Abstract
Although the past epidemic of Zika virus (ZIKV) resulted in severe neurological consequences for infected infants and adults, there are still no approved drugs to treat ZIKV infection. In this study, we applied computational approaches to screen an in-house database of 77 natural and semi-synthetic compounds against ZIKV NS5 RNA-dependent RNA-polymerase (NS5 RdRp), an essential protein for viral RNA elongation during the replication process. For this purpose, we integrated computational approaches such as binding-site conservation, chemical space analysis and molecular docking. As a result, we prioritized nine virtual hits for experimental evaluation. Enzymatic assays confirmed that pedalitin and quercetin inhibited ZIKV NS5 RdRp with IC50 values of 4.1 and 0.5 µM, respectively. Moreover, pedalitin also displayed antiviral activity on ZIKV infection with an EC50 of 19.28 µM cell-based assays, with low toxicity in Vero cells (CC50 = 83.66 µM) and selectivity index of 4.34. These results demonstrate the potential of the natural compounds pedalitin and quercetin as candidates for structural optimization studies towards the discovery of new anti-ZIKV drug candidates.
1. Introduction
Zika Virus (ZIKV) is an arthropod-borne flavivirus that circulates globally and caused a worldwide concern due to its exponential spread in the Americas in 2015–2016 [1] and its association with severe congenital effects in pregnant women infected with the virus. The congenital ZIKV syndrome is characterized by neurological and neuropsychomotor complications, ophthalmological and hearing problems, craniofacial disproportion, epilepsy, cerebral palsy and microcephaly [2]. In adults, ZIKV can cause the Guillain-Barre syndrome [3]. Recently, researchers suggested that ZIKV strains with enhanced transmissibility and pathogenicity can reemerge [4].
ZIKV is constituted by a single-strand negative RNA which encodes three structural proteins, membrane (M), envelope (E) and capsid protein (C), arranged on a lipidic membrane, and seven non-structural (NS) proteins: NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5 [5]. Among the NS proteins, the NS5 RNA-dependent RNA-polymerase (RdRp) is an essential protein, catalyzing the replication of viral RNA from the RNA template [6], and has been considered a promising target for ZIKV drug discovery.
The nucleoside and nucleotide inhibitors (NI) of RdRp bind to the catalytic and RNA binding sites [7], whereas the non-nucleosides inhibitors (NNI) bind to the N-pocket (allosteric site) [8]. The NI antiviral drug sofosbuvir has been successfully used against Hepatitis C virus (HCV) and depends on the activation by host kinases [9]. Sofosbuvir was also tested against ZIKV RdRp presenting an IC50 value of 0.38 ± 0.03 µM [10].
Computer-Assisted Drug Design (CADD) [11] techniques rationally promote the discovery, prioritization and optimization of drug candidates, using computational resources, such as databases, algorithms, programs and web servers. Compared to experimental approaches, such as high-throughput screening (HTS), computational techniques have been shown to be faster and presented higher success rates [12].
The present study aimed to discover new potential ZIKV NS5 RdRp inhibitors guided by computational and experimental approaches. DENV and ZIKV NS5 RdRp primary and tertiary sequences share high similarities. Due to this fact, DENV NS5 RdRp known inhibitors were used to search for new potential ZIKV NS5 RdRp hits. Docking calculations were performed to prioritize virtual hits, and enzymatic assays validated these computational predictions, showing that pedalitin and quercetin, two natural compounds, inhibited ZIKV NS5 RdRp. Moreover, both hits presented anti-ZIKV activity in in vitro antiviral assays, with low cytotoxicity. These results demonstrate that integrated in silico and in vitro approaches can be used to accelerate the discovery of new ZIKV antiviral candidates.
2. Results and Discussion
A general workflow of the computational and experimental steps applied in this study is presented in Figure 1.
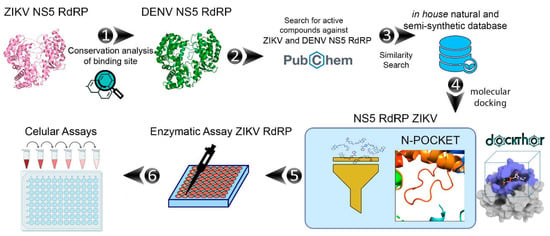
Figure 1.
General workflow applied in this work to identify of ZIKV NS5 RdRp inhibitors: (1) conservation analysis of ZIKV NS5 RdRp; (2) collection of compounds with experimental data against DENV and ZIKV RdRp in the PubChem database; (3) similarity analysis between known RdRp inhibitors and in-house collection of untested compounds available on the Laboratory of Antibiotics and Chemotherapeutics (LAC); (4) molecular docking of prioritized compounds at the ZIKV NS5 RdRp N-pocket binding site; (5) enzymatic and (6) cellular assays.
2.1. Binding Site Conservation Analysis
The binding-site conservation can provide an invaluable resource to understanding the affinity and binding mode of small molecules between homologs. In theory, proteins sharing a high similarity have the probability of sharing the same ligands [13]. Here, we employed the ConSurf analysis [14,15,16] to predict the evolutionary conservation profile of ZIKV RdRp amino acids based on the phylogenetic relations between homologous sequences such as DENV RdRp. All the polymerases resemble a right hand, with the three main regions (Figure 2a): fingers (residues 321–488 and 542–608), palm (residues 489–541 and 609–714), and thumb (residues 715–903). The RdRp domain is composed of three binding sites: the RNA site, the N-pocket (allosteric site) and the catalytic binding site [7,8,17]. The RNA site is a tunnel that single-stranded RNA enters and serves as a template for the formation of double-stranded RNA. The N-pocket is a tunnel through which the nucleotides enter. At this site, the initiation loop regulates template RNA binding and nucleotide entry. Finally, the catalytic site performs double-stranded RNA catalysis [18].
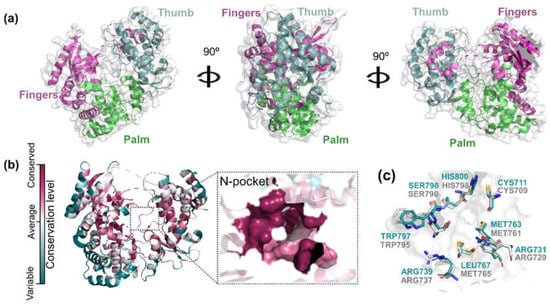
Figure 2.
Structural analysis of RdRp proteins from ZIKV (PDB ID: 6LD4) and DENV (PDB ID: 5I3Q). (a) Cartoon backbone diagram showing the front, side and back views of ZIKV RdRp. The fingers, thumb and palm domains are colored in magenta, cyan and green, respectively. (b) ConSurf analysis of ZIKV RdRp and corresponding N-pocket site. The magenta color indicates high conservation while white and turquoise colors indicate average and very low conservation, respectively. (c) Structural overlap of ZIKV and DENV N-pocket sites. ZIKV and DENV residues are colored in cyan and gray, respectively.
Although the DENV and ZIKV RdRp proteins show 64.59% of sequential identity, the evolutionary analysis of viral RdRps shows that ZIKV N-pocket is highly conserved (Figure 2b). These results suggest that the amino acid composition of the N-pocket is strongly associated with its structural and functional importance. As we can see in Figure 2c, all DENV (highlighted in gray, PDB ID: 5I3Q [18]) and ZIKV (highlighted in cyan, PDB ID: 6LD4 [19]) N-pocket residues are conserved, except for Leu767 in ZIKV, replaced by Met765 in DENV RdRp. It is important to point out that these two amino acids share similar volumes and electronic properties, and thus should not promote significative changes in the binding of small molecules to the N-pocket. The high conservation state of N-pockets corroborates with a high probability of ZIKV and DENV RdRps to share the same ligands. Based on these findings, an unsupervised cheminformatics approach using known DEV RdRp inhibitors was performed to find ZIKV RdRp hits from an in-house collection of natural and semi-synthetic compounds.
2.2. Chemical Space Analysis of RdRp Inhibitors
A chemical space analysis was conducted in order to select compounds from our in-house library that are similar to known RdRp inhibitors. Therefore, we compiled a dataset of known DENV RdRp inhibitors from PubChem and the literature. In total, 94 compounds were obtained from several bioassays on PubChem AID: 441537 [20], 642356 [21], 663478 [22], 1277364 [23], 1301573 [24], 1401288 [25], 1401306 [25], 1497239 [26], 1655471 [27], 1674514 [28], 1728708 [29] and 30 compounds were manually collected from published studies [17,20,21,23,25,27,28,30,31,32,33,34,35,36,37,38,39,40,41], providing a dataset of 124 DENV RdRp inhibitors. The in-house dataset from the Laboratory of Antibiotics and Chemotherapeutics (LAC), at São Paulo State University (UNESP), presents 77 natural and semi-synthetic compounds was merged to the publicly available dataset and a chemical space analysis was conducted using the dimensionality reduction method t-Distributed Stochastic Neighbor Embedding (t-SNE) [42].
