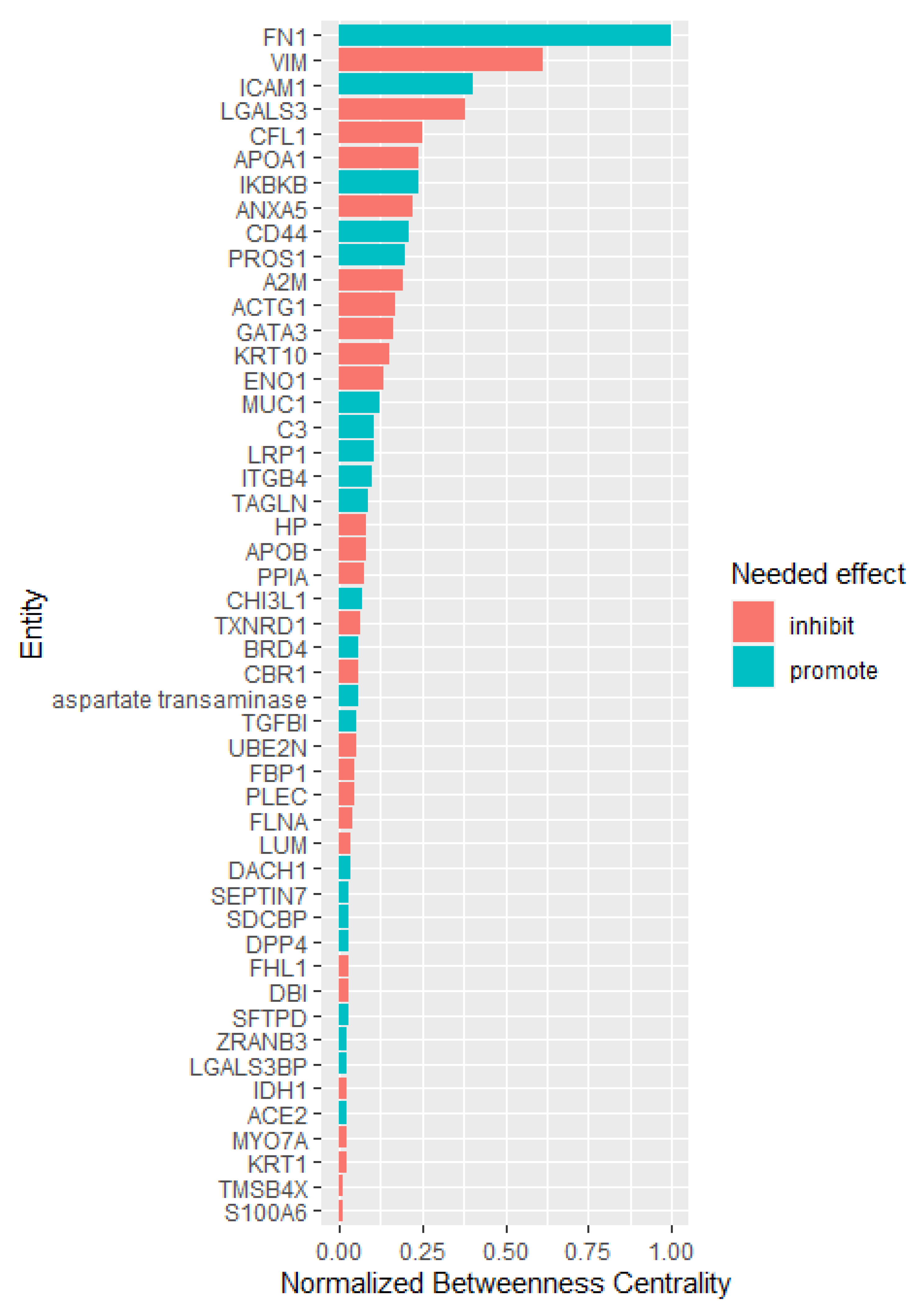
2.5.2. Network Topological Analysis
3. Discussion
3.1. Limitations and Strengths
3.2. Comparison to Previously Published Studies
4. Methods
4.1. Recruitment of Subjects
4.2. Ethics Statement
4.3. Inclusion/Exclusion Criteria
4.4. Bronchoscopy and BALF Sample Preparation
4.5. Protein Identification/Quantification
4.6. Long Gradient Nano-RPLC/Mass Spectrometry
4.7. Bioinformatics Analyses
4.7.1. Manually Curated Pathway Analysis
4.7.2. Literature Informed Protein-Protein and Protein-Drug Interaction Network
4.7.3. Shotgun Multiscale Drug Discovery Platform
4.7.4. Network Topological Analysis
4.7.5. Literature Based Drug Enrichment Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BALF | bronchoalveolar lavage fluid |
| BANDOCK | bioanalytical docking |
| CANDO | computational analysis of novel drug opportunities |
| COPD | chronic obstructive pulmonary disease |
| DAVID | database for annotation visualization and integrated discovery |
| GO | gene ontology |
| IPA | ingenuity pathway analysis |
| LTQ Orbitrap | linear ion trap combined with an orbitrap analyzer mass spectrometer |
References
- Brown, D.W.; Croft, J.B.; Greenlund, K.J.; Giles, W.H. Deaths From Chronic Obstructive Pulmonary Disease--United States, 2000–2005. J. Am. Med. Assoc. 2009, 301, 1331. [Google Scholar]
- Croft, J.B.; Wheaton, A.G.; Liu, Y.; Xu, F.; Lu, H.; Matthews, K.; Cunningham, T.J.; Wang, Y.; Holt, J.B. Urban-Rural County and State Differences in Chronic Obstructive Pulmonary Disease—United States, 2015. Morb. Mortal. Wkly. Rep. 2018, 67, 205–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- GBD 2015 Chronic Respiratory Disease Collaborators. Global, regional, and national deaths, prevalence, disability-adjusted life years, and years lived with disability for chronic obstructive pulmonary disease and asthma, 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet Respir. Med. 2017, 5, 691. [Google Scholar] [CrossRef] [Green Version]
- US Preventive Services Task Force (USPSTF). Screening for chronic obstructive pulmonary disease: US Preventive Services Task Force recommendation statement. JAMA 2016, 315, 1372–1377. [Google Scholar] [CrossRef] [Green Version]
- From the Global Strategy for the Diagnosis, Management and Prevention of COPD. Global Initiative for Chronic Obstructive Lung Disease (GOLD). 2022. Available online: http://www.goldcopd.org (accessed on 1 April 2022).
- Gartman, E.J.; Mulpuru, S.S.; Mammen, M.J.; Alexander, P.E.; Nici, L.; Aaron, S.D.; Ruminjo, J.K.; Thomson, C.C. Summary for Clinicians: Clinical Practice Guideline on Pharmacologic Management of Chronic Obstructive Pulmonary Disease. Ann. Am. Thorac. Soc. 2021, 18, 11–16. [Google Scholar] [CrossRef]
