m6A RNA Methylation in Systemic Autoimmune Diseases—A New Target for Epigenetic-Based Therapy?
Abstract
1. Introduction—A Link between Epigenetics and Autoimmunity
Role of Epigenetic Alterations in the Pathogenesis of Autoimmune Diseases
2. New Epigenetic Player—m6A Modifications of RNA
2.1. Methods for m6A Detection
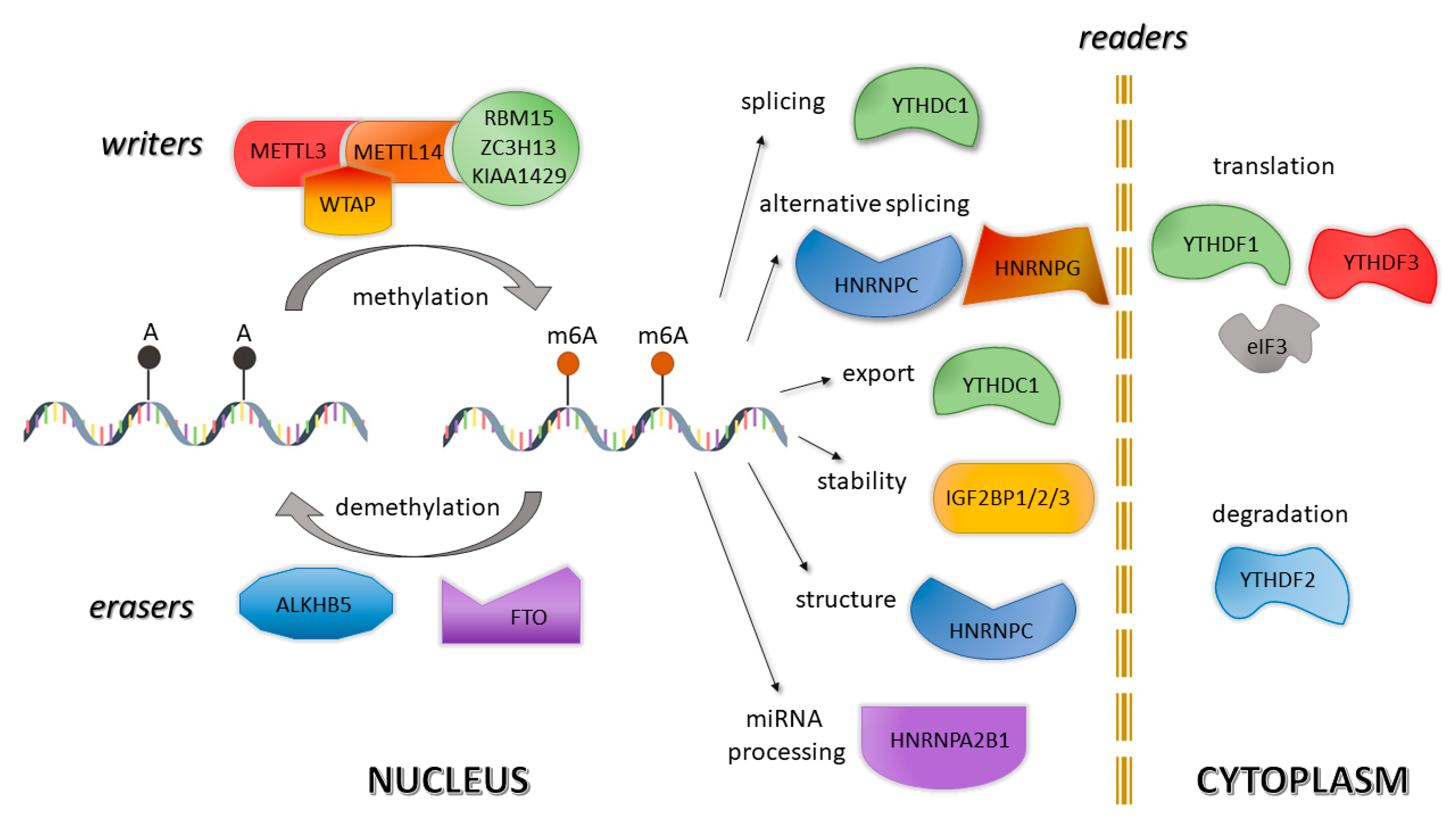
2.2. m6A in Health
2.3. m6A in Disease—Alterations in Autoimmune and Related Disorders
2.3.1. Systemic Lupus Erythematosus
2.3.2. Rheumatoid Arthritis
2.3.3. Psoriasis
2.3.4. Multiple Sclerosis
3. Epigenetic-Based Therapy in ADs—m6A as a New Target
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Finelli, R.; Leisegang, K.; Finocchi, F.; De Masi, S.; Agarwal, A.; Damiani, G. The impact of autoimmune systemic inflammation and associated medications on male reproductive health in patients with chronic rheumatological, dermatological, and gastroenterological diseases: A systematic review. Am. J. Reprod. Immunol. 2021. [Google Scholar] [CrossRef]
- Rosenblum, M.D.; Remedios, K.A.; Abbas, A.K. Mechanisms of human autoimmunity. J. Clin. Investig. 2015, 125, 2228–2233. [Google Scholar] [CrossRef]
- Ganesan, A.; Arimondo, P.B.; Rots, M.G.; Jeronimo, C.; Berdasco, M. The timeline of epigenetic drug discovery: From reality to dreams. Clin. Epigenetics 2019, 11, 174. [Google Scholar] [CrossRef]
- de Oliveira, D.T.; Guerra-Sá, R. Uncovering epigenetic landscape: A new path for biomarkers identification and drug development. Mol. Biol. Rep. 2020, 47, 9097–9122. [Google Scholar] [CrossRef] [PubMed]
- Wee, S.; Dhanak, D.; Li, H.; Armstrong, S.A.; Copeland, R.A.; Sims, R.; Baylin, S.B.; Liu, X.S.; Schweizer, L. Targeting epigenetic regulators for cancer therapy. Ann. N. Y. Acad. Sci. 2014, 1309, 30–36. [Google Scholar] [CrossRef] [PubMed]
- Juo, Y.-Y.; Gong, X.-J.; Mishra, A.; Cui, X.; Baylin, S.B.; Azad, N.S.; Ahuja, N. Epigenetic therapy for solid tumors: From bench science to clinical trials. Epigenomics 2015, 7, 215–235. [Google Scholar] [CrossRef] [PubMed]
- Bird, A. Perceptions of epigenetics. Nature 2007, 447, 396–398. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, R.; Zwergel, C.; Artico, M.; Taurone, S.; Ralli, M.; Greco, A.; Mai, A. The emerging role of epigenetics in human autoimmune disorders. Clin. Epigenetics 2019, 11, 34. [Google Scholar] [CrossRef] [PubMed]
- Moosavi, A.; Motevalizadeh Ardekani, A. Role of Epigenetics in Biology and Human Diseases. Iran. Biomed. J. 2016, 20, 246–258. [Google Scholar]
- Karagianni, P.; Tzioufas, A.G. Epigenetic perspectives on systemic autoimmune disease. J. Autoimmun. 2019, 104, 102315. [Google Scholar] [CrossRef] [PubMed]
- Wardowska, A. The epigenetic face of lupus: Focus on antigen-presenting cells. Int. Immunopharmacol. 2020, 81, 106262. [Google Scholar] [CrossRef]
- Wang, Z.; Lu, Q.; Wang, Z. Epigenetic Alterations in Cellular Immunity: New Insights into Autoimmune Diseases. Cell. Physiol. Biochem. 2017, 41, 645–660. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Yin, H.; Wang, L.; Gershwin, M.E.; Lu, Q. The critical role of epigenetics in systemic lupus erythematosus and autoimmunity. J. Autoimmun. 2016, 74, 118–138. [Google Scholar] [CrossRef] [PubMed]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef]
- Jurkowska, R.Z.; Jeltsch, A. Mechanisms and Biological Roles of DNA Methyltransferases and DNA Methylation: From Past Achievements to Future Challenges. Adv. Exp. Med. Biol. 2016, 945, 1–17. [Google Scholar]
- Zeng, Y.; Chen, T. DNA Methylation Reprogramming during Mammalian Development. Genes 2019, 10, 257. [Google Scholar] [CrossRef] [PubMed]
- Torres, I.O.; Fujimori, D.G. Functional coupling between writers, erasers and readers of histone and DNA methylation. Curr. Opin. Struct. Biol. 2015, 35, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef]
- Castillo, J.; López-Rodas, G.; Franco, L. Histone Post-Translational Modifications and Nucleosome Organisation in Transcriptional Regulation: Some Open Questions. In Protein Reviews; Springer: Berlin/Heidelberg, Germany, 2017; pp. 65–92. [Google Scholar]
