Epigenetic Mechanisms Involved in Cisplatin-Induced Nephrotoxicity: An Update
Abstract
1. Introduction
2. Induction Mechanism Cisplatin-Induced Nephrotoxicity
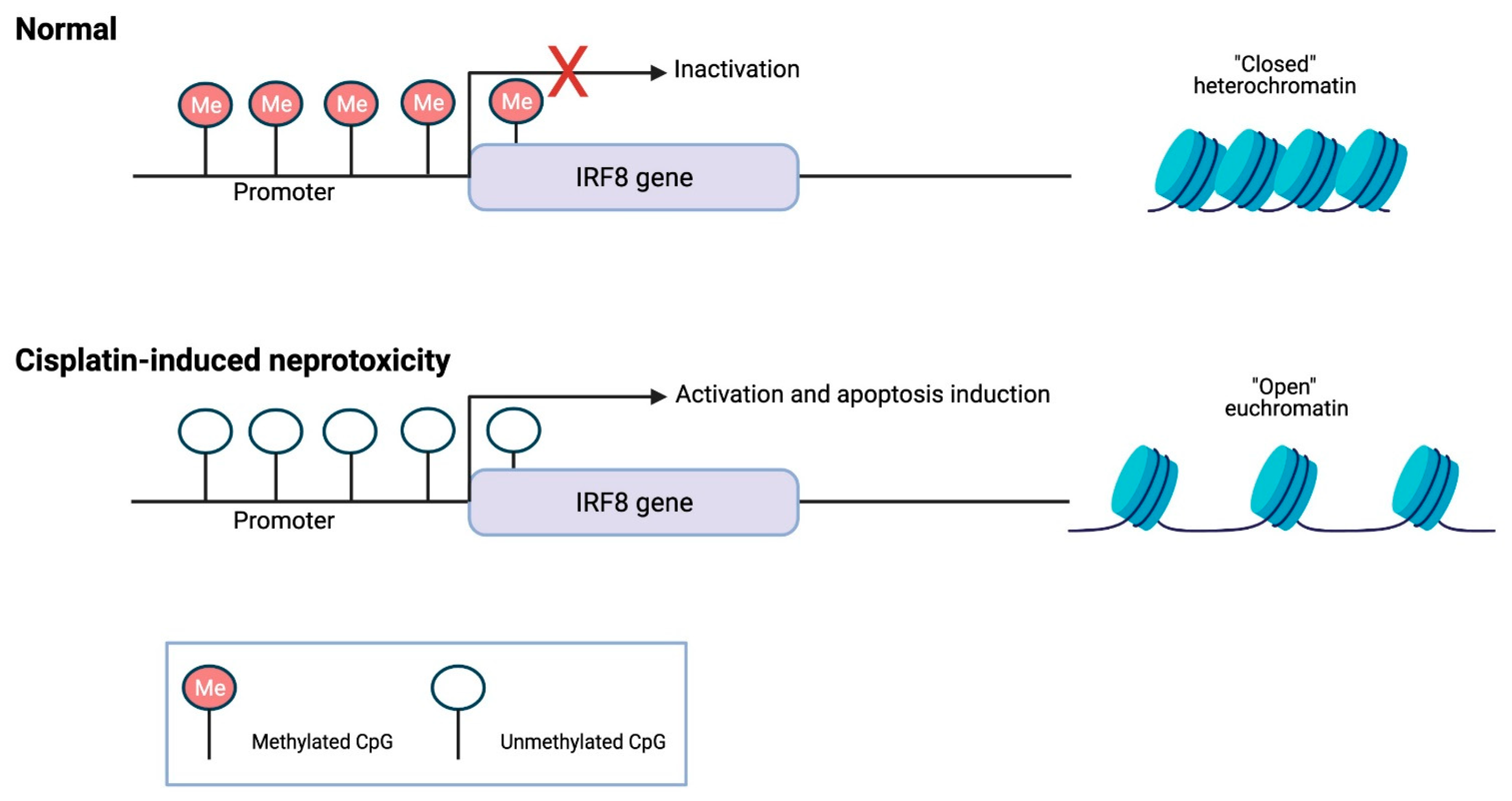
3. DNA Methylation Role in Cisplatin-Induced Renal Dysfunction
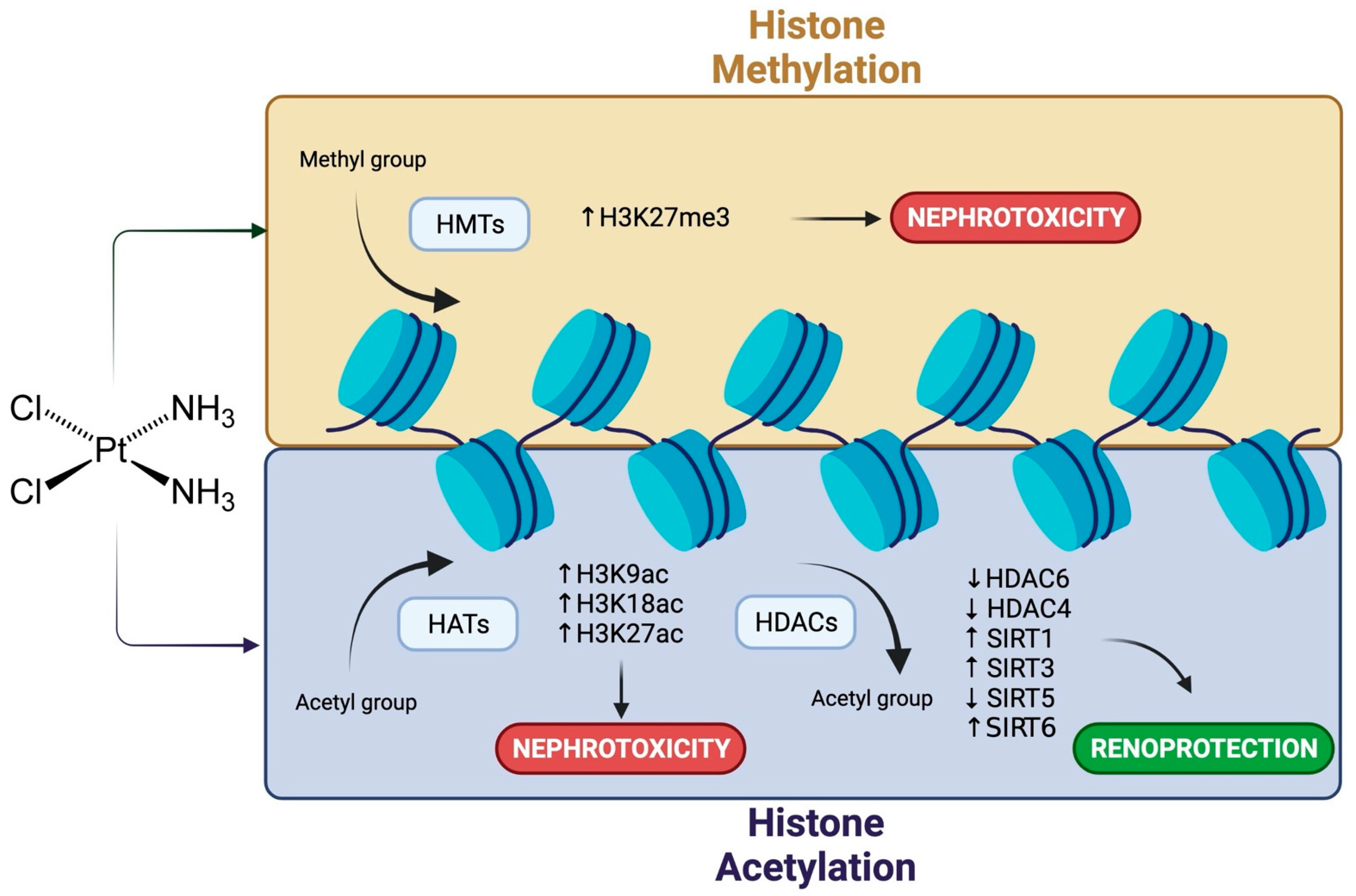
4. Cisplatin Nephrotoxicity and Histone Modifications
5. Involvement of Non-Coding RNAs in Cisplatin Nephrotoxicity
6. Potential Challenges and Future Directions
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO. World Health Statistics 2020: Monitoring Health for the SDGs; World Health Organization: Geneva, Switzerland, 2020; Volume 21, pp. 1–9. [Google Scholar] [CrossRef]
- Vermorken, J.B.; Remenar, E.; van Herpen, C.; Gorlia, T.; Mesia, R.; Degardin, M.; Stewart, J.S.; Jelic, S.; Betka, J.; Preiss, J.H.; et al. Cisplatin, fluorouracil, and docetaxel in unresectable head and neck cancer. N. Engl. J. Med. 2007, 357, 1695–1704. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Sheinfeld, J.; Mazumdar, M.; Bajorin, D.F.; Bosl, G.J.; Herr, H.; Lyn, P.; Vlamis, V. Etoposide and cisplatin adjuvant therapy for patients with pathologic stage II germ cell tumors. J. Clin. Oncol. 1995, 13, 2700–2704. [Google Scholar] [CrossRef]
- Li, D.; Zhang, Y.; Xie, Y.; Xiang, J.; Zhu, Y.; Yang, J. Enhanced tumor suppression by adenoviral PTEN gene therapy combined with cisplatin chemotherapy in small-cell lung cancer. Cancer Gene Ther. 2013, 20, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Magali, L.; Pascal, F.; Serge, A.; Mathieu, B.; Ayoube, Z.; Claire, T.; Christiane, M. Better survival in impaired renal function patients with metastatic non-small cell lung cancer treated by cisplatin-pemetrexed. Eur. J. Clin. Pharmacol. 2020, 76, 1573–1580. [Google Scholar] [CrossRef]
- Armstrong, D.K.; Bundy, B.; Wenzel, L.; Huang, H.Q.; Baergen, R.; Lele, S.; Copeland, L.J.; Walker, J.L.; Burger, R.A. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N. Engl. J. Med. 2006, 354, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Moore, K.N.; Herzog, T.J.; Lewin, S.; Giuntoli, R.L.; Armstrong, D.K.; Rocconi, R.P.; Spannuth, W.A.; Gold, M.A. A comparison of cisplatin/paclitaxel and carboplatin/paclitaxel in stage IVB, recurrent or persistent cervical cancer. Gynecol. Oncol. 2007, 105, 299–303. [Google Scholar] [CrossRef]
- Coppin, C.M.; Gospodarowicz, M.K.; James, K.; Tannock, I.F.; Zee, B.; Carson, J.; Pater, J.; Sullivan, L.D. Improved local control of invasive bladder cancer by concurrent cisplatin and preoperative or definitive radiation. The National Cancer Institute of Canada Clinical Trials Group. J. Clin. Oncol. 1996, 14, 2901–2907. [Google Scholar] [CrossRef]
- Miller, R.P.; Tadagavadi, R.K.; Ramesh, G.; Reeves, W.B. Mechanisms of Cisplatin nephrotoxicity. Toxins 2010, 2, 2490–2518. [Google Scholar] [CrossRef]
- Arany, I.; Safirstein, R.L. Cisplatin nephrotoxicity. Semin. Nephrol. 2003, 23, 460–464. [Google Scholar] [CrossRef]
- Al-Naimi, M.; Rasheed, H.; Hussien, N.; Al-Kuraishy, H.; Al-Gareeb, A. Nephrotoxicity: Role and significance of renal biomarkers in the early detection of acute renal injury. J. Adv. Pharm. Technol. Res. 2019, 10, 95–99. [Google Scholar] [CrossRef]
- Griffin, B.R.; Faubel, S.; Edelstein, C.L. Biomarkers of drug-induced kidney toxicity. Ther. Drug Monit. 2019, 41, 213–226. [Google Scholar] [CrossRef]
- Alfieri, A.B.; Cubeddu, L.X. Role of NK1 receptors on cisplatin-induced nephrotoxicity in the rat. Naunyn Schmiedebergs Arch. Pharmacol. 2000, 361, 334–338. [Google Scholar] [CrossRef]
- Zhu, S.; Pabla, N.; Tang, C.; He, L.; Dong, Z. DNA damage response in cisplatin-induced nephrotoxicity. Arch. Toxicol. 2015, 89, 2197–2205. [Google Scholar] [CrossRef]
- Cao, Y.; Mi, X.; Zhang, D.; Wang, Z.; Zuo, Y.; Tang, W. Transcriptome sequencing of circular RNA reveals a novel circular RNA-has_circ_0114427 in the regulation of inflammation in acute kidney injury. Clin. Sci. 2020, 134, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Morris, J.R. Genes, genetics, and epigenetics: A correspondence. Science 2001, 293, 1103–1105. [Google Scholar] [CrossRef]
- Calcagno, D.Q.; Gigek, C.O.; Chen, E.S.; Burbano, R.R.; Smith Mde, A. DNA and histone methylation in gastric carcinogenesis. World J. Gastroenterol. 2013, 19, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.K.; Ensing, B. Hydrolysis of cisplatin--a first-principles metadynamics study. Phys. Chem. Chem. Phys. 2010, 12, 10348–10355. [Google Scholar] [CrossRef]
- Galgamuwa, R. An Investigation Into The Prevention Of Cisplatin-Induced Nephrotoxicity By Dichloroacetate. Australian National University. Available online: https://openresearch-repository.anu.edu.au/ (accessed on 1 May 2021).
- Pabla, N.; Huang, S.; Mi, Q.S.; Daniel, R.; Dong, Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J. Biol. Chem. 2008, 283, 6572–6583. [Google Scholar] [CrossRef]
- Bhatt, K.; Zhou, L.; Mi, Q.S.; Huang, S.; She, J.X.; Dong, Z. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol. Med. 2010, 16, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Du, B.; Dai, X.M.; Li, S.; Qi, G.L.; Cao, G.X.; Zhong, Y.; Yin, P.D.; Yang, X.S. MiR-30c regulates cisplatin-induced apoptosis of renal tubular epithelial cells by targeting Bnip3L and Hspa5. Cell Death Dis. 2017, 8, e2987. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Ni, J.; Chen, S.; Bai, M.; Lin, J.; Ding, G.; Zhang, Y.; Sun, P.; Jia, Z.; Huang, S.; et al. MicroRNA-709 Mediates Acute Tubular Injury through Effects on Mitochondrial Function. J. Am. Soc. Nephrol. 2018, 29, 449–461. [Google Scholar] [CrossRef]