- Nici, L.; Mammen, M.J.; Charbek, E.; Alexander, P.E.; Au, D.H.; Boyd, C.M.; Criner, G.J.; Donaldson, G.C.; Dreher, M.; Fan, V.S.; et al. Pharmacologic Management of Chronic Obstructive Pulmonary Disease. An Official American Thoracic Society Clinical Practice Guideline. Am. J. Respir. Crit. Care Med. 2020, 201, e56–e69. [Google Scholar] [CrossRef] [Green Version]
- Geitona, M.; Hatzikou, M.; Steiropoulos, P.; Alexopoulos, E.C.; Bouros, D. The cost of COPD exacerbations: A university hospital—Based study in Greece. Respir. Med. 2011, 105, 402–409. [Google Scholar] [CrossRef] [Green Version]
- Hutchinson, A.F.; Brand, C.; Irving, L.; Roberts, C.; Thompson, P.; Campbell, D. Acute care costs of patients admitted for management of chronic obstructive pulmonary disease exacerbations: Contribution of disease severity, infection and chronic heart failure. Intern. Med. J. 2010, 40, 364–371. [Google Scholar] [CrossRef]
- Ciapponi, A.; Alison, L.; Agustina, M.; Demián, G.; Silvana, C.; Edgardo, S. The Epidemiology and Burden of COPD in Latin America and the Caribbean: Systematic Review and Meta-Analysis. COPD J. Chronic Obstr. Pulm. Dis. 2013, 11, 339–350. [Google Scholar] [CrossRef]
- Rennard, S.I.; Vestbo, J. COPD: The dangerous underestimate of 15%. Lancet 2006, 367, 1216–1219. [Google Scholar] [CrossRef]
- Rennard, S.I.; Vestbo, J. Natural Histories of Chronic Obstructive Pulmonary Disease. Proc. Am. Thorac. Soc. 2008, 5, 878–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schubert, C. Anti-tobacco efforts going up in smoke. Nat. Med. 2006, 12, 866. [Google Scholar] [CrossRef]
- Mammen, M.; Sethi, S. COPD and the microbiome. Respirology 2016, 21, 590–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rutgers, S.R.; Postma, D.S.; Ten Hacken, N.H.; Kauffman, H.F.; van der Mark, T.W.; Koëter, G.H.; Timens, W. Ongoing airway inflammation in patients with COPD who do not currently smoke. Thorax 2000, 55, 12–18. [Google Scholar] [CrossRef] [Green Version]
- Kunz, L.I.Z.; Lapperre, T.S.; Snoeck-Stroband, J.B.; Budulac, S.E.; Timens, W.; Van Wijngaarden, S.; Schrumpf, J.A.; Rabe, K.F.; Postma, D.S.; Sterk, P.J.; et al. Smoking status and anti-inflammatory macrophages in bronchoalveolar lavage and induced sputum in COPD. Respir. Res. 2011, 12, 34. [Google Scholar] [CrossRef] [Green Version]
- Hogg, J.C. Pathophysiology of airflow limitation in chronic obstructive pulmonary disease. Lancet 2004, 364, 709–721. [Google Scholar] [CrossRef]
- Hogg, J.C.; Chu, F.; Utokaparch, S.; Woods, R.; Elliott, W.M.; Buzatu, L.; Cherniack, R.M.; Rogers, R.M.; Sciurba, F.; Coxson, H.O.; et al. The Nature of Small-Airway Obstruction in Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2004, 350, 2645–2653. [Google Scholar] [CrossRef]