- Picascia, A.; Grimaldi, V.; Pignalosa, O.; De Pascale, M.R.; Schiano, C.; Napoli, C. Epigenetic control of autoimmune diseases: From bench to bedside. Clin. Immunol. 2015, 157, 1–15. [Google Scholar] [CrossRef]
- Guo, P.; Chen, W.; Li, H.; Li, M.; Li, L. The Histone Acetylation Modifications of Breast Cancer and their Therapeutic Implications. Pathol. Oncol. Res. 2018, 24, 807–813. [Google Scholar] [CrossRef]
- Rahman, M.M.; Brane, A.C.; Tollefsbol, T.O. MicroRNAs and Epigenetics Strategies to Reverse Breast Cancer. Cells 2019, 8, 1214. [Google Scholar] [CrossRef]
- Hammaker, D.; Firestein, G.S. Epigenetics of inflammatory arthritis. Curr. Opin. Rheumatol. 2018, 30, 188–196. [Google Scholar] [CrossRef]
- Zhao, C.-N.; Mao, Y.-M.; Liu, L.-N.; Li, X.-M.; Wang, D.-G.; Pan, H.-F. Emerging role of lncRNAs in systemic lupus erythematosus. Biomed. Pharmacother. 2018, 106, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.G.; Satpathy, A.T.; Chang, H.Y. Gene regulation in the immune system by long noncoding RNAs. Nat. Immunol. 2017, 18, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Li, X.; Xu, C.; Zeng, L.; Ye, J.; Guo, Y.; Huang, Z.; Li, J. Integrative analysis of long non-coding RNAs and messenger�RNA expression profiles in systemic lupus erythematosus. Mol. Med. Rep. 2017, 17, 3489–3496. [Google Scholar] [CrossRef]
- Wiener, D.; Schwartz, S. The epitranscriptome beyond m6A. Nat. Rev. Genet. 2020, 13, 1–3. [Google Scholar]
- Yang, C.; Hu, Y.; Zhou, B.; Bao, Y.; Li, Z.; Gong, C.; Yang, H.; Wang, S.; Xiao, Y. The role of m6A modification in physiology and disease. Cell Death Dis. 2020, 11, 960. [Google Scholar] [CrossRef]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m6A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m6A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Bokar, J.A.; Rath-Shambaugh, M.E.; Ludwiczak, R.; Narayan, P.; Rottman, F. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994, 269, 17697–17704. [Google Scholar] [CrossRef]
- Ping, X.-L.; Sun, B.-F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.-J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.-S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef]
- Horiuchi, K.; Kawamura, T.; Iwanari, H.; Ohashi, R.; Naito, M.; Kodama, T.; Hamakubo, T. Identification of Wilms’ Tumor 1-associating Protein Complex and Its Role in Alternative Splicing and the Cell Cycle. J. Biol. Chem. 2013, 288, 33292–33302. [Google Scholar] [CrossRef]
- Li, F.; Wang, H.; Huang, H.; Zhang, L.; Wang, D.; Wan, Y. m6A RNA Methylation Regulators Participate in the Malignant Progression and Have Clinical Prognostic Value in Lung Adenocarcinoma. Front. Genet. 2020, 11, 994. [Google Scholar] [CrossRef]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.S.; Liu, Y.; White, C.C.; Feitosa, M.; Smith, A.V.; Heard-Costa, N.; Lohman, K.; Johnson, A.D.; Foster, M.C.; Greenawalt, D.M.; et al. Genome-Wide Association for Abdominal Subcutaneous and Visceral Adipose Reveals a Novel Locus for Visceral Fat in Women. PLoS Genet. 2012, 8, e1002695. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 Is a Mammalian RNA Demethylase that Impacts RNA Metabolism and Mouse Fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bögler, O.; et al. m6A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e6. [Google Scholar] [CrossRef]
- Yao, W.; Han, X.; Ge, M.; Chen, C.; Xiao, X.; Li, H.; Hei, Z. N6-methyladenosine (m6A) methylation in ischemia–reperfusion injury. Cell Death Dis. 2020, 11, 478. [Google Scholar] [CrossRef] [PubMed]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.-S.; Hao, Y.-J.; Sun, B.-F.; Sun, H.-Y.; Li, A.; Ping, X.-L.; Lai, W.-Y.; et al. Nuclear m6A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef]
- Bi, Z.; Liu, Y.; Zhao, Y.; Yao, Y.; Wu, R.; Liu, Q.; Wang, Y.; Wang, X. A dynamic reversible RNA N6-methyladenosine modification: Current status and perspectives. J. Cell. Physiol. 2019, 234, 7948–7956. [Google Scholar] [CrossRef]
- Fu, Y.; Dominissini, D.; Rechavi, G.; He, C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat. Rev. Genet. 2014, 15, 293–306. [Google Scholar] [CrossRef]