- Zou, D.; Ganugula, R.; Arora, M.; Nabity, M.B.; Sheikh-Hamad, D.; Kumar, M. Oral delivery of nanoparticle urolithin A normalizes cellular stress and improves survival in mouse model of cisplatin-induced AKI. Am. J. Physiol. Renal Physiol. 2019, 317, F1255–F1264. [Google Scholar] [CrossRef]
- Yang, Y.; Li, Z.L.; Wang, F.M.; Tang, R.N.; Tu, Y.; Liu, H. MicroRNA26a inhibits cisplatin-induced renal tubular epithelial cells apoptosis through suppressing the expression of transient receptor potential channel 6 mediated dynamin-related protein 1. Cell Biochem. Funct. 2020, 38, 384–391. [Google Scholar] [CrossRef]
- Harrill, A.H.; Lin, H.; Tobacyk, J.; Seely, J.C. Mouse population-based evaluation of urinary protein and miRNA biomarker performance associated with cisplatin renal injury. Exp. Biol. Med. 2018, 243, 237–247. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Z.; Chen, Z.; Li, C.; Lei, L.; Wu, X.; Li, Y. Numb ameliorates necrosis and inflammation in acute kidney injury induced by cisplatin. Chem. Biol. Interact. 2020, 330, 109251. [Google Scholar] [CrossRef]
- Ramesh, G.; Reeves, W.B. TNFR2-mediated apoptosis and necrosis in cisplatin-induced acute renal failure. Am. J. Physiol. Renal Physiol. 2003, 285, F610–F618. [Google Scholar] [CrossRef]
- Landau, S.I.; Guo, X.; Velazquez, H.; Torres, R.; Olson, E.; Garcia-Milian, R.; Moeckel, G.W.; Desir, G.V.; Safirstein, R. Regulated necrosis and failed repair in cisplatin-induced chronic kidney disease. Kidney Int. 2019, 95, 797–814. [Google Scholar] [CrossRef] [PubMed]
- Nosaka, K.; Nakada, J.; Endou, H. Cisplatin-induced alterations in renal structure, ammoniagenesis and gluconeogenesis of rats. Kidney Int. 1992, 41, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Qian, W.; Nishikawa, M.; Haque, A.M.; Hirose, M.; Mashimo, M.; Sato, E.; Inoue, M. Mitochondrial density determines the cellular sensitivity to cisplatin-induced cell death. Am. J. Physiol. Cell Physiol. 2005, 289, C1466–C1475. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Xie, W.; Yang, X.; Xia, N.; Yang, K. Inhibiting microRNA-449 Attenuates Cisplatin-Induced Injury in NRK-52E Cells Possibly via Regulating the SIRT1/P53/BAX Pathway. Med. Sci. Monit. 2016, 22, 818–823. [Google Scholar] [CrossRef]
- Zhu, H.Y.; Liu, M.Y.; Hong, Q.; Zhang, D.; Geng, W.J.; Xie, Y.S.; Chen, X.M. Role of microRNA-181a in the apoptosis of tubular epithelial cell induced by cisplatin. Chin. Med. J. 2012, 125, 523–526. [Google Scholar] [CrossRef]
- McDonald, E.S.; Windebank, A.J. Cisplatin-induced apoptosis of DRG neurons involves bax redistribution and cytochrome c release but not fas receptor signaling. Neurobiol. Dis. 2002, 9, 220–233. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Hou, X.; Wang, X.; Shi, Y.; Xu, L.; Zheng, X.; Liu, N.; Qiu, A.; Zhuang, S. 3-deazaneplanocin A protects against cisplatin-induced renal tubular cell apoptosis and acute kidney injury by restoration of E-cadherin expression. Cell Death Dis. 2019, 10, 355. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.G.; Kim, J.G.; Kim, H.J.; Kwon, H.K.; Cho, I.J.; Choi, D.W.; Lee, W.H.; Kim, W.D.; Hwang, S.J.; Choi, S.; et al. Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int. 2014, 86, 943–953. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tao, X.; Yin, L.; Xu, L.; Xu, Y.; Qi, Y.; Han, X.; Song, S.; Zhao, Y.; Lin, Y.; et al. Protective effects of dioscin against cisplatin-induced nephrotoxicity via the microRNA-34a/sirtuin 1 signalling pathway. Br. J. Pharmacol. 2017, 174, 2512–2527. [Google Scholar] [CrossRef]
- Yang, A.; Liu, F.; Guan, B.; Luo, Z.; Lin, J.; Fang, W.; Liu, L.; Zuo, W. p53 induces miR-199a-3p to suppress mechanistic target of rapamycin activation in cisplatin-induced acute kidney injury. J. Cell Biochem. 2019, 120, 17625–17634. [Google Scholar] [CrossRef] [PubMed]
- Suter-Dick, L.; Mauch, L.; Ramp, D.; Caj, M.; Vormann, M.K.; Hutter, S.; Lanz, H.L.; Vriend, J.; Masereeuw, R.; Wilmer, M.J. Combining Extracellular miRNA Determination with Microfluidic 3D Cell Cultures for the Assessment of Nephrotoxicity: A Proof of Concept Study. AAPS J. 2018, 20, 86. [Google Scholar] [CrossRef]
- Yang, C.; Kaushal, V.; Haun, R.S.; Seth, R.; Shah, S.V.; Kaushal, G.P. Transcriptional activation of caspase-6 and -7 genes by cisplatin-induced p53 and its functional significance in cisplatin nephrotoxicity. Cell Death Differ. 2008, 15, 530–544. [Google Scholar] [CrossRef]
- Sun, L.; Liu, J.; Yuan, Y.; Zhang, X.; Dong, Z. Protective effect of the BET protein inhibitor JQ1 in cisplatin-induced nephrotoxicity. Am. J. Physiol. Renal Physiol. 2018, 315, F469–F478. [Google Scholar] [CrossRef]
- Sun, C.Y.; Nie, J.; Zheng, Z.L.; Zhao, J.; Wu, L.M.; Zhu, Y.; Su, Z.Q.; Zheng, G.J.; Feng, B. Renoprotective effect of scutellarin on cisplatin-induced renal injury in mice: Impact on inflammation, apoptosis, and autophagy. Biomed. Pharmacother. 2019, 112, 108647. [Google Scholar] [CrossRef]
- Li, Z.; Xu, K.; Zhang, N.; Amador, G.; Wang, Y.; Zhao, S.; Li, L.; Qiu, Y.; Wang, Z. Overexpressed SIRT6 attenuates cisplatin-induced acute kidney injury by inhibiting ERK1/2 signaling. Kidney Int. 2018, 93, 881–892. [Google Scholar] [CrossRef]
- Almaghrabi, O.A. Molecular and biochemical investigations on the effect of quercetin on oxidative stress induced by cisplatin in rat kidney. Saudi J. Biol. Sci. 2015, 22, 227–231. [Google Scholar] [CrossRef] [PubMed]
- Kruidering, M.; Van de Water, B.; de Heer, E.; Mulder, G.J.; Nagelkerke, J.F. Cisplatin-induced nephrotoxicity in porcine proximal tubular cells: Mitochondrial dysfunction by inhibition of complexes I to IV of the respiratory chain. J. Pharmacol. Exp. Ther. 1997, 280, 638–649. [Google Scholar]