As shown in the t-SNE plot (Figure 3A), 24 compounds from the in-house collection share the same chemical space of the known NS5 RdRp inhibitors. Most of them belong to the classes of acridones, diphenylamines, and flavonoids (Figure 3B). In view of this, these compounds were prioritized for molecular docking to assess their binding modes in ZIKV RdRp protein [19]. Since the scaffolds of the compounds are different from the nucleoside-like structure, the analysis was focused on the N-pocket site.
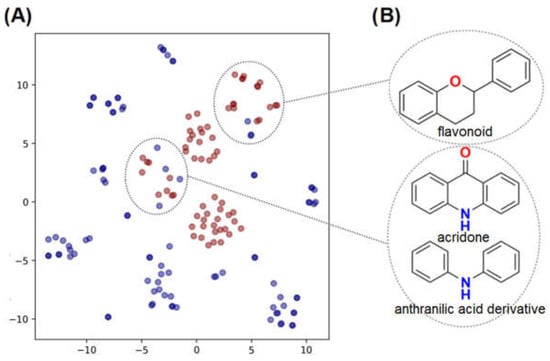
Figure 3.
t-SNE plot showing the chemical space of known DENV RdRp inhibitors and our in-house database of natural and semi-synthetic compounds. (A) DENV RdRp inhibitors collected from the literature and PubChem are shown in blue circles. The in house natural and semi-synthetic database compounds are represented in red. (B) 2D structures of the main scaffolds found in each cluster.
2.3. Docking Calculations at the ZIKV NS5 RdRp (N-Pocket)
The prioritized compounds from chemical space analysis were submitted to docking calculations to rank the most promising hits as well as to predict the binding affinities. All docking poses were analyzed according to the following parameters: (i) docking score and intermolecular interactions at the N-pocket binding site; (ii) overlap and binding mode similarity with the ZIKV RdRp NNI co-crystallized ligand 5-(3-fluorothiophen-2-yl)-2-hydroxy-4-methoxy-N-[4-(trifluoromethyl)benzenesulfonyl]benzamide and (iii) ligand efficiency. The redocking of the co-crystallized ligand was performed to verify the accuracy of the docking protocol in predicting the position of the ligands within the binding site (Supplementary Figure S2). Redocking also gives us a reference value of the docking score to consider in the compounds’ priorization. The redocked pose showed an RMSD value of 0.84 Å and a docking score value of −8.73 Kcal·mol−1.
Almost all ligands presented acceptable docking scores, close to the redocking score of the co-crystalized compound (docking score −8.73 Kcal·mol−1). Moreover, analyzing the binding modes and interactions, 16 ligands were prioritized for in vitro experimental validation. Thirteen of them had similar binding modes with the known ZIKV RdRp NNI. From them, 12 compounds presented ligand efficiency (LE) greater than 0.3 Kcal·mol−1·non-hydrogen atom−1. LE is value that normalizes the binding affinity (ΔG) or docking score with respect to the number of non-hydrogen atoms (n) [43,44,45]. The normalization of molecular weights influences the likelihood that a hit compound can be further optimized into prospective hit-to-lead investigations, as larger compounds tend to show greater docking scores due to the larger number of interactions [46,47].
A medicinal chemistry-based inspection was conducted [47,48], considering favorable scores for a higher number of hydrogen bonds between ligand and protein residues; salt bridges; π-cation and π-stacking interactions; and unfavorable scores for nonpolar regions of the ligand exposed to solvent. After this inspection, nine compounds were prioritized for the experimental evaluation (Table 1).

Table 1.
Virtual hits prioritized based on the computational approaches.
Four out of the nine virtual hits are naturally-occurring flavonoids (chrysin (6), sorbifolin (7), pedalitin (8) and quercetin (9)). Flavonoids have already been described in the literature as inhibitors of the RdRp domains of DENV and ZIKV [49]. Three compounds belong to the class of acridones, a class already described by some authors due to their antiviral activity and capability of inhibition of DNA and RNA viruses [50,51]. A potent activity of N-substituted acridones has already been demonstrated against DENV-2, blocking its multiplication in vitro [52]. ARORA and coworkers [53] demonstrated that compounds containing the diphenylamine subunit were able to inhibit the RdRp domain of DENV including the compound bis-chloro-diphenylamine, 2-aminoindan-2-carboxyl derivative NITD-434 (13) (Figure 4) that interacts with residues Thr795 and Thr796 of the N-pocket site. Three of the nine hits are diphenylamines.
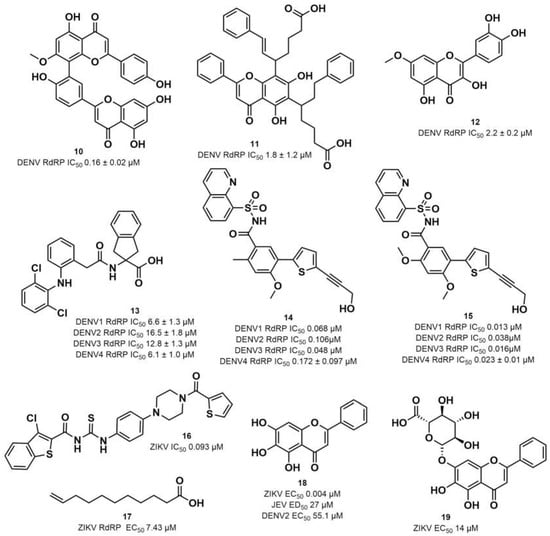
Figure 4.
DENV and ZIKV RdRp known inhibitors and their respective IC50 values at RdRp: (10) podocarpusflavone A [31]; (11) chartaceone D [21]; (12) rhamnetin [30]; (13) bis-chloro-diphenylamine, 2-aminoindan-2-carboxyl derivative [49]; (14) 5-(5-(3-Hydroxyprop-1-yn-1-yl)thiophen-2-yl)-2,4-dimethoxy-N-((3-methoxyphenyl)sulfonyl)benzamide [8]; (15) 5-(5-(3-Hydroxyprop-1-yn-1-yl)thiophen-2-yl)-4-methoxy-2-methyl-N-(quinolin-8-ylsulfonyl)benzamide [24]; (16) TPB [57]; (17) undecylenic acid [38]. Flavones (18) baicalein [59,60,61] and (19) baicalin [59] with antiviral activity (EC50) against several flaviviruses.
RdRp inhibitors have been classified as NI and NNI. The NIs present a structural similarity to nucleosides and have to be converted into triphosphate forms by host kinases to be incorporated into viral DNA or RNA, acting as chain terminators [54]. On the other hand, the NNIs interact directly with viral polymerase and present different scaffolds, such as flavonoids, alkaloids, acetylenic acids, terpenes, steroids, benzothiazine 2,2-dioxide analogs, pyrazole-5-phenylamine analogs, thiophene-based analogs, N-sulfonylpyrazoles and N-sulfonylanthranilic acids, thiazolidinone-thiadiazole and pyridobenzothiazole analogs [49]. NNIs act into the RdRp allosteric site and, in general, display fewer side effects since they are more selective for viral than host polymerase targets [55]. Among the DENV NNIs, there are natural products including flavonoids 10, 11 and 12 (Figure 4). Furthermore, another DENV RdRp NNI, the bis-chloro-diphenylamine, 2-aminoindan-2-carboxyl derivative compound (13) or NITD-434 (Figure 4), occupies the template RNA site and performs interactions with conserved residues between the four serotypes of DENV and ZIKV [53]. The synthetic co-crystallized DENV RdRp NNIs acylsulfonamide derivatives compounds (14) and (15) [8] (Figure 4), occupy the N-pocket site and had IC50 values ranging from 0.172 to 5.46 μM for compound 14 and 0.023 to 0.427 μM for compound 15 [10,56]. Among the ZIKV NNIs, there are few natural compounds such as chalcones and alkaloids, as well as synthetic compounds undecylenic acid compound 17 (Figure 4) and thienylcarbonyl-piperazinyl-benzothiophene (TBP), compound 16 (Figure 4) that act to inhibit ZIKV NS5 RdRp [57].