- Liao, S.; Sun, H.; Xu, C. YTH Domain: A Family of N6-methyladenosine (m6A) Readers. Genomics. Proteomics Bioinformatics 2018, 16, 99–107. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 Is a Mediator of m6A-Dependent Nuclear RNA Processing Events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine-dependent RNA structural switches regulate RNA–protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.-B.; Jaffrey, S.R. 5′ UTR m6A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Sun, W.; Qin, X.; Shi, H.; Wu, H.; Zhao, B.S.; Mesquita, A.; Liu, C.; Yuan, C.L.; et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018, 20, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Dubin, D.T.; Taylor, R.H. The methylation state of poly A-containing-messenger RNA from cultured hamster cells. Nucleic Acids Res. 1975, 2, 1653–1668. [Google Scholar] [CrossRef] [PubMed]
- Amos, H.; Korn, M. 5-Methyl cytosine in the RNA of Escherichia coli. Biochim. Biophys. Acta 1958, 29, 444–445. [Google Scholar] [CrossRef]
- Dunn, D.B. The occurence of 1-methyladenine in ribonucleic acid. Biochim. Biophys. Acta 1961, 46, 198–200. [Google Scholar] [CrossRef]
- Zhu, W.; Wang, J.; Xu, Z.; Cao, M.; Hu, Q.; Pan, C.; Guo, M.; Wei, J.; Yang, H. Detection of N6-methyladenosine modification residues (Review). Int. J. Mol. Med. 2019, 43, 2267–2278. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Yue, M.; Wang, J.; Kumar, S.; Wechsler-Reya, R.J.; Zhang, Z.; Ogawa, Y.; Kellis, M.; Duester, G.; et al. N6-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat. Neurosci. 2018, 21, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Imanishi, M.; Tsuji, S.; Suda, A.; Futaki, S. Detection of N6-methyladenosine based on the methyl-sensitivity of MazF RNA endonuclease. Chem. Commun. 2017, 53, 12930–12933. [Google Scholar] [CrossRef]
- Mishima, E.; Jinno, D.; Akiyama, Y.; Itoh, K.; Nankumo, S.; Shima, H.; Kikuchi, K.; Takeuchi, Y.; Elkordy, A.; Suzuki, T.; et al. Immuno-Northern Blotting: Detection of RNA Modifications by Using Antibodies against Modified Nucleosides. PLoS ONE 2015, 10, e0143756. [Google Scholar] [CrossRef]
- Mishima, E.; Abe, T. Immuno-Northern Blotting: Detection of Modified RNA Using Gel Separation and Antibodies to Modified Nucleosides. In Epitranscriptomics; Humana Press: New York, NY, USA, 2019; pp. 179–187. [Google Scholar]
- Arguello, A.E.; DeLiberto, A.N.; Kleiner, R.E. RNA Chemical Proteomics Reveals the N6-Methyladenosine (m6A)-Regulated Protein–RNA Interactome. J. Am. Chem. Soc. 2017, 139, 17249–17252. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Feng, P.; Ding, H.; Lin, H. Identifying N6-methyladenosine sites in the Arabidopsis thaliana transcriptome. Mol. Genet. Genomics 2016, 291, 2225–2229. [Google Scholar] [CrossRef]
- Luo, G.-Z.; MacQueen, A.; Zheng, G.; Duan, H.; Dore, L.C.; Lu, Z.; Liu, J.; Chen, K.; Jia, G.; Bergelson, J.; et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014, 5, 5630. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Salmon-Divon, M.; Amariglio, N.; Rechavi, G. Transcriptome-wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013, 8, 176–189. [Google Scholar] [CrossRef]
- Wei, W.; Ji, X.; Guo, X.; Ji, S. Regulatory Role of N6-methyladenosine (m6A) Methylation in RNA Processing and Human Diseases. J. Cell. Biochem. 2017, 118, 2534–2543. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Parisien, M.; Dai, Q.; Zheng, G.; He, C.; Pan, T. Probing N6-methyladenosine RNA modification status at single nucleotide resolution in mRNA and long noncoding RNA. RNA 2013, 19, 1848–1856. [Google Scholar] [CrossRef] [PubMed]
- Molinie, B.; Wang, J.; Lim, K.S.; Hillebrand, R.; Lu, Z.; Van Wittenberghe, N.; Howard, B.D.; Daneshvar, K.; Mullen, A.C.; Dedon, P.; et al. m6A-LAIC-seq reveals the census and complexity of the m6A epitranscriptome. Nat. Methods 2016, 13, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Roberts, J.T.; Porman, A.M.; Johnson, A.M. Identification of m6A residues at single-nucleotide resolution using eCLIP and an accessible custom analysis pipeline. RNA 2020, 29. [Google Scholar] [CrossRef]