- Hao, Y.; Guo, F.; Huang, Z.; Feng, Y.; Xia, Z.; Liu, J.; Li, L.; Huang, R.; Lin, L.; Ma, L.; et al. 2-Methylquinazoline derivative 23BB as a highly selective histone deacetylase 6 inhibitor alleviated cisplatin-induced acute kidney injury. Biosci. Rep. 2020, 40. [Google Scholar] [CrossRef]
- Xiang, X.; Guo, C.; Tang, C.; Cai, J.; Dong, Z. Epigenetic Regulation in Kidney Toxicity: Insights From Cisplatin Nephrotoxicity. Semin. Nephrol. 2019, 39, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Saldanha, S.N.; Tollefsbol, T.O. Epigenetic Approaches to Cancer Therapy. In Epigenetics in Human Disease; Tollefsbol, T.O., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 6, pp. 219–247. [Google Scholar]
- Guo, C.; Dong, G.; Liang, X.; Dong, Z. Epigenetic regulation in AKI and kidney repair: Mechanisms and therapeutic implications. Nat. Rev. Nephrol. 2019, 15, 220–239. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Pei, L.; Xiao, X.; Wei, Q.; Chen, J.K.; Ding, H.F.; Huang, S.; Fan, G.; Shi, H.; Dong, Z. DNA methylation protects against cisplatin-induced kidney injury by regulating specific genes, including interferon regulatory factor 8. Kidney Int. 2017, 92, 1194–1205. [Google Scholar] [CrossRef] [PubMed]
- Scholpa, N.E.; Kolli, R.T.; Moore, M.; Arnold, R.D.; Glenn, T.C.; Cummings, B.S. Nephrotoxicity of epigenetic inhibitors used for the treatment of cancer. Chem. Biol. Interact. 2016, 258, 21–29. [Google Scholar] [CrossRef]
- Tikoo, K.; Ali, I.Y.; Gupta, J.; Gupta, C. 5-Azacytidine prevents cisplatin induced nephrotoxicity and potentiates anticancer activity of cisplatin by involving inhibition of metallothionein, pAKT and DNMT1 expression in chemical induced cancer rats. Toxicol. Lett. 2009, 191, 158–166. [Google Scholar] [CrossRef]
- Qian, C.Y.; Zheng, Y.; Wang, Y.; Chen, J.; Liu, J.Y.; Zhou, H.H.; Yin, J.Y.; Liu, Z.Q. Associations of genetic polymorphisms of the transporters organic cation transporter 2 (OCT2), multidrug and toxin extrusion 1 (MATE1), and ATP-binding cassette subfamily C member 2 (ABCC2) with platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Chin. J. Cancer 2016, 35, 85. [Google Scholar] [CrossRef]
- Aoki, M.; Terada, T.; Kajiwara, M.; Ogasawara, K.; Ikai, I.; Ogawa, O.; Katsura, T.; Inui, K. Kidney-specific expression of human organic cation transporter 2 (OCT2/SLC22A2) is regulated by DNA methylation. Am. J. Physiol. Renal Physiol. 2008, 295, F165–F170. [Google Scholar] [CrossRef] [PubMed]
- Mercurio, C.; Plyte, S.; Minucci, S. Chapter 4—Alterations of Histone Modifications in Cancer. In Epigenetics in Human Disease; Translational Epigenetics; Elsevier Inc.: Amsterdam, The Netherlands, 2012; pp. 53–87. [Google Scholar] [CrossRef]
- Cheng, L.; Li, C.; Xi, Z.; Wei, K.; Yuan, S.; Arnesano, F.; Natile, G.; Liu, Y. Cisplatin reacts with histone H1 and the adduct forms a ternary complex with DNA. Metallomics 2019, 11, 556–564. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Q.; Li, J.Q.; Wang, X.Q.; Lei, W.J.; Li, H.; Wan, J.; Hu, Z.; Zou, Y.W.; Wu, X.Y.; Niu, H.X. EZH2-inhibitor DZNep enhances apoptosis of renal tubular epithelial cells in presence and absence of cisplatin. Cell Div. 2020, 15, 8. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, A.; Minucci, S. Chapter 6—Alterations of Histone Modifications in Cancer. In Epigenetics in Human Disease; Tollefsbol, T.O., Ed.; Academic Press: Cambridge, MA, USA; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 6, pp. 141–217. [Google Scholar]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef]
- Chen, L.F.; Mu, Y.; Greene, W.C. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J. 2002, 21, 6539–6548. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Jo, J.; Leem, J.; Park, K.K. Inhibition of p300 by Garcinol Protects against Cisplatin-Induced Acute Kidney Injury through Suppression of Oxidative Stress, Inflammation, and Tubular Cell Death in Mice. Antioxidants 2020, 9, 1271. [Google Scholar] [CrossRef]
- Jiang, W.; Yuan, X.; Zhu, H.; He, C.; Ge, C.; Tang, Q.; Xu, C.; Hu, B.; Huang, C.; Ma, T. Inhibition of Histone H3K27 Acetylation Orchestrates Interleukin-9-Mediated and Plays an Anti-Inflammatory Role in Cisplatin-Induced Acute Kidney Injury. Front. Immunol. 2020, 11, 231. [Google Scholar] [CrossRef]
- Ma, T.; Huang, C.; Xu, Q.; Yang, Y.; Liu, Y.; Meng, X.; Li, J.; Ye, M.; Liang, H. Suppression of BMP-7 by histone deacetylase 2 promoted apoptosis of renal tubular epithelial cells in acute kidney injury. Cell Death Dis. 2017, 8, e3139. [Google Scholar] [CrossRef] [PubMed]
- Sakao, Y.; Kato, A.; Tsuji, T.; Yasuda, H.; Togawa, A.; Fujigaki, Y.; Kahyo, T.; Setou, M.; Hishida, A. Cisplatin induces Sirt1 in association with histone deacetylation and increased Werner syndrome protein in the kidney. Clin. Exp. Nephrol. 2011, 15, 363–372. [Google Scholar] [CrossRef][Green Version]
- Tang, J.; Shi, Y.; Liu, N.; Xu, L.; Zang, X.; Li, P.; Zhang, J.; Zheng, X.; Qiu, A.; Zhuang, S. Blockade of histone deacetylase 6 protects against cisplatin-induced acute kidney injury. Clin. Sci 2018, 132, 339–359. [Google Scholar] [CrossRef]