Recently, the flavonoids luteolin and quercetin were tested against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RdRp and presented IC50s values of 4.6 µM and 6.9 µM, respectively [58]. The authors also performed docking and molecular dynamic simulations of both ligands at the N-pocket and RNA binding sites, suggesting that they may properly bind to both sites.
2.4. Pedalitin and Quercetin Inhibits ZIKV RdRp Activity
Nine prioritized hits were submitted to endpoint assay at 20 µM to verify their inhibitory activity against ZIKV RdRp. Most of the compounds evaluated did not obtain significant inhibition results. Pedalitin and quercetin were the only ones with activity greater than 80%, with inhibitory activity of 97% and 99%, respectively, and consequently were selected for the concentration-response assays.
We then investigated ZIKV RdRp activity in the presence of pedalitin and quercetin. A concentration-response assay was performed at concentrations ranging from 80 µM to 0.156 µM to determine the inhibitory concentration of 50% (IC50). From this range of concentrations, it was determined that pedalitin and quercetin had IC50 values of 4.1 ± 0.3 µM and 0.5 ± 0.1 µM, respectively (Supplementary Figure S1). The enzymatic activities obtained are in agreement with those described for other flavonoids, as shown in Figure 4.
2.5. Pedalitin and Quercetin Binding Modes Predicted by Docking
From the enzymatic data, the two flavonoids pedalitin and quercetin were highlighted as promising ZIKV RdRp inhibitors. In Figure 5A,B we show the binding mode of quercetin and pedalitin, predicted by our docking calculations. Quercetin presented four interactions highlighted with an asterisk (*) (Figure 5A), that are the same interactions performed by the co-crystallized 5-(3-fluorothiophen-2-yl)-2-hydroxy-4-methoxy-N-[4-(trifluoromethyl)benzenesulfonyl]benzamide compound. These interactions are hydrogen bonds with residues Ser712, Arg731, Trp797, Ser798 and Asp666 (catalytic triad residue). In the same way, pedalitin presented four hydrogen bonds, with Ser712, Ser798, Trp797 and Thr796 and a cation–π interaction with the residue Arg731. Moreover, it also interacts with Asp666 via a hydrogen bond.
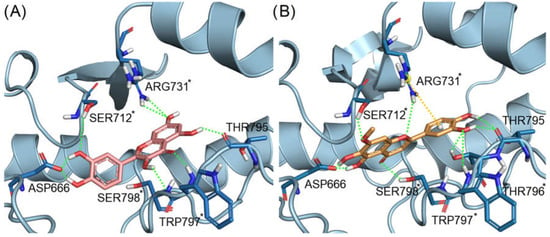
Figure 5.
Docking poses of (A) quercetin (carbon atoms in pink sticks representation) and (B) pedalitin (carbon atoms in orange sticks) at the N-pocket of ZIKV RdRp. Hydrogen bonds are represented as green dotted lines and cation–π interactions in yellow dotted lines. The interactions of residues highlighted with an asterisk (*) are the same observed with the co-crystallized ligand.
The binding modes of the flavonoids quercetin and pedalitin predicted by docking with ZIKV RdRp suggested a promising binding affinity with the allosteric binding site of the protein, corroborating the enzymatic assays results.
2.6. Pedalitin and Quercetin Inhibits ZIKV Replication In Vitro
The anti-ZIKV activities of the pedalitin and quercetin were further investigated through the employment of Vero cells infected with ZIKV wild type (ZIKVBR) (Figure 6). For this, a concentration-response assay was performed to determine the effective concentration of 50% (EC50) and cytotoxicity of 50% (CC50), and to calculate the Selective Index (SI = CC50/EC50). Vero cells were infected with ZIKVBR and simultaneously treated with pedalitin or quercetin at concentrations ranging from 200 µM to 0.005 µM for 72 h when viral replication rates were assessed (Figure 6). Cell viability analysis was performed in parallel (Figure 6).
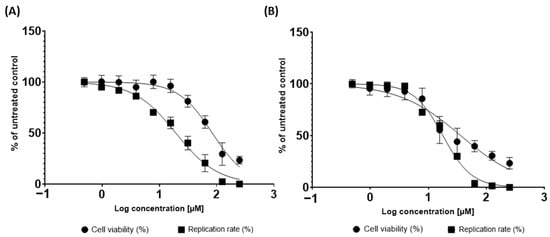
Figure 6.
Inhibition of replicative viral cycle by activity of pedalitin (A) and quercetin (B) against ZIKVBR infection. ZIKVBR replication was evaluated by measuring focus formation units using an immunofluorescence assay (indicated by ▄) after 72 h.p.i.. Cellular viability was measured in parallel using an MTT assay (indicated by ●). Mean values of three independent experiments each measured in quadruplicate including the standard deviation are shown.
From this range of concentrations, the treatment of ZIKV-infected cells with pedalitin demonstrated an EC50 value of 19.28, CC50 value of 83.66, and SI value of 4.34, and quercetin demonstrated an EC50 value of 17.74, CC50 value of 35.99, and SI value of 2.03 (Table 2).

Table 2.
Summary of the computational and experimental results for the best two compounds found in this study.
Summarizing computational and experimental data (Table 2), both pedalitin and quercetin bound to the N-pocket site of ZIKV RdRp, presenting good docking scores, compared to the redocking calculations, and binding site interactions similar with the co-crystalized ligand. Agreeing with docking calculations, enzymatic assays showed that both flavonoids inhibited ZIKV RdRp activity. Moreover, infection assays demonstrated that both compounds presented in vitro antiviral activity, and pedalitin presented a higher selectivity index (SI), representing a more promising hit.
Other flavonoid compounds have already demonstrated anti-ZIKV effects on Vero cells [62], such as the flavones baicalein (18) and baicalin (19) (Figure 4), which showed an EC50 of 0.004 µM and 14 µM, respectively [59]. Baicalein was also tested against other flaviviruses, such as Japanese encephalitis virus (JEV) and DENV-2, displaying an EC50 values of 27 ± 4 µM [52] and 55.1 µM, respectively [60,61]. Quercetin, identified in our study, also demonstrated anti-viral activity against DENV-2 virus in the study of Zandi and coworkers [63]. They used DENV-2 infected Vero cells and tested different concentrations of quercetin. At a concentration of 165.4 µM, the replication was reduced by 67%. Concentration-response curves were performed with administration after viral adsorption to the cells, obtaining an EC50 value of 95.6 µM [63].
Alternatively, there is a crucial role in virus-host cell interactions that provide important targets for the development of non-specific acting antivirals [64]. Non-specific antivirals can interfere with viral infection by acting on cellular signaling pathways or by modulating the differentiation and function of several immune cells [65]. This antiviral effect might contribute to a lower probability to develop viral resistance due to their reliance on host cell components [64]. Additionally, the intervention of virus-host interactions can include a broader range of activity with the immune system, especially for unknown emergent viral infections, where replication mechanisms are not elucidated [66].
Flavivirus polymerases have been reported to antagonize the interferon (IFN) signaling pathway via numerous mechanisms, including STAT2 degradation, inhibition of RIG-I, and suppression of IFNAR1 maturation [67,68,69]. Combating flavivirus infections by modulating the signaling pathway could be a factor to improve the infection outcome. In this case, non-specific antivirals are particularly desirable to be used combined with direct-acting antivirals and prepare the scientific community for future epidemics [70].
3. Materials and Methods
3.1. Computational
3.1.1. DENV and ZIKV NS5 RdRp Similarity Analysis
The 3D structure of ZIKV NS5 RdRp (PDB ID: 6LD4 [19]) was submitted to the ConSurf server [14,15,16] for estimating the evolutionary conservation of amino acids, based on their phylogenetic relations with homologues. Initially, 150 homologue sequences were imported from UNIREF-90 database [71]. The sequences with sequential identity <35% or >95% were ignored. A multiple sequence alignment (MSA) of the homologous sequences was built using the MAFFT-L-INS-I method [72] and the phylogenetic tree was built using the neighbor-joining algorithm [73]. Position-specific conservation scores were then computed using the empirical Bayesian method [74]. At the end of this analysis, the 3D structures and FASTA sequences of ZIKV (PDB ID: 5I3Q [8]) and DENV (PDB ID: 6LD4 [19]) were aligned using the PyMol v. 2.4 [75] and UniProt [76], respectively. The root-mean-square deviation (RMSD) was calculated for the distances of the conserved residues.