- Zheng, H.; Zhang, X.; Sui, N. Advances in the profiling of N6-methyladenosine (m6A) modifications. Biotechnol. Adv. 2020, 45, 107656. [Google Scholar] [CrossRef]
- Capitanchik, C.; Toolan-Kerr, P.; Luscombe, N.M.; Ule, J. How Do You Identify m6 A Methylation in Transcriptomes at High Resolution? A Comparison of Recent Datasets. Front. Genet. 2020, 11, 398. [Google Scholar] [CrossRef]
- Li, X.; Tang, J.; Huang, W.; Wang, F.; Li, P.; Qin, C.; Qin, Z.; Zou, Q.; Wei, J.; Hua, L.; et al. The M6A methyltransferase METTL3: Acting as a tumor suppressor in renal cell carcinoma. Oncotarget 2017, 8, 96103–96116. [Google Scholar] [CrossRef]
- Chen, M.; Wei, L.; Law, C.-T.; Tsang, F.H.-C.; Shen, J.; Cheng, C.L.-H.; Tsang, L.-H.; Ho, D.W.-H.; Chiu, D.K.-C.; Lee, J.M.-F.; et al. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology 2018, 67, 2254–2270. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Campos, M.A.; Edelheit, S.; Toth, U.; Safra, M.; Shachar, R.; Viukov, S.; Winkler, R.; Nir, R.; Lasman, L.; Brandis, A.; et al. Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling. Cell 2019, 178, 731–747.e16. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, L.-Q.; Zhao, Y.-L.; Yang, C.-G.; Roundtree, I.A.; Zhang, Z.; Ren, J.; Xie, W.; He, C.; Luo, G.-Z. Single-base mapping of m6A by an antibody-independent method. Sci. Adv. 2019, 5, eaax0250. [Google Scholar] [CrossRef]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Ke, S.; Alemu, E.A.; Mertens, C.; Gantman, E.C.; Fak, J.J.; Mele, A.; Haripal, B.; Zucker-Scharff, I.; Moore, M.J.; Park, C.Y.; et al. A majority of m6A residues are in the last exons, allowing the potential for 3′ UTR regulation. Genes Dev. 2015, 29, 2037–2053. [Google Scholar] [CrossRef]
- Wang, Y.; Xiao, Y.; Dong, S.; Yu, Q.; Jia, G. Antibody-free enzyme-assisted chemical approach for detection of N6-methyladenosine. Nat. Chem. Biol. 2020, 16, 896–903. [Google Scholar] [CrossRef]
- Maity, A.; Das, B. N6-methyladenosine modification in mRNA: Machinery, function and implications for health and diseases. FEBS J. 2016, 283, 1607–1630. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, Y.; Toth, J.I.; Petroski, M.D.; Zhang, Z.; Zhao, J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell Biol. 2014, 16, 191–198. [Google Scholar] [CrossRef]
- Ignatova, V.V.; Stolz, P.; Kaiser, S.; Gustafsson, T.H.; Lastres, P.R.; Sanz-Moreno, A.; Cho, Y.-L.; Amarie, O.V.; Aguilar-Pimentel, A.; Klein-Rodewald, T.; et al. The rRNA m6A methyltransferase METTL5 is involved in pluripotency and developmental programs. Genes Dev. 2020, 34, 715–729. [Google Scholar] [CrossRef]
- Du, K.; Zhang, L.; Lee, T.; Sun, T. m6A RNA Methylation Controls Neural Development and Is Involved in Human Diseases. Mol. Neurobiol. 2019, 56, 1596–1606. [Google Scholar] [CrossRef]
- Hoernes, T.P.; Clementi, N.; Faserl, K.; Glasner, H.; Breuker, K.; Lindner, H.; Hüttenhofer, A.; Erlacher, M.D. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res. 2016, 44, 852–862. [Google Scholar] [CrossRef]
- Zhou, Y.; Kong, Y.; Fan, W.; Tao, T.; Xiao, Q.; Li, N.; Zhu, X. Principles of RNA methylation and their implications for biology and medicine. Biomed. Pharmacother. 2020, 131, 110731. [Google Scholar] [CrossRef]
- Ma, C.; Chang, M.; Lv, H.; Zhang, Z.-W.; Zhang, W.; He, X.; Wu, G.; Zhao, S.; Zhang, Y.; Wang, D.; et al. RNA m6A methylation participates in regulation of postnatal development of the mouse cerebellum. Genome Biol. 2018, 19, 68. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, X.; Weng, Y.-L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.E.; Hess, S.; Meyer, K.D.; Verhagen, L.A.W.; Koch, L.; Brönneke, H.S.; Dietrich, M.O.; Jordan, S.D.; Saletore, Y.; Elemento, O.; et al. The fat mass and obesity associated gene (Fto) regulates activity of the dopaminergic midbrain circuitry. Nat. Neurosci. 2013, 16, 1042–1048. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zang, L.; Zhang, F.; Chen, J.; Shen, H.; Shu, L.; Liang, F.; Feng, C.; Chen, D.; Tao, H.; et al. Fat mass and obesity-associated (FTO) protein regulates adult neurogenesis. Hum. Mol. Genet. 2017, 26, 2398–2411. [Google Scholar] [CrossRef]