- Mikami, D.; Kobayashi, M.; Uwada, J.; Yazawa, T.; Kamiyama, K.; Nishimori, K.; Nishikawa, Y.; Morikawa, Y.; Yokoi, S.; Takahashi, N.; et al. beta-Hydroxybutyrate, a ketone body, reduces the cytotoxic effect of cisplatin via activation of HDAC5 in human renal cortical epithelial cells. Life Sci. 2019, 222, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Dong, G.; Luo, J.; Kumar, V.; Dong, Z. Inhibitors of histone deacetylases suppress cisplatin-induced p53 activation and apoptosis in renal tubular cells. Am. J. Physiol. Renal Physiol. 2010, 298, F293–F300. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhuang, S. Upregulation of AMWAP: A novel mechanism for HDAC inhibitors to protect against cisplatin nephrotoxicity. Kidney Int. 2016, 89, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Ranganathan, P.; Hamad, R.; Mohamed, R.; Jayakumar, C.; Muthusamy, T.; Ramesh, G. Histone deacetylase-mediated silencing of AMWAP expression contributes to cisplatin nephrotoxicity. Kidney Int. 2016, 89, 317–326. [Google Scholar] [CrossRef]
- Zhu, H.; Jiang, W.; Zhao, H.; He, C.; Tang, X.; Xu, S.; Xu, C.; Feng, R.; Li, J.; Ma, T.; et al. PSTPIP2 inhibits cisplatin-induced acute kidney injury by suppressing apoptosis of renal tubular epithelial cells. Cell Death Dis. 2020, 11, 1057. [Google Scholar] [CrossRef]
- Arany, I.; Herbert, J.; Herbert, Z.; Safirstein, R.L. Restoration of CREB function ameliorates cisplatin cytotoxicity in renal tubular cells. Am. J. Physiol. Renal Physiol. 2008, 294, F577–F581. [Google Scholar] [CrossRef]
- Liu, J.; Livingston, M.J.; Dong, G.; Tang, C.; Su, Y.; Wu, G.; Yin, X.M.; Dong, Z. Histone deacetylase inhibitors protect against cisplatin-induced acute kidney injury by activating autophagy in proximal tubular cells. Cell Death Dis. 2018, 9, 322. [Google Scholar] [CrossRef]
- Jung, Y.J.; Lee, J.E.; Lee, A.S.; Kang, K.P.; Lee, S.; Park, S.K.; Lee, S.Y.; Han, M.K.; Kim, D.H.; Kim, W. SIRT1 overexpression decreases cisplatin-induced acetylation of NF-kappaB p65 subunit and cytotoxicity in renal proximal tubule cells. Biochem. Biophys. Res. Commun. 2012, 419, 206–210. [Google Scholar] [CrossRef]
- Kim, D.H.; Jung, Y.J.; Lee, J.E.; Lee, A.S.; Kang, K.P.; Lee, S.; Park, S.K.; Han, M.K.; Lee, S.Y.; Ramkumar, K.M.; et al. SIRT1 activation by resveratrol ameliorates cisplatin-induced renal injury through deacetylation of p53. Am. J. Physiol. Renal Physiol. 2011, 301, F427–F435. [Google Scholar] [CrossRef]
- Kim, J.Y.; Jo, J.; Kim, K.; An, H.J.; Gwon, M.G.; Gu, H.; Kim, H.J.; Yang, A.Y.; Kim, S.W.; Jeon, E.J.; et al. Pharmacological Activation of Sirt1 Ameliorates Cisplatin-Induced Acute Kidney Injury by Suppressing Apoptosis, Oxidative Stress, and Inflammation in Mice. Antioxidants 2019, 8, 322. [Google Scholar] [CrossRef]
- Jung, Y.J.; Park, W.; Kang, K.P.; Kim, W. SIRT2 is involved in cisplatin-induced acute kidney injury through regulation of mitogen-activated protein kinase phosphatase-1. Nephrol. Dial. Transplant. 2020, 35, 1145–1156. [Google Scholar] [CrossRef]
- Yoon, S.P.; Kim, J. Poly(ADP-ribose) polymerase 1 contributes to oxidative stress through downregulation of sirtuin 3 during cisplatin nephrotoxicity. Anat. Cell Biol. 2016, 49, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Park, W.; Lee, S.; Kim, W.; Park, S.K.; Kang, K.P. Absence of Sirt3 aggravates cisplatin nephrotoxicity via enhanced renal tubular apoptosis and inflammation. Mol. Med. Rep. 2018, 18, 3665–3672. [Google Scholar] [CrossRef]
- Chiba, T.; Peasley, K.D.; Cargill, K.R.; Maringer, K.V.; Bharathi, S.S.; Mukherjee, E.; Zhang, Y.; Holtz, A.; Basisty, N.; Yagobian, S.D.; et al. Sirtuin 5 Regulates Proximal Tubule Fatty Acid Oxidation to Protect against AKI. J. Am. Soc. Nephrol. 2019, 30, 2384–2398. [Google Scholar] [CrossRef] [PubMed]
- Bertrand-Lehouillier, V.; Legault, L.-M.; McGraw, S. Endocrine Epigenetics, Epigenetic Profiling and Biomarker Identification. In Encyclopedia of Endocrine Diseases; Huhtaniemi, I., Martini, L., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 31–35. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Pavkovic, M.; Robinson-Cohen, C.; Chua, A.S.; Nicoara, O.; Cardenas-Gonzalez, M.; Bijol, V.; Ramachandran, K.; Hampson, L.; Pirmohamed, M.; Antoine, D.J.; et al. Detection of Drug-Induced Acute Kidney Injury in Humans Using Urinary KIM-1, miR-21, -200c, and -423. Toxicol. Sci. 2016, 152, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Zhu, Q.; Yan, L.; Shao, F. Mesenchymal stem cells alleviate acute kidney injury via miR-107-mediated regulation of ribosomal protein S19. Ann. Transl. Med. 2019, 7, 765. [Google Scholar] [CrossRef]
- Wu, J.; Li, D.D.; Li, J.Y.; Yin, Y.C.; Li, P.C.; Qiu, L.; Chen, L.M. Identification of microRNA-mRNA networks involved in cisplatin-induced renal tubular epithelial cells injury. Eur. J. Pharmacol. 2019, 851, 1–12. [Google Scholar] [CrossRef]
- Hao, J.; Lou, Q.; Wei, Q.; Mei, S.; Li, L.; Wu, G.; Mi, Q.S.; Mei, C.; Dong, Z. MicroRNA-375 Is Induced in Cisplatin Nephrotoxicity to Repress Hepatocyte Nuclear Factor 1-beta. J. Biol. Chem. 2017, 292, 4571–4582. [Google Scholar] [CrossRef] [PubMed]
- Khandia, R.; Dadar, M.; Munjal, A.; Dhama, K.; Karthik, K.; Tiwari, R.; Yatoo, M.I.; Iqbal, H.M.N.; Singh, K.P.; Joshi, S.K.; et al. A Comprehensive Review of Autophagy and Its Various Roles in Infectious, Non-Infectious, and Lifestyle Diseases: Current Knowledge and Prospects for Disease Prevention, Novel Drug Design, and Therapy. Cells 2019, 8, 674. [Google Scholar] [CrossRef]
- Yang, C.; Kaushal, V.; Shah, S.V.; Kaushal, G.P. Autophagy is associated with apoptosis in cisplatin injury to renal tubular epithelial cells. Am. J. Physiol. Renal Physiol. 2008, 294, F777–F787. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Kimura, T.; Takabatake, Y.; Namba, T.; Kaimori, J.; Kitamura, H.; Matsui, I.; Niimura, F.; Matsusaka, T.; Fujita, N.; et al. Autophagy guards against cisplatin-induced acute kidney injury. Am. J. Pathol. 2012, 180, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yu, J.; Yin, L.; Zhou, Y.; Sun, Z.; Jia, H.; Tao, Y.; Liu, W.; Zhang, B.; Zhang, J.; et al. MicroRNA-146b, a Sensitive Indicator of Mesenchymal Stem Cell Repair of Acute Renal Injury. Stem Cells Transl. Med. 2016, 5, 1406–1415. [Google Scholar] [CrossRef]