3.1.2. Collection of DENV RdRp Inhibitors
Initially, a search was performed in PubChem [77,78,79,80] databases for inhibitors of the NS5 RdRp of DENV. The activity IC50 threshold for component selection was 50 µM defined by PubChem. The bioassays selected for RdRp data collection were: PubChem AID: 441537 [20], 642356 [21], 663478 [22], 1277364 [23], 1301573 [24], 1401288 [25], 1401306 [25], 1497239 [26], 1655471 [27], 1674514 [28] and 1728708 [29]. Moreover, RdRp inhibitors from articles were manually collected from the literature [17,20,21,23,25,27,28,30,31,32,33,34,35,36,37,38,39,40,41] and added to our database.
3.1.3. Chemical Space Analysis of RdRp Inhibitors
The chemical space of known DENV RdRp inhibitors and an in-house collection of natural and semi-synthetic compounds was performed using t-Distributed Stochastic Neighbor Embedding (t-SNE) [42]. The t-SNE dimensionality reduction was performed using scikit-learn v. 1.0.2 [81] and extended connectivity fingerprints (ECFP6) with 2048 bits available on RDkit package v. 2022.03.2 [82].
3.1.4. Protein and Ligand Preparation
The 3D structure of ZIKV NS5 RdRp (PDB ID: 6LD4; resolution: 1.5 Å [19]) complexed with the compound 5-(3-fluorothiophen-2-yl)-2-hydroxy-4-methoxy-N-[4-(trifluoromethyl)benzenesulfonyl]benzamide was imported into Maestro workspace v.9.3 (Schrödinger, LCC, New York, NY, USA, 2012) and processed using the Protein Preparation Wizard [83,84]. In this step, bond orders and formal charges were adjusted, while hydrogen atoms were added to the protein. The protonation states (pKa) of polar amino acid residues were predicted by the Epik program [85,86] at pH = 7.4 ± 0.5, whereas the OPLS-2005 force field was used to minimize the energy of the 3D structure. In parallel, protonation states and 3D geometric optimization of prioritized compounds were predicted using LigPrep software [84,87] at pH = 7.4 ± 0.5.
3.1.5. Molecular Docking
Molecular docking studies were performed using the DockThor VS webserver [88,89]. The grid box was centered at the x, y, and z coordinates of the co-crystallized ligand bound to the N-pocket site. The search algorithm precision mode was set up as the standard configuration of genetic algorithm parameters, and the soft docking mode was activated. At the end of the docking procedure, we used the PLIP server [90] to analyze the intermolecular interaction patterns of the docking poses (hydrogen bonds, hydrophobic interaction, cation-π, π-stacking, water and salt bridge interactions). Finally, the binding mode of the ligands obtained was compared to that of co-crystallized ligand (PDB ID: 6LD4). Then, the Pymol software v. 2.4 [75] was used for visual inspection and to render the pose images.
3.2. Experimental
3.2.1. Quercetin and Pedalitin
Quercetin and pedalitin were obtained from Pterogyne nitens, a medicinal Brazilian tree, according to our previous phytochemical procedures [91].
3.2.2. Protein Cloning, Expression and Purification
ZIKV NS5 RdRp polymerase was cloned at pETTRX by the LIC method and expressed and purified according to the protocol described in [92]. Briefly, NS5 RdRp polymerase was expressed in ZYM 5052 auto-induction medium and purified in four steps: (i) a HisTrap HP 5.0 mL with a Ni Sepharose resin (GE Healthcare, Sao Carlos, Brazil); (ii) a buffer exchanged by dialysis and a concomitant TEV protease cleavage from 6His-TRX-tag; (iii) an inverse HisTrap HP 5.0 mL to separate protein from 6His-TRX-tag and (iv) a size-exclusion chromatography at a XK 16/60 Superdex 75 column (GE Healthcare, Sao Carlos, Brazil).
3.2.3. NS5 RdRp Activity Assays
ZIKV NS5-RdRp activity assays were performed as described by Fernandes and coworkers [93]. The endpoint assays were performed at 20 μM, and the compounds that inhibited more than 80% of activity in this assay were submitted to a concentration-response test. The concentration-responses assays were performed as described in [94]. In all cases, the percentage inhibition values were calculated based on a control reaction, containing only DMSO in the same concentrations used for the tested compounds. The results were analyzed and plotted using the GraphPad Prism v. 8.0 program [95].
3.3. Cell Culture
Vero cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Sigma–Aldrich, MO, USA) supplemented with 100 U/mL penicillin (Gibco Life Technologies, Paisley, UK), 100 mg/mL streptomycin (Gibco Life Technologies, Paisley, UK), 1% (v/v) non-essential amino acids (Gibco Life Technologies, Paisley, UK) and 10% (v/v) fetal bovine serum (FBS; Hyclone, UT, USA) at 37 °C in a humidified 5% CO2 incubator.
3.4. Virus Rescue and Titration
A wild-type ZIKV isolate from a clinical patient in Brazil (ZIKVBR, PA, Brazil) was provided by the Evandro Chagas Institute in Belém, Pará [96]. The virus was amplified employing Vero cells in a 175 cm2 flask. To determine viral titers, 1 × 104 Vero cells were seeded in each of 24 wells plate 24 h prior to the infection. Cells were infected with 10-fold serially dilutions of ZIKVBR for 1 h at 37 °C and then supplemented with medium containing 1% penicillin, 1% streptomycin, 2% FBS and 1% carboxymethyl cellulose (CMC). Infected cells were incubated for seven days in a humidified 5% CO2 incubator at 37 °C, followed by fixation with 4% formaldehyde and staining with 0.5% violet crystal. The viral foci were counted to determine viral titers which were expressed in plaque formation unit per milliliters (PFU/mL).
3.5. Cell Viabillity
Cell viability was measured by the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] (Sigma–Aldrich) method. Vero cells were seeded in a 96-well plate at a density of 5 × 103 cells per well and incubated overnight at 37 °C in a humidified 5% CO2 incubator. A drug-containing medium at concentrations ranging from 200 to 0.005 µM was added to the cell culture. After 72 h at 37 °C, the media was removed and a solution containing MTT at the final concentration of 1 mg/mL was added to each well and incubated for 30 min at 37 °C in a humidified 5% CO2 incubator, after which media was replaced with 100 μL of DMSO to solubilize the formazan crystals. Absorbance was measured by the optical density (OD) of each well at 490 nm, using a spectrophotometer. Cell viability was calculated according to the equation (T/C) × 100%, where T and C represent the mean optical density of the treated group and vehicle control group, respectively. The cytotoxic concentration of 50% (CC50) was calculated using Graph Pad Prism v. 8 [95].
3.6. Antiviral Assays
Vero cells were seeded at density of 5 × 103 cells per well into 96-well plates 24 h prior to the infection. ZIKV-WTBR at a multiplicity of infection (MOI) of 0.1 and compound at concentrations ranging from 200 to 0.005 µM were simultaneously added to cells. 72 h post-infection (h.p.i.), cells were fixed with paraformaldehyde 4%, washed with PBS and blocking buffer (BB) containing: 0.1% Triton X-100 (Vetec Labs, PR, BR), 0.2% bovine albumin (BSA) and PBS for 30 min. Then, cells were incubated with primary rabbit polyclonal anti-NS3 antibody diluted in BB for 1 h. Alexa Fluor 488 conjugated anti-rabbit IgG was used as secondary antibody (Abcam, Cambridge, UK). Images were analyzed by EVO cell imaging systems fluorescence microscopy (Thermo Fisher Scientific, OH, USA) and foci of infection were counted. The antiviral activity was calculated according to the equation (T/C) × 100%, where T and C represent the mean of the treated group and mean of the last concentration, respectively. The effective concentration of 50% inhibition (EC50) was calculated using Graph Pad Prism v. 8. The values of CC50 and EC50 were used to calculate the selectivity index (SI = CC50/EC50).