- Lin, Z.; Hsu, P.J.; Xing, X.; Fang, J.; Lu, Z.; Zou, Q.; Zhang, K.-J.; Zhang, X.; Zhou, Y.; Zhang, T.; et al. Mettl3-/Mettl14-mediated mRNA N6-methyladenosine modulates murine spermatogenesis. Cell Res. 2017, 27, 1216–1230. [Google Scholar] [CrossRef]
- Xu, K.; Yang, Y.; Feng, G.-H.; Sun, B.-F.; Chen, J.-Q.; Li, Y.-F.; Chen, Y.-S.; Zhang, X.-X.; Wang, C.-X.; Jiang, L.-Y.; et al. Mettl3-mediated m6A regulates spermatogonial differentiation and meiosis initiation. Cell Res. 2017, 27, 1100–1114. [Google Scholar] [CrossRef]
- Hsu, P.J.; Zhu, Y.; Ma, H.; Guo, Y.; Shi, X.; Liu, Y.; Qi, M.; Lu, Z.; Shi, H.; Wang, J.; et al. Ythdc2 is an N6-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 2017, 27, 1115–1127. [Google Scholar] [CrossRef]
- Huang, T.; Guo, J.; Lv, Y.; Zheng, Y.; Feng, T.; Gao, Q.; Zeng, W. Meclofenamic acid represses spermatogonial proliferation through modulating m6A RNA modification. J. Anim. Sci. Biotechnol. 2019, 10, 63. [Google Scholar] [CrossRef]
- Li, H.-B.; Tong, J.; Zhu, S.; Batista, P.J.; Duffy, E.E.; Zhao, J.; Bailis, W.; Cao, G.; Kroehling, L.; Chen, Y.; et al. m6A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature 2017, 548, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.; Cao, G.; Zhang, T.; Sefik, E.; Amezcua Vesely, M.C.; Broughton, J.P.; Zhu, S.; Li, H.; Li, B.; Chen, L.; et al. m6A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018, 28, 253–256. [Google Scholar] [CrossRef]
- Wang, H.; Hu, X.; Huang, M.; Liu, J.; Gu, Y.; Ma, L.; Zhou, Q.; Cao, X. Mettl3-mediated mRNA m6A methylation promotes dendritic cell activation. Nat. Commun. 2019, 10, 1898. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Shi, Q.; Zheng, L.; Li, Q.; Yang, L.; Zhang, Y. Gene signatures of m5C regulators may predict prognoses of patients with head and neck squamous cell carcinoma. Am. J. Transl. Res. 2020, 12, 6841–6852. [Google Scholar] [PubMed]
- Xing, L.; Cai, Y.; Yang, T.; Yu, W.; Gao, M.; Chai, R.; Ding, S.; Wei, J.; Pan, J.; Chen, G. Epitranscriptomic m6A regulation following spinal cord injury. J. Neurosci. Res. 2020, 99, 843–857. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Lu, Y.; Duan, D.; Wang, H.; Man, G.; Kang, C.; Abulimiti, K.; Li, Y. Systematic Investigation of mRNA N6-Methyladenosine Machinery in Primary Prostate Cancer. Dis. Markers 2020, 2020, 1–21. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Q.; Zheng, Q.; Yu, X.; Guo, W. Distinct 5-methylcytosine profiles of circular RNA in human hepatocellular carcinoma. Am. J. Transl. Res. 2020, 12, 5719–5729. [Google Scholar] [PubMed]
- Han, X.; Wang, M.; Zhao, Y.-L.; Yang, Y.; Yang, Y.-G. RNA methylations in human cancers. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef]
- Zhao, W.; Qi, X.; Liu, L.; Liu, Z.; Ma, S.; Wu, J. Epigenetic Regulation of m6A Modifications in Human Cancer. Mol. Ther. Nucleic Acids 2020, 19, 405–412. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, L.; Qiang, P. The Potential Role of N6-Methyladenosine (m6A) Demethylase Fat Mass and Obesity-Associated Gene (FTO) in Human Cancers. Onco. Targets. Ther. 2020, 13, 12845–12856. [Google Scholar] [CrossRef]
- Xie, S.; Chen, W.; Chen, K.; Chang, Y.; Yang, F.; Lin, A.; Shu, Q.; Zhou, T.; Yan, X. Emerging roles of RNA methylation in gastrointestinal cancers. Cancer Cell Int. 2020, 20, 585. [Google Scholar] [CrossRef]
- Yang, X.; Hu, X.; Liu, J.; Wang, R.; Zhang, C.; Han, F.; Chen, Y.; Ma, D. N6-methyladenine modification in noncoding RNAs and its function in cancer. Biomark. Res. 2020, 8, 61. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Du, Y.; Zhou, M.; Hu, Y.; Zhang, S. Emerging roles of N6-methyladenosine (m6A) modification in breast cancer. Cell Biosci. 2020, 10, 136. [Google Scholar] [CrossRef]
- Xu, R.; Pang, G.; Zhao, Q.; Yang, L.; Chen, S.; Jiang, L.; Shen, Y.; Shao, W. The momentous role of N6-methyladenosine in lung cancer. J. Cell. Physiol. 2020, 236, 3244–3256. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.-L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.H.; et al. Epitranscriptomic m6A Regulation of Axon Regeneration in the Adult Mammalian Nervous System. Neuron 2018, 97, 313–325.e6. [Google Scholar] [CrossRef] [PubMed]