- El Magdoub, H.M.; Schaalan, M.F.; Rahmo, R.M.; Farag, D.B.; Khedr, L.H. Implications of miRNAs on TGF-beta/TAK1/mTOR pathway in mediating the renoprotective effects of pentoxifylline against cisplatin-induced nephrotoxicity in rats. Toxicol. Appl. Pharmacol. 2020, 404, 115184. [Google Scholar] [CrossRef]
- Jung, C.H.; Ro, S.H.; Cao, J.; Otto, N.M.; Kim, D.H. mTOR regulation of autophagy. FEBS Lett. 2010, 584, 1287–1295. [Google Scholar] [CrossRef]
- De Almeida, D.C.; Bassi, E.J.; Azevedo, H.; Anderson, L.; Origassa, C.S.; Cenedeze, M.A.; de Andrade-Oliveira, V.; Felizardo, R.J.; da Silva, R.C.; Hiyane, M.I.; et al. A Regulatory miRNA-mRNA Network Is Associated with Tissue Repair Induced by Mesenchymal Stromal Cells in Acute Kidney Injury. Front. Immunol. 2016, 7, 645. [Google Scholar] [CrossRef]
- Liao, W.; Fu, Z.; Zou, Y.; Wen, D.; Ma, H.; Zhou, F.; Chen, Y.; Zhang, M.; Zhang, W. MicroRNA-140-5p attenuated oxidative stress in Cisplatin induced acute kidney injury by activating Nrf2/ARE pathway through a Keap1-independent mechanism. Exp. Cell Res. 2017, 360, 292–302. [Google Scholar] [CrossRef]
- Loboda, A.; Damulewicz, M.; Pyza, E.; Jozkowicz, A.; Dulak, J. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: An evolutionarily conserved mechanism. Cell Mol. Life Sci. 2016, 73, 3221–3247. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; Ji, J.A.; Jiang, Z.Y.; You, Q.D. The Keap1-Nrf2-ARE Pathway As a Potential Preventive and Therapeutic Target: An Update. Med. Res. Rev. 2016, 36, 924–963. [Google Scholar] [CrossRef]
- Xu, Y.; Ma, H.; Shao, J.; Wu, J.; Zhou, L.; Zhang, Z.; Wang, Y.; Huang, Z.; Ren, J.; Liu, S.; et al. A Role for Tubular Necroptosis in Cisplatin-Induced AKI. J. Am. Soc. Nephrol. 2015, 26, 2647–2658. [Google Scholar] [CrossRef] [PubMed]
- Najafov, A.; Chen, H.; Yuan, J. Necroptosis and Cancer. Trends Cancer 2017, 3, 294–301. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Kong, W.; Zhang, B. Functional role of microRNA-500a-3P-loaded liposomes in the treatment of cisplatin-induced AKI. IET Nanobiotechnol. 2020, 14, 465–469. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Liu, X.Q.; Ma, Q.; Yang, Q.; Gao, L.; Li, H.D.; Wang, J.N.; Wei, B.; Wen, J.; Li, J.; et al. hsa-miR-500a-3P alleviates kidney injury by targeting MLKL-mediated necroptosis in renal epithelial cells. FASEB J. 2019, 33, 3523–3535. [Google Scholar] [CrossRef]
- Pellegrini, K.L.; Han, T.; Bijol, V.; Saikumar, J.; Craciun, F.L.; Chen, W.W.; Fuscoe, J.C.; Vaidya, V.S. MicroRNA-155 deficient mice experience heightened kidney toxicity when dosed with cisplatin. Toxicol. Sci. 2014, 141, 484–492. [Google Scholar] [CrossRef]
- Zhou, X.; Jiang, K.; Luo, H.; Wu, C.; Yu, W.; Cheng, F. Novel lncRNA XLOC_032768 alleviates cisplatin-induced apoptosis and inflammatory response of renal tubular epithelial cells through TNF-alpha. Int. Immunopharmacol. 2020, 83, 106472. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fan, X.; Wang, Q.; Gong, Y.; Guo, L. Long Noncoding RNA PRNCR1 Reduces Renal Epithelial Cell Apoptosis in Cisplatin-Induced AKI by Regulating miR-182-5p/EZH1. Kidney Blood Press. Res. 2021, 46, 162–172. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, P.M.; Niu, Y.; Garcia Cordoba, C.A.; Huang, X.R.; Yu, C.; Lan, H.Y. Long Non-coding RNA LRNA9884 Promotes Acute Kidney Injury via Regulating NF-kB-Mediated Transcriptional Activation of MIF. Front. Physiol. 2020, 11, 590027. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Bi, H.; Wang, S.; Sun, X.; Li, Y. Long non-coding RNA GAS5 aggravate renal epithelial cell apoptosis in cisplatin-induced AKI by regulating miR-205-5p. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Huang, S.; Yang, B.; Chen, B.J.; Bliim, N.; Ueberham, U.; Arendt, T.; Janitz, M. The emerging role of circular RNAs in transcriptome regulation. Genomics 2017, 109, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, K.L.; Gerlach, C.V.; Craciun, F.L.; Ramachandran, K.; Bijol, V.; Kissick, H.T.; Vaidya, V.S. Application of small RNA sequencing to identify microRNAs in acute kidney injury and fibrosis. Toxicol. Appl. Pharmacol. 2016, 312, 42–52. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Li, C.; Li, Q.; Li, J.; Lu, X. Puerarin alleviates cisplatin-induced acute renal damage and upregulates microRNA-31-related signaling. Exp. Ther. Med. 2020, 20, 3122–3129. [Google Scholar] [CrossRef]
- Joo, M.S.; Lee, C.G.; Koo, J.H.; Kim, S.G. miR-125b transcriptionally increased by Nrf2 inhibits AhR repressor, which protects kidney from cisplatin-induced injury. Cell Death Dis. 2013, 4, e899. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, X.; Yuan, X.; Wang, L.; Xiao, Y. MicroRNA-205 inhibits renal cells apoptosis via targeting CMTM4. Iran. J. Basic Med. Sci. 2015, 18, 1020–1026. [Google Scholar] [PubMed]
- Quintanilha, J.; Cursino, M.; Borges, J.; Torso, N.; Bastos, L.; Oliveira, J.; Cobaxo, T.; Pincinato, E.; Hirata, M.; Geraldo, M.; et al. MiR-3168, miR-6125, and miR-4718 as Potential Predictors of Cisplatin-Induced Nephrotoxicity in Patients with Head and Neck Cancer. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Pavkovic, M.; Riefke, B.; Ellinger-Ziegelbauer, H. Urinary microRNA profiling for identification of biomarkers after cisplatin-induced kidney injury. Toxicology 2014, 324, 147–157. [Google Scholar] [CrossRef]