- Cosio, M.G.; Saetta, M.; Agusti, A. Immunologic Aspects of Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2009, 360, 2445–2454. [Google Scholar] [CrossRef]
- Sethi, S. Infection as a comorbidity of COPD. Eur. Respir. J. 2010, 35, 1209–1215. [Google Scholar] [CrossRef] [Green Version]
- Tu, C.; Mammen, M.J.; Li, J.; Shen, X.; Jiang, X.; Hu, Q.; Wang, J.; Sethi, S.; Qu, J. Large-Scale, Ion-Current-Based Proteomics Investigation of Bronchoalveolar Lavage Fluid in Chronic Obstructive Pulmonary Disease Patients. J. Proteome Res. 2013, 13, 627–639. [Google Scholar] [CrossRef] [Green Version]
- Da Huang, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.-K.; Da Huang, W.; Bt, S.; Ra, L. Faculty Opinions recommendation of Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, S.D. Proteolysis in the lung. Eur. Respir. J. Suppl. 2003, 44 (Suppl. 44), 30s–32s. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Annoni, R.; Lanças, T.; Yukimatsu Tanigawa, R.; de Medeiros Matsushita, M.D.; de Morais Fernezlian, S.D.; Bruno, A.; da Fernando Ferraz Silva, L.; Roughley, P.J.; Battaglia, S.; Dolhnikoff, M.; et al. Extracellular matrix composition in COPD. Eur. Respir. J. 2012, 40, 1362–1373. [Google Scholar] [CrossRef] [Green Version]
- Riise, G.; Larsson, S.; Löfdahl, C.-G.; Andersson, B. Circulating cell adhesion molecules in bronchial lavage and serum in COPD patients with chronic bronchitis. Eur. Respir. J. 1994, 7, 1673–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ri, A.; Hagiyama, M.; Inoué, T.; Yoneshige, A.; Kimura, R.; Murakami, Y.; Ito, A. Progression of Pulmonary Emphysema and Continued Increase in Ectodomain Shedding of Cell Adhesion Molecule 1 After Cessation of Cigarette Smoke Exposure in Mice. Front. Cell Dev. Biol. 2018, 6, 52. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Kohler, M.; Heyder, T.; Forsslund, H.; Garberg, H.K.; Karimi, R.; Grunewald, J.; Berven, F.S.; Nyren, S.; Sköld, C.M.; et al. Proteomic profiling of lung immune cells reveals dysregulation of phagocytotic pathways in female-dominated molecular COPD phenotype. Respir. Res. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Sethi, S. Bacterial Infection and the Pathogenesis of COPD. Chest 2000, 117, 286S–291S. [Google Scholar] [CrossRef] [Green Version]
- Zuo, L.; He, F.; Sergakis, G.G.; Koozehchian, M.S.; Stimpfl, J.N.; Rong, Y.; Diaz, P.T.; Best, T.M. Interrelated role of cigarette smoking, oxidative stress, and immune response in COPD and corresponding treatments. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2014, 307, L205–L218. [Google Scholar] [CrossRef] [Green Version]