4. Conclusions
Despite the severe neurological consequences caused by ZIKV infection, there is still no antiviral for the treatment of ZIKV, and only a few ZIKV NS5 RdRp inhibitors have been described in the literature. In our study, guided by known DENV NS5 RdRp inhibitors, through binding site conservation and chemical space analysis as well as docking calculations we prioritized and identified the flavonoids pedalitin and quercetin as new inhibitors of ZIKV NS5 RdRp. Enzymatic assays reinforced the computational results, and both compounds presented antiviral activity against ZIKV in infected cell cultures. Therefore, quercetin and pedalitin may be promising candidates for hit-to-lead optimization, boosting the discovery of new anti-ZIKV drug candidates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ph15121493/s1. Figure S1: ZIKV NS5 RdRp enzymatic assays. Concentration-response curves adjusted with Hill to determine IC50 ± Δ IC50 values for (A) pedalitin and (B) quercetin. Figure S2: Superposition of the co-crystallized 5-(3-fluorothiophen-2-yl)-2-hydroxy-4-methoxy-N-[4-(trifluoromethyl)benzenesulfonyl]benzamide compound in crystal (C atoms are represented in blue) and the redocking pose (C atoms are represented in green) at the ZIKV NS5 RdRP structure (PDB ID 6LD4 [21]). Table S1: Docking results for all compounds selected by chemical space analysis.
Author Contributions
P.R.P.d.S.R. and M.M. prepared proteins, ligands, binding sites, and submitted docking calculations. P.R.P.d.S.R. and M.M. performed the dockings. V.A.F.C. and B.J.N. performed the binding site conservation analysis. P.R.P.d.S.R., J.V.V.B.B., M.M. and C.H.A. analyzed and curated the data, as well as selected the compounds. C.S.L. and L.R.A. were responsible for isolation and identification of quercetin of pedalitin from Pterogyne nitens. N.C.d.M.R.M., K.Z.d.O., R.V.C.G. and G.O. performed and analyzed biophysical, enzymatic assays. N.M.C., I.A.S. and A.C.G.J. performed cytotoxic and cell-based antiviral assays. The manuscript was written by P.R.P.d.S.R. and M.M. with contributions of all authors. Final editing was accomplished by P.R.P.d.S.R., M.M., L.O.R., B.J.N. and C.H.A. All authors have read and agreed to the published version of the manuscript.
Funding
MM and CHA thank the support of CNPq and FAPEG (grants 300508/2017-4, 471129/2013-5, 306251/2016-7, 429322/2018-6 and 20171026700006). This work has been funded by CNPq (grants 300508/2017-4 and 150759/2017-7), CNPq BRICS STI COVID-19 (grant 441038/2020-4), FAPEG (grants 20171026700006 and 202010267000272), FAPESP (CEPID CIBFar grant 2013/07600-3, 2014/18330-0, 2018/15083-2, 2019/01762-8 and 2020/12904-5), CAPES (Finance Code 001). NMC thanks Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq; scholarships # 133956/2020-2 and # 142495/2020–4, respectively). IAS thanks to CNPq for the scholarship # 142495/2020-4 and CAPES.Print scholarship # 88887.700246/2022-00. ACGJ is grateful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)—Brazil -Prevention and Combat of Outbreaks, Endemics, Epidemics and Pandemics—Finance Code #88881.506794/2020–01 and to FAPEMIG (Minas Gerais Research Foundation APQ-03385–18 and APQ-01487-22).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article and Supplementary Material.
Acknowledgments
The authors would like to acknowledge Brazilian funding agencies, FAPEG, FAPESP, CNPq, FAPEMIG, and CAPES for the financial support and fellowships. CHA also thanks the “L’Oréal-UNESCO-ABC Para Mulheres na Ciência” and “L’Oréal-UNESCO International Rising Talents” for the awards and fellowships received, which partially funded this work. CHA, RVCG, ACGJ, and GO are CNPq research fellows. We sincerely thank the OpenZika project, the collaborators, and volunteers on this project as well as the support from IBM’s World Community Grid team.
Conflicts of Interest
The authors declare no competing interests.
Abbreviations
DENV: Dengue virus; HCV, Hepatitis C virus; NI, nucleoside and nucleotide inhibitor; NNI, non-nucleoside inhibitor; capsid; M, membrane; E, envelope; NS, nonstructural proteins; RdRp, RNA-dependent RNA-polymerase; ZIKV, Zika virus.
References
- World Health Organization Zika Virus Outbreaks in the Americas. Relevé Épidémiologique Hebdomadaire/Section D’hygiène du Secrétariat de La Société des Nations = Weekly Epidemiological Record/Health Section of the Secretariat of the League of Nations; World Health Organization: Washington, DC, USA, 2015; Volume 90, pp. 609–610. [Google Scholar]
- Teixeira, G.A.; Dantas, D.N.A.; Carvalho, G.A.F.d.L.; da Silva, A.N.; Lira, A.L.B.d.C.; Enders, B.C. Analysis of the Concept of the Zika Virus Congenital Syndrome. Cienc. Saude Coletiva 2020, 25, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Dirlikov, E.; Major, C.G.; Mayshack, M.; Medina, N.; Matos, D.; Ryff, K.R.; Torres-Aponte, J.; Alkis, R.; Munoz-Jordan, J.; Colon-Sanchez, C.; et al. Guillain-Barré Syndrome during Ongoing Zika Virus Transmission—Puerto Rico, January 1–July 31, 2016. Morb. Mortal. Wkly. Rep. 2016, 65, 910–914. [Google Scholar] [CrossRef] [PubMed]
- Regla-Nava, J.A.; Wang, Y.-T.; Fontes-Garfias, C.R.; Liu, Y.; Syed, T.; Susantono, M.; Gonzalez, A.; Viramontes, K.M.; Verma, S.K.; Kim, K.; et al. A Zika Virus Mutation Enhances Transmission Potential and Confers Escape from Protective Dengue Virus Immunity. Cell Rep. 2022, 39, 110655. [Google Scholar] [CrossRef] [PubMed]
- Mottin, M.; Borba, J.V.V.B.; Braga, R.C.; Torres, P.H.M.; Martini, M.C.; Proenca-Modena, J.L.; Judice, C.C.; Costa, F.T.M.; Ekins, S.; Perryman, A.L.; et al. The A–Z of Zika Drug Discovery. Drug Discov. Today 2018, 23, 1833–1847. [Google Scholar] [CrossRef] [PubMed]
- Malet, H.; Massé, N.; Selisko, B.; Romette, J.-L.; Alvarez, K.; Guillemot, J.C.; Tolou, H.; Yap, T.L.; Vasudevan, S.G.; Lescar, J.; et al. The Flavivirus Polymerase as a Target for Drug Discovery. Antiviral Res. 2008, 80, 23–35. [Google Scholar] [CrossRef]
- Noble, C.G.; Lim, S.P.; Chen, Y.-L.; Liew, C.W.; Yap, L.; Lescar, J.; Shi, P.-Y. Conformational Flexibility of the Dengue Virus RNA-Dependent RNA Polymerase Revealed by a Complex with an Inhibitor. J. Virol. 2013, 87, 5291–5295. [Google Scholar] [CrossRef]
- Lim, S.P.; Noble, C.G.; Seh, C.C.; Soh, T.S.; El Sahili, A.; Chan, G.K.Y.; Lescar, J.; Arora, R.; Benson, T.; Nilar, S.; et al. Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling. PLoS Pathog. 2016, 12, e1005737. [Google Scholar] [CrossRef]
- Asselah, T. Sofosbuvir for the Treatment of Hepatitis C Virus. Expert Opin. Pharmacother. 2014, 15, 121–130. [Google Scholar] [CrossRef]