- Richard, E.M.; Polla, D.L.; Assir, M.Z.; Contreras, M.; Shahzad, M.; Khan, A.A.; Razzaq, A.; Akram, J.; Tarar, M.N.; Blanpied, T.A.; et al. Bi-allelic Variants in METTL5 Cause Autosomal-Recessive Intellectual Disability and Microcephaly. Am. J. Hum. Genet. 2019, 105, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Frayling, T.M.; Timpson, N.J.; Weedon, M.N.; Zeggini, E.; Freathy, R.M.; Lindgren, C.M.; Perry, J.R.B.; Elliott, K.S.; Lango, H.; Rayner, N.W.; et al. A Common Variant in the FTO Gene Is Associated with Body Mass Index and Predisposes to Childhood and Adult Obesity. Science 2007, 316, 889–894. [Google Scholar] [CrossRef]
- Samaan, Z.; Anand, S.; Zhang, X.; Desai, D.; Rivera, M.; Pare, G.; Thabane, L.; Xie, C.; Gerstein, H.; Engert, J.C.; et al. The protective effect of the obesity-associated rs9939609 A variant in fat mass- and obesity-associated gene on depression. Mol. Psychiatry 2013, 18, 1281–1286. [Google Scholar] [CrossRef]
- Milaneschi, Y.; Lamers, F.; Mbarek, H.; Hottenga, J.-J.; Boomsma, D.I.; Penninx, B.W.J.H. The effect of FTO rs9939609 on major depression differs across MDD subtypes. Mol. Psychiatry 2014, 19, 960–962. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, Z.; Sengupta, S.M.; Grizenko, N.; Thakur, G.A.; Fortier, M.-E.; Schmitz, N.; Joober, R. Association between obesity-related gene FTO and ADHD. Obesity 2013, 21, E738–E744. [Google Scholar] [CrossRef]
- Angelova, M.T.; Dimitrova, D.G.; Dinges, N.; Lence, T.; Worpenberg, L.; Carré, C.; Roignant, J.-Y. The Emerging Field of Epitranscriptomics in Neurodevelopmental and Neuronal Disorders. Front. Bioeng. Biotechnol. 2018, 6, 46. [Google Scholar] [CrossRef]
- Han, M.; Liu, Z.; Xu, Y.; Liu, X.; Wang, D.; Li, F.; Wang, Y.; Bi, J. Abnormality of m6A mRNA Methylation Is Involved in Alzheimer’s Disease. Front. Neurosci. 2020, 14, 98. [Google Scholar] [CrossRef]
- Bai, L.; Tang, Q.; Zou, Z.; Meng, P.; Tu, B.; Xia, Y.; Cheng, S.; Zhang, L.; Yang, K.; Mu, S.; et al. m6A Demethylase FTO Regulates Dopaminergic Neurotransmission Deficits Caused by Arsenite. Toxicol. Sci. 2018, 165, 431–446. [Google Scholar] [CrossRef]
- Chen, X.; Yu, C.; Guo, M.; Zheng, X.; Ali, S.; Huang, H.; Zhang, L.; Wang, S.; Huang, Y.; Qie, S.; et al. Down-Regulation of m6A mRNA Methylation Is Involved in Dopaminergic Neuronal Death. ACS Chem. Neurosci. 2019, 10, 2355–2363. [Google Scholar] [CrossRef]
- Wu, J.; Frazier, K.; Zhang, J.; Gan, Z.; Wang, T.; Zhong, X. Emerging role of m6A RNA methylation in nutritional physiology and metabolism. Obes. Rev. 2020, 21. [Google Scholar] [CrossRef] [PubMed]
- Fustin, J.-M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-Methylation-Dependent RNA Processing Controls the Speed of the Circadian Clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef]
- Chen, X.; Hua, W.; Huang, X.; Chen, Y.; Zhang, J.; Li, G. Regulatory Role of RNA N6-Methyladenosine Modification in Bone Biology and Osteoporosis. Front. Endocrinol. (Lausanne). 2020, 10, 911. [Google Scholar] [CrossRef]
- Yan, G.; Yuan, Y.; He, M.; Gong, R.; Lei, H.; Zhou, H.; Wang, W.; Du, W.; Ma, T.; Liu, S.; et al. m6A Methylation of Precursor-miR-320/RUNX2 Controls Osteogenic Potential of Bone Marrow-Derived Mesenchymal Stem Cells. Mol. Ther. Nucleic Acids 2020, 19, 421–436. [Google Scholar] [CrossRef]
- Dorn, L.E.; Lasman, L.; Chen, J.; Xu, X.; Hund, T.J.; Medvedovic, M.; Hanna, J.H.; van Berlo, J.H.; Accornero, F. The N6-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019, 139, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Kmietczyk, V.; Riechert, E.; Kalinski, L.; Boileau, E.; Malovrh, E.; Malone, B.; Gorska, A.; Hofmann, C.; Varma, E.; Jürgensen, L.; et al. m6A-mRNA methylation regulates cardiac gene expression and cellular growth. Life Sci. Alliance 2019, 2, e201800233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fu, J.; Zhou, Y. A Review in Research Progress Concerning m6A Methylation and Immunoregulation. Front. Immunol. 2019, 10, 922. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Liu, J.; Chen, C.; Dong, L.; Liu, Y.; Chang, R.; Huang, X.; Liu, Y.; Wang, J.; Dougherty, U.; et al. Anti-tumour immunity controlled through mRNA m6A methylation and YTHDF1 in dendritic cells. Nature 2019, 566, 270–274. [Google Scholar] [CrossRef]