- Stockley, R.A. Neutrophils and the Pathogenesis of COPD. Chest 2002, 121, 151S–155S. [Google Scholar] [CrossRef] [Green Version]
- Davies, J.C.; Moskowitz, S.M.; Brown, C.; Horsley, A.; Mall, M.A.; McKone, E.F.; Plant, B.J.; Prais, D.; Ramsey, B.W.; Taylor-Cousar, J.L.; et al. VX-659–Tezacaftor–Ivacaftor in Patients with Cystic Fibrosis and One or Two Phe508del Alleles. N. Engl. J. Med. 2018, 379, 1599–1611. [Google Scholar] [CrossRef] [PubMed]
- Frick, M.H.; Elo, O.; Haapa, K.; Heinonen, O.P.; Heinsalmi, P.; Helo, P.; Huttunen, J.K.; Kaitaniemi, P.; Koskinen, P.; Manninen, V.; et al. Helsinki Heart Study: Primary-Prevention Trial with Gemfibrozil in Middle-Aged Men with Dyslipidemia. N. Engl. J. Med. 1987, 317, 1237–1245. [Google Scholar] [CrossRef] [PubMed]
- Ferraro, R.A.; Leucker, T.; Martin, S.S.; Banach, M.; Jones, S.R.; Toth, P.P. Contemporary Management of Dyslipidemia. Drugs 2022, 82, 559–576. [Google Scholar] [CrossRef] [PubMed]
- Eckland, D.; Danhof, M. Clinical pharmacokinetics of pioglitazone. Exp. Clin. Endocrinol. Diabet. 2000, 108 (Suppl. 2), 234–242. [Google Scholar] [CrossRef]
- Kamdar, M.R.; Stanley, C.E.; Carroll, M.; Wogulis, L.; Dowling, W.; Deus, H.F.; Samarasinghe, M. Text Snippets to Corroborate Medical Relations: An Unsupervised Approach using a Knowledge Graph and Embeddings. AMIA Summits Transl. Sci. Proc. 2020, 2020, 288–297. [Google Scholar]
- Walter, R.E.; Wilk, J.B.; Larson, M.; Vasan, R.S.; Keaney, J.; Lipinska, I.; O’Connor, G.; Benjamin, E. Systemic Inflammation and COPD: The Framingham Heart Study. Chest 2008, 133, 19–25. [Google Scholar] [CrossRef]
- Mammen, M.J.; Sands, M.F.; Abou-Jaoude, E.; Aalinkeel, R.; Reynolds, J.L.; Parikh, N.U.; Sharma, U.; Schwartz, S.A.; Mahajan, S.D. Role of Galectin-3 in the pathophysiology underlying allergic lung inflammation in a tissue inhibitor of metalloproteinases 1 knockout model of murine asthma. Immunology 2017, 153, 387–396. [Google Scholar] [CrossRef]
- Muro, A.F.; Moretti, F.A.; Moore, B.B.; Yan, M.; Atrasz, R.G.; Wilke, C.A.; Flaherty, K.R.; Martinez, F.J.; Tsui, J.L.; Sheppard, D.; et al. An Essential Role for Fibronectin Extra Type III Domain A in Pulmonary Fibrosis. Am. J. Respir. Crit. Care Med. 2008, 177, 638–645. [Google Scholar] [CrossRef] [Green Version]
- To, W.S.; Midwood, K.S. Plasma and cellular fibronectin: Distinct and independent functions during tissue repair. Fibrogen. Tissue Repair 2011, 4, 21. [Google Scholar] [CrossRef] [Green Version]
- Li, F.J.; Surolia, R.; Li, H.; Wang, Z.; Kulkarni, T.; Liu, G.; De Andrade, J.A.; Kass, D.J.; Thannickal, V.J.; Duncan, S.R.; et al. Autoimmunity to Vimentin Is Associated with Outcomes of Patients with Idiopathic Pulmonary Fibrosis. J. Immunol. 2017, 199, 1596–1605. [Google Scholar] [CrossRef] [Green Version]