- Sacramento, C.Q.; de Melo, G.R.; de Freitas, C.S.; Rocha, N.; Hoelz, L.V.B.; Miranda, M.; Fintelman-Rodrigues, N.; Marttorelli, A.; Ferreira, A.C.; Barbosa-Lima, G.; et al. The Clinically Approved Antiviral Drug Sofosbuvir Inhibits Zika Virus Replication. Sci. Rep. 2017, 7, 40920. [Google Scholar] [CrossRef]
- Marshall, G.R. Computer-Aided Drug Design. Annu. Rev. Pharmacol. Toxicol. 1987, 27, 193–213. [Google Scholar] [CrossRef]
- Reynolds, C. Impact of Computational Structure-Based Methods on Drug Discovery. Curr. Pharm. Des. 2014, 20, 3380–3386. [Google Scholar] [CrossRef] [PubMed]
- Andrade, C.H.; Neves, B.J.; Melo-Filho, C.C.; Rodrigues, J.; Silva, D.C.; Braga, R.C.; Cravo, P.V.L. In Silico Chemogenomics Drug Repositioning Strategies for Neglected Tropical Diseases. Curr. Med. Chem. 2019, 26, 4355–4379. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazy, H.; Erez, E.; Martz, E.; Pupko, T.; Ben-Tal, N. ConSurf 2010: Calculating Evolutionary Conservation in Sequence and Structure of Proteins and Nucleic Acids. Nucleic Acids Res. 2010, 38, W529–W533. [Google Scholar] [CrossRef] [PubMed]
- Celniker, G.; Nimrod, G.; Ashkenazy, H.; Glaser, F.; Martz, E.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Isr. J. Chem. 2013, 53, 199–206. [Google Scholar] [CrossRef]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T.; Ben-Tal, N. ConSurf 2016: An Improved Methodology to Estimate and Visualize Evolutionary Conservation in Macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Noble, C.G.; Lim, S.P.; Arora, R.; Yokokawa, F.; Nilar, S.; Seh, C.C.; Wright, S.K.; Benson, T.E.; Smith, P.W.; Shi, P.-Y. A Conserved Pocket in the Dengue Virus Polymerase Identified through Fragment-Based Screening. J. Biol. Chem. 2016, 291, 8541–8548. [Google Scholar] [CrossRef]
- Duan, W.; Song, H.; Wang, H.; Chai, Y.; Su, C.; Qi, J.; Shi, Y.; Gao, G.F. The Crystal Structure of Zika Virus NS 5 Reveals Conserved Drug Targets. EMBO J. 2017, 36, 919–933. [Google Scholar] [CrossRef]
- Gharbi-Ayachi, A.; Santhanakrishnan, S.; Wong, Y.H.; Chan, K.W.K.; Tan, S.T.; Bates, R.W.; Vasudevan, S.G.; el Sahili, A.; Lescar, J. Non-Nucleoside Inhibitors of Zika Virus RNA-Dependent RNA Polymerase. J. Virol. 2020, 94, e00794-20. [Google Scholar] [CrossRef]
- Yin, Z.; Chen, Y.-L.; Kondreddi, R.R.; Chan, W.L.; Wang, G.; Ng, R.H.; Lim, J.Y.H.; Lee, W.Y.; Jeyaraj, D.A.; Niyomrattanakit, P.; et al. N-Sulfonylanthranilic Acid Derivatives as Allosteric Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase. J. Med. Chem. 2009, 52, 7934–7937. [Google Scholar] [CrossRef]
- Allard, P.-M.; Dau, E.T.H.; Eydoux, C.; Guillemot, J.-C.; Dumontet, V.; Poullain, C.; Canard, B.; Guéritte, F.; Litaudon, M. Alkylated Flavanones from the Bark of Cryptocarya Chartacea as Dengue Virus NS5 Polymerase Inhibitors. J. Nat. Prod. 2011, 74, 2446–2453. [Google Scholar] [CrossRef]
- Bourjot, M.; Leyssen, P.; Eydoux, C.; Guillemot, J.-C.; Canard, B.; Rasoanaivo, P.; Guéritte, F.; Litaudon, M. Flacourtosides A-F, Phenolic Glycosides Isolated from Flacourtia Ramontchi. J. Nat. Prod. 2012, 75, 752–758. [Google Scholar] [CrossRef] [PubMed]
- Benmansour, F.; Eydoux, C.; Querat, G.; de Lamballerie, X.; Canard, B.; Alvarez, K.; Guillemot, J.-C.; Barral, K. Novel 2-Phenyl-5-[(E)-2-(Thiophen-2-Yl)Ethenyl]-1,3,4-Oxadiazole and 3-Phenyl-5-[(E)-2-(Thiophen-2-Yl)Ethenyl]-1,2,4-Oxadiazole Derivatives as Dengue Virus Inhibitors Targeting NS5 Polymerase. Eur. J. Med. Chem. 2016, 109, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Yokokawa, F.; Nilar, S.; Noble, C.G.; Lim, S.P.; Rao, R.; Tania, S.; Wang, G.; Lee, G.; Hunziker, J.; Karuna, R.; et al. Discovery of Potent Non-Nucleoside Inhibitors of Dengue Viral RNA-Dependent RNA Polymerase from a Fragment Hit Using Structure-Based Drug Design. J. Med. Chem. 2016, 59, 3935–3952. [Google Scholar] [CrossRef] [PubMed]
- Cannalire, R.; Tarantino, D.; Astolfi, A.; Barreca, M.L.; Sabatini, S.; Massari, S.; Tabarrini, O.; Milani, M.; Querat, G.; Mastrangelo, E.; et al. Functionalized 2,1-Benzothiazine 2,2-Dioxides as New Inhibitors of Dengue NS5 RNA-Dependent RNA Polymerase. Eur. J. Med. Chem. 2018, 143, 1667–1676. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Lim, S.P.; Chen, Y.-L.; Hunziker, J.; Rao, R.; Gu, F.; Seh, C.C.; Ghafar, N.A.; Xu, H.; Chan, K.; et al. Structure-Activity Relationship of Uridine-Based Nucleoside Phosphoramidate Prodrugs for Inhibition of Dengue Virus RNA-Dependent RNA Polymerase. Bioorganic Med. Chem. Lett. 2018, 28, 2324–2327. [Google Scholar] [CrossRef]
- Cannalire, R.; Chan, K.W.K.; Burali, M.S.; Gwee, C.P.; Wang, S.; Astolfi, A.; Massari, S.; Sabatini, S.; Tabarrini, O.; Mastrangelo, E.; et al. Pyridobenzothiazolones Exert Potent Anti-Dengue Activity by Hampering Multiple Functions of NS5 Polymerase. ACS Med. Chem. Lett. 2020, 11, 773–782. [Google Scholar] [CrossRef]
- Peyrat, L.-A.; Eparvier, V.; Eydoux, C.; Guillemot, J.-C.; Litaudon, M.; Stien, D. Carneic Acids from an Endophytic Phomopsis sp. as Dengue Virus Polymerase Inhibitors. J. Nat. Prod. 2020, 83, 2330–2336. [Google Scholar] [CrossRef]
- Felicetti, T.; Burali, M.S.; Gwee, C.P.; Ki Chan, K.W.; Alonso, S.; Massari, S.; Sabatini, S.; Tabarrini, O.; Barreca, M.L.; Cecchetti, V.; et al. Sustainable, Three-Component, One-Pot Procedure to Obtain Active Anti-Flavivirus Agents. Eur. J. Med. Chem. 2021, 210, 112992. [Google Scholar] [CrossRef]
- Coulerie, P.; Nour, M.; Maciuk, A.; Eydoux, C.; Guillemot, J.-C.; Lebouvier, N.; Hnawia, E.; Leblanc, K.; Lewin, G.; Canard, B.; et al. Structure-Activity Relationship Study of Biflavonoids on the Dengue Virus Polymerase DENV-NS5 RdRp. Planta Med. 2013, 79, 1313–1318. [Google Scholar] [CrossRef]
- Coulerie, P.; Eydoux, C.; Hnawia, E.; Stuhl, L.; Maciuk, A.; Lebouvier, N.; Canard, B.; Figadère, B.; Guillemot, J.-C.; Nour, M. Biflavonoids of Dacrydium Balansae with Potent Inhibitory Activity on Dengue 2 NS5 Polymerase. Planta Med. 2012, 78, 672–677. [Google Scholar] [CrossRef]
- Bourjot, M.; Leyssen, P.; Eydoux, C.; Guillemot, J.-C.; Canard, B.; Rasoanaivo, P.; Guéritte, F.; Litaudon, M. Chemical Constituents of Anacolosa Pervilleana and Their Antiviral Activities. Fitoterapia 2012, 83, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Manvar, D.; Küçükgüzel, İ.; Erensoy, G.; Tatar, E.; Deryabaşoğulları, G.; Reddy, H.; Talele, T.T.; Cevik, O.; Kaushik-Basu, N. Discovery of Conjugated Thiazolidinone-Thiadiazole Scaffold as Anti-Dengue Virus Polymerase Inhibitors. Biochem. Biophys. Res. Commun. 2016, 469, 743–747. [Google Scholar] [CrossRef] [PubMed]
- Niyomrattanakit, P.; Chen, Y.-L.; Dong, H.; Yin, Z.; Qing, M.; Glickman, J.F.; Lin, K.; Mueller, D.; Voshol, H.; Lim, J.Y.H.; et al. Inhibition of Dengue Virus Polymerase by Blocking of the RNA Tunnel. J. Virol. 2010, 84, 5678–5686. [Google Scholar] [CrossRef] [PubMed]