- Paramasivam, A.; Priyadharsini, J.V.; Raghunandhakumar, S. Implications of m6A modification in autoimmune disorders. Cell. Mol. Immunol. 2020, 17, 550–551. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, G.C. Autoimmunity and organ damage in systemic lupus erythematosus. Nat. Immunol. 2020, 21, 605–614. [Google Scholar] [CrossRef]
- Kiriakidou, M.; Ching, C.L. Systemic Lupus Erythematosus. Ann. Intern. Med. 2020, 172, ITC81–ITC96. [Google Scholar] [CrossRef] [PubMed]
- Drehmel, K.R.; Erickson, A.R.; England, B.R.; Michaud, K.D.; Sayles, H.R.; Hearth-Holmes, M.P. Applying SLICC and ACR/EULAR systemic lupus erythematosus classification criteria in a cohort of patients with undifferentiated connective tissue disease. Lupus 2020, 30. [Google Scholar] [CrossRef]
- Porter, D.; Basu, N.; Siebert, S. Classification criteria: Time for a rethink? Ann. Rheum. Dis. 2020, 79, 1264–1266. [Google Scholar] [CrossRef] [PubMed]
- Farivar, S.; Shaabanpour Aghamaleki, F. Effects of Major Epigenetic Factors on Systemic Lupus Erythematosus. Iran. Biomed. J. 2018, 22, 294–302. [Google Scholar] [CrossRef]
- Guo, Y.; Sawalha, A.H.; Lu, Q. Epigenetics in the treatment of systemic lupus erythematosus: Potential clinical application. Clin. Immunol. 2014, 155, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Coit, P.; Renauer, P.; Jeffries, M.A.; Merrill, J.T.; McCune, W.J.; Maksimowicz-McKinnon, K.; Sawalha, A.H. Renal involvement in lupus is characterized by unique DNA methylation changes in naïve CD4+ T cells. J. Autoimmun. 2015, 61, 29–35. [Google Scholar] [CrossRef]
- Honarpisheh, M.; Köhler, P.; von Rauchhaupt, E.; Lech, M. The Involvement of MicroRNAs in Modulation of Innate and Adaptive Immunity in Systemic Lupus Erythematosus and Lupus Nephritis. J. Immunol. Res. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Hu, N.; Qiu, X.; Luo, Y.; Yuan, J.; Li, Y.; Lei, W.; Zhang, G.; Zhou, Y.; Su, Y.; Lu, Q. Abnormal histone modification patterns in lupus CD4+ T cells. J. Rheumatol. 2008, 35, 804–810. [Google Scholar] [PubMed]
- Javierre, B.M.; Fernandez, A.F.; Richter, J.; Al-Shahrour, F.; Martin-Subero, J.I.; Rodriguez-Ubreva, J.; Berdasco, M.; Fraga, M.F.; O’Hanlon, T.P.; Rider, L.G.; et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010, 20, 170–179. [Google Scholar] [CrossRef]
- Luo, Q.; Fu, B.; Zhang, L.; Guo, Y.; Huang, Z.; Li, J. Decreased Peripheral Blood ALKBH5 Correlates with Markers of Autoimmune Response in Systemic Lupus Erythematosus. Dis. Markers 2020, 2020, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Rao, J.; Zhang, L.; Fu, B.; Guo, Y.; Huang, Z.; Li, J. The study of METTL14, ALKBH5, and YTHDF2 in peripheral blood mononuclear cells from systemic lupus erythematosus. Mol. Genet. Genomic Med. 2020, 8, e1298. [Google Scholar] [CrossRef]
- Furlan, M.; Galeota, E.; De Pretis, S.; Caselle, M.; Pelizzola, M. m6A-Dependent RNA Dynamics in T Cell Differentiation. Genes 2019, 10, 28. [Google Scholar] [CrossRef]
- Luo, S.; Wang, Y.; Zhao, M.; Lu, Q. The important roles of type I interferon and interferon-inducible genes in systemic lupus erythematosus. Int. Immunopharmacol. 2016, 40, 542–549. [Google Scholar] [CrossRef]
- McInnes, I.B.; Schett, G. The Pathogenesis of Rheumatoid Arthritis. N. Engl. J. Med. 2011, 365, 2205–2219. [Google Scholar] [CrossRef]
- Littlejohn, E.A.; Monrad, S.U. Early Diagnosis and Treatment of Rheumatoid Arthritis. Prim. Care Clin. Off. Pract. 2018, 45, 237–255. [Google Scholar] [CrossRef]
- Blum, A.; Adawi, M. RETRACTED: Rheumatoid arthritis (RA) and cardiovascular disease. Autoimmun. Rev. 2019, 18, 679–690. [Google Scholar] [CrossRef]
- Brandt, B.; Rashidiani, S.; Bán, Á.; Rauch, T.A. DNA Methylation-Governed Gene Expression in Autoimmune Arthritis. Int. J. Mol. Sci. 2019, 20, 5646. [Google Scholar] [CrossRef]