- Ouhtit, A.; Thouta, R.; Zayed, H.; Gaur, R.L.; Fernando, A.; Rahman, M.; Welsh, D.A. CD44 mediates stem cell mobilization to damaged lung via its novel transcriptional targets, Cortactin and Survivin. Int. J. Med Sci. 2020, 17, 103–111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mills, J.S.; Bowers, A.; Djerassi, C.; Ringold, H.J. Steroids. CXXXVII.1 Synthesis of a New Class of Potent Cortical Hormones. 6α,9α-Difluoro-16α-hydroxyprednisolone and its Acetonide. J. Am. Chem. Soc. 1960, 82, 3399–3404. [Google Scholar] [CrossRef]
- Campochiaro, P.A.; Nguyen, Q.D.; Hafiz, G.; Bloom, S.; Brown, D.M.; Busquets, M.; Ciulla, T.; Feiner, L.; Sabates, N.; Billman, K.; et al. Aqueous Levels of Fluocinolone Acetonide after Administration of Fluocinolone Acetonide Inserts or Fluocinolone Acetonide Implants. Ophthalmology 2013, 120, 583–587. [Google Scholar] [CrossRef] [PubMed]
- Wiseman, L.R.; Spencer, C.M. Dexrazoxane. A review of its use as a cardioprotective agent in patients receiving anthracycline-based chemotherapy. Drugs 1998, 56, 385–403. [Google Scholar] [CrossRef] [PubMed]
- Cvetković, R.S.; Scott, L.J. Dexrazoxane. Drugs 2005, 65, 1005–1024. [Google Scholar] [CrossRef] [PubMed]
- Hara, E.S.; Ono, M.; Pham, H.T.; Sonoyama, W.; Kubota, S.; Takigawa, M.; Matsumoto, T.; Young, M.F.; Olsen, B.R.; Kuboki, T. Fluocinolone acetonide is a potent synergistic factor of TGF-β3–associated chondrogenesis of bone marrow–derived mesenchymal stem cells for articular surface regeneration. J. Bone Miner. Res. 2015, 30, 1585–1596. [Google Scholar] [CrossRef] [Green Version]
- Di Stefano, A.; Sangiorgi, C.; Gnemmi, I.; Casolari, P.; Brun, P.; Ricciardolo, F.L.; Contoli, M.; Papi, A.; Maniscalco, P.; Ruggeri, P.; et al. TGF-beta Signaling Pathways in Different Compartments of the Lower Airways of Patients With Stable COPD. Chest 2018, 153, 851–862. [Google Scholar] [CrossRef] [Green Version]
- Kirkham, P.A.; Barnes, P.J. Oxidative Stress in COPD. Chest 2013, 144, 266–273. [Google Scholar] [CrossRef]
- Moisieieva, N.V.; Burya, L.V.; A Kapustianskaya, A.; A Kolenko, I.; A Rumyantseva, M.; Shumeiko, O.H. Comprehensive patterns of comorbidity: Copd and depression. Asp. Treatment. Wiadomości. Lek. 2018, 71, 588–591. [Google Scholar]
- Morris, M.C.; Richman, S.; Lyman, C.A.; Qu, J.; Mammen, M.J.; Sethi, S.; Broderick, G. Hacking the Immune Response to Infection in Chronic Obstructive Pulmonary Disease. In Proceedings of the 2020 IEEE 20th International Conference on Bioinformatics and Bioengineering (BIBE)2020, Cincinnati, OH, USA, 26–28 October 2020. [Google Scholar]