- Wolenski, F.S.; Shah, P.; Sano, T.; Shinozawa, T.; Bernard, H.; Gallacher, M.J.; Wyllie, S.D.; Varrone, G.; Cicia, L.A.; Carsillo, M.E.; et al. Identification of microRNA biomarker candidates in urine and plasma from rats with kidney or liver damage. J. Appl. Toxicol. 2017, 37, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Glineur, S.F.; Hanon, E.; Dremier, S.; Snelling, S.; Berteau, C.; De Ron, P.; Nogueira da Costa, A. Assessment of a Urinary Kidney MicroRNA Panel as Potential Nephron Segment-Specific Biomarkers of Subacute Renal Toxicity in Preclinical Rat Models. Toxicol. Sci. 2018, 166, 409–419. [Google Scholar] [CrossRef]
- Kanki, M.; Moriguchi, A.; Sasaki, D.; Mitori, H.; Yamada, A.; Unami, A.; Miyamae, Y. Identification of urinary miRNA biomarkers for detecting cisplatin-induced proximal tubular injury in rats. Toxicology 2014, 324, 158–168. [Google Scholar] [CrossRef]
- Kagawa, T.; Zarybnicky, T.; Omi, T.; Shirai, Y.; Toyokuni, S.; Oda, S.; Yokoi, T. A scrutiny of circulating microRNA biomarkers for drug-induced tubular and glomerular injury in rats. Toxicology 2019, 415, 26–36. [Google Scholar] [CrossRef]
- Zhang, W.; Chen, C.; Jing, R.; Liu, T.; Liu, B. Remote Ischemic Preconditioning Protects Cisplatin-Induced Acute Kidney Injury through the PTEN/AKT Signaling Pathway. Oxid. Med. Cell Longev. 2019, 2019, 7629396. [Google Scholar] [CrossRef]
- Cho, Y.E.; Kim, S.H.; Lee, B.H.; Baek, M.C. Circulating Plasma and Exosomal microRNAs as Indicators of Drug-Induced Organ Injury in Rodent Models. Biomol. Ther. 2017, 25, 367–373. [Google Scholar] [CrossRef]
- Huang, S.J.; Huang, J.; Yan, Y.B.; Qiu, J.; Tan, R.Q.; Liu, Y.; Tian, Q.; Guan, L.; Niu, S.S.; Zhang, Y.; et al. The renoprotective effect of curcumin against cisplatin-induced acute kidney injury in mice: Involvement of miR-181a/PTEN axis. Ren. Fail. 2020, 42, 350–357. [Google Scholar] [CrossRef]

| miRNA | Assay | Model/Cell Line | Source | CDDP Dose | Time | Effect | Exp. | Induced by | Gene Target | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| miR-9-3p | in vitro | HK-2 cell line | PTECs | 2, 10 or 50 µM | 24 h | Toxic | Down | - | CASK | [84] |
| miR-18a-5p | in vitro | HPTEC cell line | PTECs | 40 µM | 6 and 24 h | Toxic | Down | - | - | [106] |
| miR-21 | in vivo | Patients with malignant mesothelioma | Urine and kidney | CDDP chemotherapy | 4–144 h | Toxic | Up | - | P21, BCL2 | [82] |
| in vitro | HPTEC cell line | PTECs | 85 µM | 24 h | Down | - | P21, BCL2 | [82] | ||
| in vitro | ciPTEC-OAT1 | Cell culture media | 5 to 30 µM | 48 h | Toxic | Down | - | - | [39] | |
| miR-26a | in vitro | HK-2 cell line | PTECs | 4 µmol/L | 24 h | Toxic | Down | - | TRPC6, DRP1 | [25] |
| miR-29a | in vitro | ciPTEC-OAT1 | Cell culture media | 5–30 µM | 48 h | Toxic | Up | - | - | [39] |
| miR-30c | in vitro | HK-2 cell line | PTECs | 10 µM | 24–72 h | Toxic | Down | - | BNIP3L, HSPA5 | [22] |
| miR-31-5p | in vitro | HK-2 cell line | PTECs | 40 µM | 0–48 h | Toxic | Up | - | NUMB | [107] |
| miR-34a | in vitro | HK-2 cell line | PTECs | 9 µg/mL | 24 h | Toxic | Up | - | SIRT1 | [37] |
| in vitro | ciPTEC-OAT1 | Cell culture media | 5–30 µM | 48 h | Toxic | Up | - | - | [39] | |
| miR-107 | in vitro | HK-2 cell line | PTECs | 7.5 mM | 6 h | Toxic | Up | - | RPS19 | [83] |
| miR-125b | in vitro | HepG2, HEK-293 cell line | PTECs | 30 µM | 24 h | Protective | Up | NRF2 | AhRR, MDM2 | [108] |
| miR-140-5p | in vitro | HK-2 and 293T cell lines | PTECs | 1.5–12 µM | 24 h | Toxic | Up | - | NRF2, MnSOD, LDH | [93] |
| miR-146b-5p | in vitro | HPTEC cell line | PTECs | 40 µM | 6 and 24 h | Toxic | Down | - | - | [106] |
| miR-181a | in vitro | HK-2 cell line | PTECs | 50 µmol/L | 24 h | Toxic | Up | - | BCL2, BAX | [33] |
| miR-182 | in vitro | HK-2 and 293T cell line | PTECs | 20 µmol/L | 24 h | Toxic | Down | - | FOXO1 | [15] |
| miR-192 | in vitro | ciPTEC-OAT1 | Cell culture media | 5–30 µM | 48 h | Toxic | Up | - | - | [39] |
| miR-194-5p | in vitro | HK-2 cell line | PTECs | 20 µM | 24 h | Toxic | Up | - | - | [99] |
| miR-199a-3p | in vitro | HK-2 cell line | PTECs | 100 µM | 24 h | Toxic | Up | p53 | mTOR | [38] |
| miR-200c | in vivo | Patients with malignant mesothelioma | Urine and kidney | CDDP chemotherapy | 4–144 h | Toxic | Up | - | P21, BCL2 | [82] |
| in vitro | HPTEC cell line | PTECs | 85 µM | 24 h | Down | - | P21, BCL2 | [82] | ||
| miR-203 | in vitro | HK-2 and 293T cell line | PTECs | 20 µmol/L | 24 h | Toxic | Down | - | GSK-3β | [15] |
| miR-205 | in vitro | HK-2 and HEK-293 cells | PTECs | 100 µg/mL | 6 h | Toxic | Down | - | CMTM4 | [109] |
| miR-371b-5p | in vitro | HK-2 cell line | PTECs | 2, 10 or 50 µM | 24 h | Toxic | Down | - | CDK6 | [84] |
| miR-423 | in vivo | Patients with malignant mesothelioma | Urine and kidney | CDDP chemotherapy | 4–144 h | Toxic | Up | - | P21, BCL2 | [82] |
| in vitro | HPTEC cell line | PTECs | 85 µM | 24 h | Down | - | P21, BCL2 | [82] | ||
| miR-494 | in vitro | HK-2 and 293T cell line | PTECs | 20 µmol/L | 24 h | Toxic | Down | - | ATF3 | [15] |
| miR-500a-3p | in vitro | HK-2 cell line | PTECs | 20 µM | 24 h | Toxic | Down | - | MLKL, RIPK1, RIPK3 | [99] |
| in vitro | HK-2 cell line | PTECs | 15 µM | 6–48 h | Toxic | Down | - | MLKL, RIPK3 | [98] | |
| miR-577 | in vitro | HK-2 cell line | PTECs | 20 µM | 24 h | Toxic | Up | - | - | [99] |
| miR-3168 | in vivo | Patients with primary squamous cell carcinoma of the head and neck | Blood | CDDP chemotherapy | - | Toxic | Up | - | PRKAB2, TOP2A | [110] |