- Venkatesham, A.; Saudi, M.; Kaptein, S.; Neyts, J.; Rozenski, J.; Froeyen, M.; Van Aerschot, A. Aminopurine and Aminoquinazoline Scaffolds for Development of Potential Dengue Virus Inhibitors. Eur. J. Med. Chem. 2017, 126, 101–109. [Google Scholar] [CrossRef]
- Kaushik, S.; Dar, L.; Kaushik, S.; Yadav, J.P. Identification and Characterization of New Potent Inhibitors of Dengue Virus NS5 Proteinase from Andrographis Paniculata Supercritical Extracts on in Animal Cell Culture and in Silico Approaches. J. Ethnopharmacol. 2021, 267, 113541. [Google Scholar] [CrossRef]
- Mottin, M.; Caesar, L.K.; Brodsky, D.; Mesquita, N.C.M.R.; de Oliveira, K.Z.; Noske, G.D.; Sousa, B.K.P.; Ramos, P.R.P.S.; Jarmer, H.; Loh, B.; et al. Chalcones from Angelica Keiskei (Ashitaba) Inhibit Key Zika Virus Replication Proteins. Bioorganic Chem. 2022, 120, 105649. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, H.; Song, W.; Si, S.; Han, Y.; Jiang, J. Identification and Characterization of Zika Virus NS5 RNA-Dependent RNA Polymerase Inhibitors. Int. J. Antimicrob. Agents 2019, 54, 502–506. [Google Scholar] [CrossRef]
- Yang, S.; Xu, M.; Lee, E.M.; Gorshkov, K.; Shiryaev, S.A.; He, S.; Sun, W.; Cheng, Y.-S.; Hu, X.; Tharappel, A.M.; et al. Emetine Inhibits Zika and Ebola Virus Infections through Two Molecular Mechanisms: Inhibiting Viral Replication and Decreasing Viral Entry. Cell Discov. 2018, 4, 31. [Google Scholar] [CrossRef]
- Bernatchez, J.A.; Tran, L.T.; Li, J.; Luan, Y.; Siqueira-Neto, J.L.; Li, R. Drugs for the Treatment of Zika Virus Infection. J. Med. Chem. 2020, 63, 470–489. [Google Scholar] [CrossRef]
- Vincetti, P.; Kaptein, S.J.F.; Costantino, G.; Neyts, J.; Radi, M. Scaffold Morphing Approach To Expand the Toolbox of Broad-Spectrum Antivirals Blocking Dengue/Zika Replication. ACS Med. Chem. Lett. 2019, 10, 558–563. [Google Scholar] [CrossRef]
- Hinton, G.; Roweis, S. Stochastic Neighbor Embedding. Adv. Neural Inf. Process. Syst. 2003. [Google Scholar]
- Hopkins, A.L.; Groom, C.R.; Alex, A. Ligand Efficiency: A Useful Metric for Lead Selection. Drug Discov. Today 2004, 9, 430–431. [Google Scholar] [CrossRef]
- Kenny, P.W. The Nature of Ligand Efficiency. J. Cheminform. 2019, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Abad-Zapatero, C. Ligand Efficiency Indices for Effective Drug Discovery. Expert Opin. Drug Discov. 2007, 2, 469–488. [Google Scholar] [CrossRef]
- Hevener, K.E.; Pesavento, R.; Ren, J.; Lee, H.; Ratia, K.; Johnson, M.E. Hit-to-Lead: Hit Validation and Assessment. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2018; pp. 265–309. [Google Scholar]
- Neves, B.J.; Mottin, M.; Moreira-Filho, J.T.; De, B.K.; Sousa, P.; Mendonça, S.S.; Andrade, C.H. Best Practices for Docking-Based Virtual Screening. In Molecular Docking for Computer-Aided Drug Design; Elsevier: Amsterdam, The Netherlands, 2021; pp. 75–98. ISBN 9780128223123. [Google Scholar]
- Mottin, M.; de Paula Sousa, B.K.; de Moraes Roso Mesquita, N.C.; de Oliveira, K.I.Z.; Noske, G.D.; Sartori, G.R.; de Oliveira Albuquerque, A.; Urbina, F.; Puhl, A.C.; Moreira-Filho, J.T.; et al. Discovery of New Zika Protease and Polymerase Inhibitors through the Open Science Collaboration Project OpenZika. J. Chem. Inf. Model. 2022. [Google Scholar] [CrossRef]
- Nascimento, I.J.d.S.; Santos-Júnior, P.F.d.S.; de Aquino, T.M.; de Araújo-Júnior, J.X.; da Silva-Júnior, E.F. Insights on Dengue and Zika NS5 RNA-Dependent RNA Polymerase (RdRp) Inhibitors. Eur. J. Med. Chem. 2021, 224, 113698. [Google Scholar] [CrossRef]
- Campos, G.R.F.; Bittar, C.; Jardim, A.C.G.; Shimizu, J.F.; Batista, M.N.; Paganini, E.R.; de Assis, L.R.; Bartlett, C.; Harris, M.; Bolzani, V.d.S.; et al. Hepatitis C Virus in Vitro Replication Is Efficiently Inhibited by Acridone Fac4. J. Gen. Virol. 2017, 98, 1693–1701. [Google Scholar] [CrossRef]
- Sasvari, Z.; Bach, S.; Blondel, M.; Nagy, P.D. Inhibition of RNA Recruitment and Replication of an RNA Virus by Acridine Derivatives with Known Anti-Prion Activities. PLoS ONE 2009, 4, e7376. [Google Scholar] [CrossRef][Green Version]
- Sepúlveda, C.S.; Fascio, M.L.; Mazzucco, M.B.; Palacios, M.L.D.; Pellón, R.F.; García, C.C.; D’Accorso, N.B.; Damonte, E.B. Synthesis and Evaluation of N -Substituted Acridones as Antiviral Agents against Haemorrhagic Fever Viruses. Antivir. Chem. Chemother. 2008, 19, 41–47. [Google Scholar] [CrossRef]
- Arora, R.; Liew, C.W.; Soh, T.S.; Otoo, D.A.; Seh, C.C.; Yue, K.; Nilar, S.; Wang, G.; Yokokawa, F.; Noble, C.G.; et al. Two RNA Tunnel Inhibitors Bind in Highly Conserved Sites in Dengue Virus NS5 Polymerase: Structural and Functional Studies. J. Virol. 2020, 94, e01130-20. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Yokokawa, F.; Shi, P.-Y. The Search for Nucleoside/Nucleotide Analog Inhibitors of Dengue Virus. Antivir. Res. 2015, 122, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Sofia, M.J.; Chang, W.; Furman, P.A.; Mosley, R.T.; Ross, B.S. Nucleoside, Nucleotide, and Non-Nucleoside Inhibitors of Hepatitis C Virus NS5B RNA-Dependent RNA-Polymerase. J. Med. Chem. 2012, 55, 2481–2531. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.-T.; Colby-Germinario, S.P.; Hassounah, S.A.; Fogarty, C.; Osman, N.; Palanisamy, N.; Han, Y.; Oliveira, M.; Quan, Y.; Wainberg, M.A. Evaluation of Sofosbuvir (β-D-2′-Deoxy-2′-α-Fluoro-2′-β-C-Methyluridine) as an Inhibitor of Dengue Virus Replication #. Sci. Rep. 2017, 7, 6345. [Google Scholar] [CrossRef] [PubMed]
- Pattnaik, A.; Palermo, N.; Sahoo, B.R.; Yuan, Z.; Hu, D.; Annamalai, A.S.; Vu, H.L.X.; Correas, I.; Prathipati, P.K.; Destache, C.J.; et al. Discovery of a Non-Nucleoside RNA Polymerase Inhibitor for Blocking Zika Virus Replication through in Silico Screening. Antivir. Res. 2018, 151, 78–86. [Google Scholar] [CrossRef]
- Munafò, F.; Donati, E.; Brindani, N.; Ottonello, G.; Armirotti, A.; De Vivo, M. Quercetin and Luteolin Are Single-Digit Micromolar Inhibitors of the SARS-CoV-2 RNA-Dependent RNA Polymerase. Sci. Rep. 2022, 12, 10571. [Google Scholar] [CrossRef]
- Oo, A.; Teoh, B.T.; Sam, S.S.; Bakar, S.A.; Zandi, K. Baicalein and Baicalin as Zika Virus Inhibitors. Arch. Virol. 2019, 164, 585–593. [Google Scholar] [CrossRef]
- Badshah, S.L.; Faisal, S.; Muhammad, A.; Poulson, B.G.; Emwas, A.H.; Jaremko, M. Antiviral Activities of Flavonoids. Biomed. Pharmacother. 2021, 140, 111596. [Google Scholar] [CrossRef]
- Moghaddam, E.; Teoh, B.-T.; Sam, S.-S.; Lani, R.; Hassandarvish, P.; Chik, Z.; Yueh, A.; Abubakar, S.; Zandi, K. Baicalin, a Metabolite of Baicalein with Antiviral Activity against Dengue Virus. Sci. Rep. 2015, 4, 5452. [Google Scholar] [CrossRef]
- Fong, Y.D.; Chu, J.J.H. Natural Products as Zika Antivirals. Med. Res. Rev. 2022. [Google Scholar] [CrossRef]