- Nicholas, B.L.; Skipp, P.; Barton, S.; Singh, D.; Bagmane, D.; Mould, R.; Angco, G.; Ward, J.; Guha-Niyogi, B.; Wilson, S.; et al. Identification of Lipocalin and Apolipoprotein A1 as Biomarkers of Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2010, 181, 1049–1060. [Google Scholar] [CrossRef] [Green Version]
- Ohlmeier, S.; Mazur, W.; Linja-Aho, A.; Louhelainen, N.; Rönty, M.; Toljamo, T.; Bergmann, U.; Kinnula, V.L. Sputum Proteomics Identifies Elevated PIGR levels in Smokers and Mild-to-Moderate COPD. J. Proteome Res. 2011, 11, 599–608. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; In, K.H.; Kim, J.H.; Lee, S.Y.; Shin, C.; Shim, J.J.; Kang, K.H.; Yoo, S.H.; Kim, C.H.; Kim, H.-K.; et al. Proteomic Analysis in Lung Tissue of Smokers and COPD Patients. Chest 2009, 135, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Berenson, C.S.; Garlipp, M.A.; Grove, L.J.; Maloney, J.; Sethi, S. Impaired phagocytosis of nontypeable Haemophilus influenzae by human alveolar macrophages in chronic obstructive pulmonary disease. J. Infect. Dis. 2006, 194, 1375–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, D.; Agusti, A.; Anzueto, A.; Barnes, P.J.; Bourbeau, J.; Celli, B.R.; Criner, G.J.; Frith, P.; Halpin, D.M.G.; Han, M.; et al. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Lung Disease: The GOLD science committee report 2019. Eur. Respir. J. 2019, 53, 1900164. [Google Scholar] [CrossRef]
- Berenson, C.S.; Wrona, C.T.; Grove, L.J.; Maloney, J.; Garlipp, M.A.; Wallace, P.K.; Stewart, C.C.; Sethi, S. Impaired Alveolar Macrophage Response toHaemophilusAntigens in Chronic Obstructive Lung Disease. Am. J. Respir. Crit. Care Med. 2006, 174, 31–40. [Google Scholar] [CrossRef] [Green Version]
- Tu, C.; Li, J.; Young, R.; Page, B.J.; Engler, F.; Halfon, M.S.; Canty, J.J.M.; Qu, J. Combinatorial Peptide Ligand Library Treatment Followed by a Dual-Enzyme, Dual-Activation Approach on a Nanoflow Liquid Chromatography/Orbitrap/Electron Transfer Dissociation System for Comprehensive Analysis of Swine Plasma Proteome. Anal. Chem. 2011, 83, 4802–4813. [Google Scholar] [CrossRef] [Green Version]
- Tu, C.; Li, J.; Bu, Y.; Hangauer, D.; Qu, J. An ion-current-based, comprehensive and reproducible proteomic strategy for comparative characterization of the cellular responses to novel anti-cancer agents in a prostate cell model. J. Proteom. 2012, 77, 187–201. [Google Scholar] [CrossRef] [Green Version]
- Novichkova, S.; Egorov, S.; Daraselia, N. MedScan, a natural language processing engine for MEDLINE abstracts. Bioinformatics 2003, 19, 1699–1706. [Google Scholar] [CrossRef] [Green Version]
- Cheadle, C.; Cao, H.; Kalinin, A.; Hodgkinson, J. Advanced literature analysis in a Big Data world. Ann. N. Y. Acad. Sci. 2016, 1387, 25–33. [Google Scholar] [CrossRef]