| miR-4718 | in vivo | Patients with primary squamous cell carcinoma of the head and neck | Blood | CDDP chemotherapy | - | Toxic | Down | - | PRKAB2, ERCC1 | [110] |
| miR-6125 | in vivo | Patients with primary squamous cell carcinoma of the head and neck | Blood | CDDP chemotherapy | - | Toxic | Up | - | ERCC1 | [110] |
| miRNA | Specie | Assay | Model/Cell Line | Source | CDDP Dose | Time | Effect | Exp. | Induced by | Gene Target | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|
| miR-9-3p | Rat | in vitro | NRK52E cell line | PTECs | 2, 10 or 50 µM | 24 h | Toxic | Down | - | CASK, CDK6, JNK | [84] |
| miR-15 | Rat | in vivo | Male Han Wistar rat | Blood, urine and kidney | 1 or 3 mg/kg | 0–26 d | Toxic | Up | - | - | [111] |
| miR-16 | Rat | in vivo | Male Han Wistar rat | Blood, urine and kidney | 1 or 3 mg/kg | 0–26 d | Toxic | Up | - | BTG2, PHLD3A | [111] |
| miR-20a | Rat | in vivo | Male Han Wistar rat | Blood, urine and kidney | 1 or 3 mg/kg | 0–26 d | Toxic | Up | - | p21 | [111] |
| miR-21 | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Up | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | |||||||
| miR-22 | Rat | in vivo | Male Sprague-Dawley rats | Urine | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-26a | Mouse | in vivo | Male C57BL/6 mice | Kidney | 20 mg/kg | 3 d | Toxic | Down | - | TRPC6, DRP1 | [25] |
| miR-26b | Rat | in vivo | Male Sprague-Dawley rats | Urine | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| Male Wistar rats | Blood and kidney | 5 mg/kg | 3 d | Toxic | Down | - | TGFβR-1, TAK1, mTOR, LC3-II | [90] | |||
| miR-30c | Rat | in vivo | Wistar rat | Kidney | 10 mg/kg | 1, 3 or 7 d | Toxic | Down | - | BNIP3L, HSPA5 | [22] |
| in vitro | NRK-52E cell line | PTECs | 10 µM | 24–72 h | |||||||
| miR-31-5p | Rat | in vivo | Female Sprague-Dawley rats | Blood and kidney | 20 mg/kg | 14 d | Toxic | Up | - | NUMB | [107] |
| miR-34a | Mouse | in vitro | BUMPT-306 cell line | PTECs | 40 µmol/L | 0–20 h | Toxic | Up | - | p53 | [21] |
| Mouse | in vivo | Wild-type and p53-deficientC57BL/6 mice | Blood and kidney | 30 mg/kg | 0–3 d | ||||||
| Mouse | in vivo | Male C57BL/6 mice | Kidney | 15 mg/kg | 3 d | Toxic | Up | - | SIRT1, p53, PHLDA3 | [36] | |
| Rat | in vitro | NRK-52E cell line | PTECs | 9 µg/mL | 24 h | Toxic | Up | - | SIRT1 | [37] | |
| RatMouse | in vivo | Male Wistar ratsMale C57BL-6J mice | Blood and kidney | 7 mg/kg25 mg/kg | 12 d | - | - | [37] | |||
| Rat | in vivo | Male Sprague-Dawley rats | Kidney and plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] | |
| Rat | in vivo | Male Wistar rats | Blood and kidney | 5 mg/kg | 3 d | Toxic | Down | - | TGFβR-1, TAK1, mTOR, LC3-II | [90] | |
| miR-34c | Rat | in vivo | Male Sprague-Dawley rats | Kidney and plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-34c-5p | Rat | in vivo | Male Hannover Wistar rats | Kidney and urine | 2.5 mg/kg | 0–27 d | Toxic | Down | - | - | [113] |
| miR-92a | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Down | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-92b | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-93-5p | Rat | in vivo | Male Sprague-Dawley rats | Kidney and urine | 1, 3 or 6 mg/kg | 1–7 d | Toxic | Up | - | - | [114] |
| miR-107 | Rat | in vivo | Male Sprague-Dawley rats | Kidney and blood | 6 mg/kg | 3 d | Toxic | Up | - | RPS19 | [83] |
| miR-122 | Mouse | in vivo | Male C57BL/6 mice | Kidney | 15 mg/kg | 3 d | Toxic | Down | - | FOXO3, p53, PHLDA3 | [36] |
| miR-122-5p | Rat | in vivo | Male Sprague-Dawley rats | Blood and kidney | 6 mg/kg | 1–5 d | Toxic | Down | - | - | [115] |
| miR-125b | Mouse | in vivo | Male C57BL/6 mice and Nrf2 KO | Blood and kidney | 15 mg/kg | 3 d | Protective | Up | NRF2 | AhRR, MDM2 | [108] |
| miR-128 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-130b | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-134 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-138 | Mouse | in vivo | Female Diversity Outbred mice | Urine and kidney | 5 mg/kg | 3 d | Toxic | Up | - | - | [26] |
| miR-140 | Rat | in vivo | Male Sprague-Dawley rats | Urine | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-140-5p | Mouse | in vivo | Adult male mice | Blood and kidney | 20 mg/kg | 1–14 d | Toxic | Up | - | NRF2, MnSOD, LDH | [93] |
| in vivo | Male C57BL/6J mice | Blood and kidney | 20 mg/kg | 19 d | Toxic | Down | - | NRF2, p53 | [24] | ||
| miR-141 | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Down | - | ULK2 | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-143-3p | Rat | in vivo | Male Sprague-Dawley rats | Blood and kidney | 6 mg/kg | 1–5 d | Toxic | Down | - | - | [115] |
| miR-144 | Mouse | in vivo | Male C57BL/6 mice | Blood and kidney | 20 mg/kg | 6 h | Toxic | Down | - | PTEN, ATK, GSk3β | [116] |
| Rat | in vivo | Male Sprague-Dawley rats | Kidney | 2 or 5 mg/kg | 72 h | Toxic | Down | - | - | [112] | |
| miR-146a | Mouse | in vivo | Male Balb/C mice | Blood and kidney | 10 mg/kg | 24 h | Toxic | Up | - | - | [117] |
| miR-146b | Rat | in vivo | Sprague-Dawley rats | Serum and kidney | 6 mg/kg | 5 d | Toxic | Up | - | ERBB4 | [89] |
| in vitro | NRK-52E cell line | PTECs | 7.5 µM | 48 h | - | ||||||
| in vivo | Male Sprague-Dawley rats | Kidney | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] | ||
| miR-151 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-155 | Mouse | in vivo | miR-155−/− and C57BL/6 mice | Blood and kidney | 20 mg/kg | 72 h | Toxic | Down | - | c-FOS, TNF-α, IL-1β | [100] |