- Zandi, K.; Teoh, B.-T.; Sam, S.-S.; Wong, P.-F.; Mustafa, M.R.; AbuBakar, S. Antiviral Activity of Four Types of Bioflavonoid against Dengue Virus Type-2. Virol. J. 2011, 8, 560. [Google Scholar] [CrossRef]
- Hoenen, T.; Groseth, A. Virus–Host Cell Interactions. Cells 2022, 11, 804. [Google Scholar] [CrossRef] [PubMed]
- Amsden, H.; Kourko, O.; Roth, M.; Gee, K. Antiviral Activities of Interleukin-27: A Partner for Interferons? Front. Immunol. 2022, 13, 902853. [Google Scholar] [CrossRef] [PubMed]
- Iannello, A.; Debbeche, O.; Martin, E.; Attalah, L.H.; Samarani, S.; Ahmad, A. Viral Strategies for Evading Antiviral Cellular Immune Responses of the Host. J. Leukoc. Biol. 2006, 79, 16–35. [Google Scholar] [CrossRef] [PubMed]
- Lubick, K.J.; Robertson, S.J.; McNally, K.L.; Freedman, B.A.; Rasmussen, A.L.; Taylor, R.T.; Walts, A.D.; Tsuruda, S.; Sakai, M.; Ishizuka, M.; et al. Flavivirus Antagonism of Type I Interferon Signaling Reveals Prolidase as a Regulator of IFNAR1 Surface Expression. Cell Host Microbe 2015, 18, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Laurent-Rolle, M.; Morrison, J.; Rajsbaum, R.; Macleod, J.M.L.; Pisanelli, G.; Pham, A.; Ayllon, J.; Miorin, L.; Martínez-Romero, C.; TenOever, B.R.; et al. The Interferon Signaling Antagonist Function of Yellow Fever Virus NS5 Protein Is Activated by Type I Interferon. Cell Host Microbe 2014, 16, 314–327. [Google Scholar] [CrossRef]
- De Maio, F.A.; Risso, G.; Iglesias, N.G.; Shah, P.; Pozzi, B.; Gebhard, L.G.; Mammi, P.; Mancini, E.; Yanovsky, M.J.; Andino, R.; et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016, 12, e1005841. [Google Scholar] [CrossRef]
- Shugar, D. Viral and Host-Cell Protein KinasesEnticing Antiviral Targets and Relevance of Nucleoside, and Viral Thymidine, Kinases. Pharmacol. Ther. 1999, 82, 315–335. [Google Scholar] [CrossRef] [PubMed]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H. UniRef Clusters: A Comprehensive and Scalable Alternative for Improving Sequence Similarity Searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Kuma, K.; Miyata, T.; Toh, H. Improvement in the Accuracy of Multiple Sequence Alignment Program MAFFT. Genome Inform. 2005, 16, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Pupko, T.; Bell, R.E.; Mayrose, I.; Glaser, F.; Ben-Tal, N. Rate4Site: An Algorithmic Tool for the Identification of Functional Regions in Proteins by Surface Mapping of Evolutionary Determinants within Their Homologues. Bioinformatics 2002, 18, S71–S77. [Google Scholar] [CrossRef]
- Mayrose, I. Comparison of Site-Specific Rate-Inference Methods for Protein Sequences: Empirical Bayesian Methods Are Superior. Mol. Biol. Evol. 2004, 21, 1781–1791. [Google Scholar] [CrossRef] [PubMed]
- The PyMOL Molecular Graphics System, Version 1.8; Schrödinger, LCC: New York, NY, USA, 2015.
- Bateman, A. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, J.; Suzek, T.O.; Zhang, J.; Wang, J.; Zhou, Z.; Han, L.; Karapetyan, K.; Dracheva, S.; Shoemaker, B.A.; et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012, 40, D400-12. [Google Scholar] [CrossRef] [PubMed]
- Bolton, E.E.; Wang, Y.; Thiessen, P.A.; Bryant, S.H. PubChem: Integrated Platform of Small Molecules and Biological Activities. In Annual Reports in Computational Chemistry Volume 4; American Chemical Society: Washington, DC, USA, 2008; Volume 4, pp. 217–241. [Google Scholar]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound Databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. PubChem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef] [PubMed]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Landrum, G. Open-Source Cheminformatics. Available online: Https://Www.Rdkit.Org (accessed on 10 August 2022).
- Schrödinger Release 2015-2: Protein Preparation Wizard; Schrödinger, LLC: New York, NY, USA, 2015.
- Madhavi Sastry, G.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and Ligand Preparation: Parameters, Protocols, and Influence on Virtual Screening Enrichments. J. Comput. Aided Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef]
- Shelley, J.C.; Cholleti, A.; Frye, L.L.; Greenwood, J.R.; Timlin, M.R.; Uchimaya, M. Epik: A Software Program for PK a Prediction and Protonation State Generation for Drug-like Molecules. J. Comput. Aided Mol. Des. 2007, 21, 681–691. [Google Scholar] [CrossRef]
- Schrödinger Release 2015-2: Epik; Schrödinger, LLC: New York, NY, USA, 2015.
- Schrödinger Release 2015-2: LigPrep; Schrödinger, LLC: New York, NY, USA, 2015.
- Santos, K.B.; Guedes, I.A.; Karl, A.L.M.; Dardenne, L.E. Highly Flexible Ligand Docking: Benchmarking of the DockThor Program on the LEADS-PEP Protein–Peptide Data Set. J. Chem. Inf. Model. 2020, 60, 667–683. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto, A.M.S.; Marinho, D.; Krempser, E.; Kuenemann, M.A.; Sperandio, O.; Dardenne, L.E.; Miteva, M.A. New Machine Learning and Physics-Based Scoring Functions for Drug Discovery. Sci. Rep. 2021, 11, 3198. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully Automated Protein–Ligand Interaction Profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Lima, C.S.; Mottin, M.; de Assis, L.R.; Mesquita, N.C.d.M.R.; Sousa, B.K.d.P.; Coimbra, L.D.; Santos, K.B.; Zorn, K.M.; Guido, R.V.C.; Ekins, S.; et al. Flavonoids from Pterogyne Nitens as Zika Virus NS2B-NS3 Protease Inhibitors. Bioorganic Chem. 2021, 109, 104719. [Google Scholar] [CrossRef] [PubMed]
- Godoy, A.S.; Lima, G.M.A.; Oliveira, K.I.Z.; Torres, N.U.; Maluf, F.V.; Guido, R.V.C.; Oliva, G. Crystal Structure of Zika Virus NS5 RNA-Dependent RNA Polymerase. Nat. Commun. 2017, 8, 14764. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.S.; Noske, G.D.; Gawriljuk, V.O.; de Oliveira, K.I.Z.; Godoy, A.S.; Mesquita, N.C.M.R.; Oliva, G. High-Throughput Antiviral Assays to Screen for Inhibitors of Zika Virus Replication. J. Vis. Exp. Jove 2021, e62422. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, R.S.; de Godoy, A.S.; Santos, I.A.; Noske, G.D.; de Oliveira, K.I.Z.; Gawriljuk, V.O.; Gomes Jardim, A.C.; Oliva, G. Discovery of an Imidazonaphthyridine and a Riminophenazine as Potent Anti-Zika Virus Agents through a Replicon-Based High-Throughput Screening. Virus Res. 2021, 299, 198388. [Google Scholar] [CrossRef] [PubMed]
- GraphPad Prism, Version 8.0.0 for Windows; GraphPad Software: San Diego, CA, USA. Available online: www.graphpad.com (accessed on 25 March 2021).
- Cugola, F.R.; Fernandes, I.R.; Russo, F.B.; Freitas, B.C.; Dias, J.L.M.; Guimarães, K.P.; Benazzato, C.; Almeida, N.; Pignatari, G.C.; Romero, S.; et al. The Brazilian Zika Virus Strain Causes Birth Defects in Experimental Models. Nature 2016, 534, 267–271. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).