- Falls, Z.; Mangione, W.; Schuler, J.; Samudrala, R. Exploration of interaction scoring criteria in the CANDO platform. BMC Res. Notes 2019, 12, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Schuler, J.; Samudrala, R. Fingerprinting CANDO: Increased Accuracy with Structure- and Ligand-Based Shotgun Drug Repurposing. ACS Omega 2019, 4, 17393–17403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mangione, W.; Falls, Z.; Chopra, G.; Samudrala, R. cando.py: Open Source Software for Predictive Bioanalytics of Large Scale Drug-Protein-Disease Data. J. Chem. Inf. Model 2020, 60, 4131–4136. [Google Scholar] [CrossRef] [PubMed]
- Hudson, M.; Samudrala, R. Multiscale Virtual Screening Optimization for Shotgun Drug Repurposing Using the CANDO Platform. Molecules 2021, 26, 2581. [Google Scholar] [CrossRef] [PubMed]
- Schuler, J.; Falls, Z.; Mangione, W.; Hudson, M.L.; Bruggemann, L.; Samudrala, R. Evaluating the performance of drug-repurposing technologies. Drug Discov. Today 2021, 27, 49–64. [Google Scholar] [CrossRef] [PubMed]
- Horst, J.; Pieper, U.; Sali, A.; Zhan, L.; Chopra, G.; Samudrala, R.; Featherstone, J. Strategic Protein Target Analysis for Developing Drugs to Stop Dental Caries. Adv. Dent. Res. 2012, 24, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Minie, M.; Chopra, G.; Sethi, G.; Horst, J.; White, G.; Roy, A.; Hatti, K.; Samudrala, R. CANDO and the infinite drug discovery frontier. Drug Discov. Today 2014, 19, 1353–1363. [Google Scholar] [CrossRef] [Green Version]
- Chopra, G.; Samudrala, R. Multiscale Modelling of Relationships between Protein Classes and Drug Behavior Across all Diseases Using the CANDO Platform. Mini-Rev. Med. Chem. 2015, 15, 705–717. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Roy, A.; Zhang, Y. Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 2013, 29, 2588–2595. [Google Scholar] [CrossRef]
- Dutta, S.; Burkhardt, K.; Young, J.; Swaminathan, G.J.; Matsuura, T.; Henrick, K.; Nakamura, H.; Berman, H.M. Data Deposition and Annotation at the Worldwide Protein Data Bank. Mol. Biotechnol. 2008, 42, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Gore, S.; Sanz-García, E.; Hendrickx, P.; Gutmanas, A.; Westbrook, J.D.; Yang, H.; Feng, Z.; Baskaran, K.; Berrisford, J.; Hudson, B.P.; et al. Validation of Structures in the Protein Data Bank. Structure 2017, 25, 1916–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Gragera, A.; Suppakitpaisarn, V. Semimetric properties of sørensen-dice and tversky indexes. In International Workshop on Algorithms and Computation; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Brands, U. A faster algorithm for betweenness cen-trality. J. Math. Sociol. 2001, 25, 163–177. [Google Scholar] [CrossRef]