| miR-181a | Mouse | in vivo | Male C57BL/6 mice | Blood and kidney | 20 mg/kg | 3 d | Toxic | Up | - | PTEN | [118] |
| miR-181b | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-182 | Mouse | in vivo | Male C57BL/6 mice | Blood and kidney | 30 mg/kg | 72 h | Toxic | Down | - | FOXO1 | [15] |
| miR-191a | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-191a-5p | Rat | in vivo | Male Sprague-Dawley rats | Kidney and urine | 1, 3 or 6 mg/kg | 1–7 d | Toxic | Up | - | - | [114] |
| miR-192 | Rat | in vivo | Male Han Wistar rat | Blood, urine and kidney | 1 or 3 mg/kg | 0–26 d | Toxic | Up | - | - | [111] |
| miR-192-5p | Rat | in vivo | Male Sprague-Dawley rats | Kidney and urine | 1, 3 or 6 mg/kg | 1–7 d | Toxic | Up | - | - | [114] |
| Mouse | in vivo | Male C57BL/6J mice | Kidney and blood | 20 mg/kg | 19 d | Toxic | Down | - | NRF2, p53 | [24] | |
| miR-193 | Rat | in vivo | Male Han Wistar rat | Blood, urine and kidney | 1 or 3 mg/kg | 0–26 d | Toxic | Up | - | - | [111] |
| miR-199a-3p | Mouse | in vivo | Male C57/BL mice | Kidney | 20 mg/kg | 3 d | Toxic | Up | p53 | mTOR | [38] |
| miR-203 | Mouse | in vivo | Male C57BL/6 mice | Blood and kidney | 30 mg/kg | 72 h | Toxic | Down | - | GSK-3β | [15] |
| miR-208a | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Down | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-210 | Rat | in vivo | Male Han Wistar rat | Blood, urine and kidney | 1 or 3 mg/kg | 0–26 d | Toxic | Up | - | - | [111] |
| miR-218 | Mouse | in vivo | Female Diversity Outbred mice | Urine and kidney | 5 mg/kg | 3 d | Toxic | Up | - | - | [26] |
| miR-292-3p | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Down | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-293* | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Up | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-371b-5p | Rat | in vitro | NRK52E cell line | PTECs | 2, 10 or 50 µM | 24 h | Toxic | Down | - | CASK, CDK6, JNK | [84] |
| miR-375 | Mouse | in vivo | Wild-type and PT-Dicer−/− mice | Kidney | 30 mg/kg | 3 d | Toxic | Up | p53, NF-κB | HNF-1β | [85] |
| Rat | in vitro | RPTC cell line | PTECs | 20 µM | 16 h | ||||||
| miR-377 | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Up | - | CUL1 | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-378a | Rat | in vivo | Male Sprague-Dawley rats | Urine | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-431 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-449 | Rat | in vitro | NRK-52E cell line | PTECs | 20 µg/mL | 24 h | Toxic | Up | - | SIRT1, P53, BAX | [32] |
| miR-451 | Rat | in vivo | Male Sprague-Dawley rats | Kidney | 2 or 5 mg/kg | 72 h | Toxic | Down | - | - | [112] |
| miR-463 | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Up | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-484 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-494 | Mouse | in vivo | Male C57BL/6 mice | Blood and kidney | 30 mg/kg | 72 h | Toxic | Down | - | ATF3, IL6 | [15] |
| miR-673-5p | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Down | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-675-3p | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Up | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-685 | Mouse | in vivo | Female Diversity Outbred mice | Urine and kidney | 5 mg/kg | 3 d | Toxic | Up | - | - | [26] |
| miR-702 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-709 | Mouse | in vivo | C57BL/6 mice | Blood and kidney | 20 mg/kg | 72 h | Toxic | Up | - | TFAM | [23] |
| in vitro | mPTC cell line | PTECs | 1–20 µM | 2–24 h | - | ||||||
| miR-741 | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Up | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-1190 | Mouse | in vivo | Adult C57BL/6 mice | Urine, blood and kidney | 15 mg/kg | 4 d | Toxic | Down | - | - | [92] |
| in vitro | MM55.K tubular cells | PTECs | 8 µg/mL | 48 h | - | ||||||
| miR-1839 | Rat | in vivo | Male Sprague-Dawley rats | Urine | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-6215 | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-let-7b | Rat | in vivo | Male Wistar rats | Blood and kidney | 5 mg/kg | 3 d | Toxic | Down | - | TGFβR-1, TAK1, mTOR, LC3-II | [90] |
| miR-let-7e | Rat | in vivo | Male Sprague-Dawley rats | Plasma | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-let-7g | Rat | in vivo | Male Sprague-Dawley rats | Urine | 2 or 5 mg/kg | 72 h | Toxic | Up | - | - | [112] |
| miR-let-7g-5p | Rat | in vivo | Male Sprague-Dawley rats | Kidney and urine | 1, 3 or 6 mg/kg | 1–7 d | Toxic | Up | - | - | [114] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Loren, P.; Saavedra, N.; Saavedra, K.; Zambrano, T.; Moriel, P.; Salazar, L.A. Epigenetic Mechanisms Involved in Cisplatin-Induced Nephrotoxicity: An Update. Pharmaceuticals 2021, 14, 491. https://doi.org/10.3390/ph14060491
Loren P, Saavedra N, Saavedra K, Zambrano T, Moriel P, Salazar LA. Epigenetic Mechanisms Involved in Cisplatin-Induced Nephrotoxicity: An Update. Pharmaceuticals. 2021; 14(6):491. https://doi.org/10.3390/ph14060491
Chicago/Turabian StyleLoren, Pía, Nicolás Saavedra, Kathleen Saavedra, Tomás Zambrano, Patricia Moriel, and Luis A. Salazar. 2021. "Epigenetic Mechanisms Involved in Cisplatin-Induced Nephrotoxicity: An Update" Pharmaceuticals 14, no. 6: 491. https://doi.org/10.3390/ph14060491
APA StyleLoren, P., Saavedra, N., Saavedra, K., Zambrano, T., Moriel, P., & Salazar, L. A. (2021). Epigenetic Mechanisms Involved in Cisplatin-Induced Nephrotoxicity: An Update. Pharmaceuticals, 14(6), 491. https://doi.org/10.3390/ph14060491