- Gyorgy, A. A Practical Step-by-Step Guide for Quantifying Retroactivity in Gene Networks. Synth. Gene Circuits 2021, 2229, 293–311. [Google Scholar] [CrossRef]
- RC Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Thada, V.; Jaglan, V. Comparison of jaccard, dice, cosine similarity coefficient to find best fitness value for web retrieved documents using genetic algorithm. Int. J. Innov. Eng. Technol. 2013, 2, 202–205. [Google Scholar]
| Control Subjects n = 10 | COPD Subjects (GOLD Stage 2) n = 10 | p-Value | |
|---|---|---|---|
| Age (years) | 63.4 ± 11.7 | 67.8 ± 8.5 | 0.15 |
| Sex | 0.31 | ||
| Male | 6 | 7 | |
| Female | 4 | 3 | |
| Race | 0.083 | ||
| Caucasian | 8 | 10 | |
| African-American | 2 | 0 | |
| BMI (kg/m2) | 28.5 ± 4.2 | 32 ± 9.7 | 0.32 |
| Years patient quit smoking | NA | 12.9 ± 4.4 | |
| Tobacco smoking, Pack years | NA | 56.6 ± 17.2 | <0.001 |
| FEV1 (% predicted) | 96.3 ± 14.8 | 65.9 ± 8.1 | <0.001 * |
| FVC (% predicted) | 95.6 ± 13.4 | 87.6 ± 13.1 | 0.19 |
| FEV1/FVC | 77.6 ± 3.8 | 57.8 ± 8.6 | <0.001 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mammen, M.J.; Tu, C.; Morris, M.C.; Richman, S.; Mangione, W.; Falls, Z.; Qu, J.; Broderick, G.; Sethi, S.; Samudrala, R. Proteomic Network Analysis of Bronchoalveolar Lavage Fluid in Ex-Smokers to Discover Implicated Protein Targets and Novel Drug Treatments for Chronic Obstructive Pulmonary Disease. Pharmaceuticals 2022, 15, 566. https://doi.org/10.3390/ph15050566
Mammen MJ, Tu C, Morris MC, Richman S, Mangione W, Falls Z, Qu J, Broderick G, Sethi S, Samudrala R. Proteomic Network Analysis of Bronchoalveolar Lavage Fluid in Ex-Smokers to Discover Implicated Protein Targets and Novel Drug Treatments for Chronic Obstructive Pulmonary Disease. Pharmaceuticals. 2022; 15(5):566. https://doi.org/10.3390/ph15050566
Chicago/Turabian StyleMammen, Manoj J., Chengjian Tu, Matthew C. Morris, Spencer Richman, William Mangione, Zackary Falls, Jun Qu, Gordon Broderick, Sanjay Sethi, and Ram Samudrala. 2022. "Proteomic Network Analysis of Bronchoalveolar Lavage Fluid in Ex-Smokers to Discover Implicated Protein Targets and Novel Drug Treatments for Chronic Obstructive Pulmonary Disease" Pharmaceuticals 15, no. 5: 566. https://doi.org/10.3390/ph15050566
APA StyleMammen, M. J., Tu, C., Morris, M. C., Richman, S., Mangione, W., Falls, Z., Qu, J., Broderick, G., Sethi, S., & Samudrala, R. (2022). Proteomic Network Analysis of Bronchoalveolar Lavage Fluid in Ex-Smokers to Discover Implicated Protein Targets and Novel Drug Treatments for Chronic Obstructive Pulmonary Disease. Pharmaceuticals, 15(5), 566. https://doi.org/10.3390/ph15050566