- Conrad, K.; Roggenbuck, D.; Reinhold, D.; Dörner, T. Profiling of rheumatoid arthritis associated autoantibodies. Autoimmun. Rev. 2010, 9, 431–435. [Google Scholar] [CrossRef]
- Firestein, G.S.; Budd, R.C.; Gabriel, S.E.; McInnes, I.B.; O’Dell, J.R. Front Matter. In Kelley’s Textbook of Rheumatology; Elsevier: Amsterdam, The Netherlands, 2013; pp. i–iii. [Google Scholar]
- Nemtsova, M.V.; Zaletaev, D.V.; Bure, I.V.; Mikhaylenko, D.S.; Kuznetsova, E.B.; Alekseeva, E.A.; Beloukhova, M.I.; Deviatkin, A.A.; Lukashev, A.N.; Zamyatnin, A.A. Epigenetic Changes in the Pathogenesis of Rheumatoid Arthritis. Front. Genet. 2019, 10, 570. [Google Scholar] [CrossRef] [PubMed]
- Coutant, F.; Miossec, P. Evolving concepts of the pathogenesis of rheumatoid arthritis with focus on the early and late stages. Curr. Opin. Rheumatol. 2020, 32, 57–63. [Google Scholar] [CrossRef]
- Scott, D.L.; Wolfe, F.; Huizinga, T.W. Rheumatoid arthritis. Lancet 2010, 376, 1094–1108. [Google Scholar] [CrossRef]
- Doody, K.M.; Bottini, N.; Firestein, G.S. Epigenetic alterations in rheumatoid arthritis fibroblast-like synoviocytes. Epigenomics 2017, 9, 479–492. [Google Scholar] [CrossRef] [PubMed]
- Ospelt, C. Epigenetic biomarkers in rheumatology—The future? Swiss Med. Wkly. 2016, 146, w14312. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hardy, R.S.; Hülso, C.; Liu, Y.; Gasparini, S.J.; Fong-Yee, C.; Tu, J.; Stoner, S.; Stewart, P.M.; Raza, K.; Cooper, M.S.; et al. Characterisation of fibroblast-like synoviocytes from a murine model of joint inflammation. Arthritis Res. Ther. 2013, 15, R24. [Google Scholar] [CrossRef] [PubMed]
- Karouzakis, E.; Gay, R.E.; Michel, B.A.; Gay, S.; Neidhart, M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2009, 60, 3613–3622. [Google Scholar] [CrossRef]
- Liu, C.-C.; Fang, T.-J.; Ou, T.-T.; Wu, C.-C.; Li, R.-N.; Lin, Y.-C.; Lin, C.-H.; Tsai, W.-C.; Liu, H.-W.; Yen, J.-H. Global DNA methylation, DNMT1, and MBD2 in patients with rheumatoid arthritis. Immunol. Lett. 2011, 135, 96–99. [Google Scholar] [CrossRef]
- Romano, M.; Fanelli, G.; Albany, C.J.; Giganti, G.; Lombardi, G. Past, Present, and Future of Regulatory T Cell Therapy in Transplantation and Autoimmunity. Front. Immunol. 2019, 10, 43. [Google Scholar] [CrossRef]
- Ibáñez-Cabellos, J.S.; Seco-Cervera, M.; Osca-Verdegal, R.; Pallardó, F.V.; García-Giménez, J.L. Epigenetic Regulation in the Pathogenesis of Sjögren Syndrome and Rheumatoid Arthritis. Front. Genet. 2019, 10, 1104. [Google Scholar] [CrossRef]
- Guo, S.; Xu, L.; Chang, C.; Zhang, R.; Jin, Y.; He, D. Epigenetic Regulation Mediated by Methylation in the Pathogenesis and Precision Medicine of Rheumatoid Arthritis. Front. Genet. 2020, 11, 811. [Google Scholar] [CrossRef]
- Zhang, W.; He, L.; Liu, Z.; Ren, X.; Qi, L.; Wan, L.; Wang, W.; Tu, C.; Li, Z. Multifaceted Functions and Novel Insight Into the Regulatory Role of RNA N6-Methyladenosine Modification in Musculoskeletal Disorders. Front. Cell Dev. Biol. 2020, 8, 870. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.-B.; Zhang, Y.-H.; Lei, S.-F. Genome-Wide Identification of N6-Methyladenosine (m6A) SNPs Associated with Rheumatoid Arthritis. Front. Genet. 2018, 9, 299. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yan, S.; Lu, H.; Wang, S.; Xu, D. METTL3 Attenuates LPS-Induced Inflammatory Response in Macrophages via NF-κB Signaling Pathway. Mediators Inflamm. 2019, 2019, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Gao, Y.; Zhang, L.; Rao, J.; Guo, Y.; Huang, Z.; Li, J. Decreased ALKBH5, FTO, and YTHDF2 in Peripheral Blood Are as Risk Factors for Rheumatoid Arthritis. Biomed Res. Int. 2020, 2020, 1–9. [Google Scholar] [CrossRef]
- Honma, M.; Hayashi, K. Psoriasis: Recent progress in molecular-targeted therapies. J. Dermatol. 2021. [Google Scholar] [CrossRef]
- Wang, Y.-N.; Jin, H.-Z. Transcriptome-Wide m6A Methylation in Skin Lesions from Patients with Psoriasis Vulgaris. Front. Cell Dev. Biol. 2020, 8, 591629. [Google Scholar] [CrossRef]
- Perera, G.K.; Di Meglio, P.; Nestle, F.O. Psoriasis. Annu. Rev. Pathol. Mech. Dis. 2012, 7, 385–422. [Google Scholar] [CrossRef]
- Caputo, V.; Strafella, C.; Termine, A.; Dattola, A.; Mazzilli, S.; Lanna, C.; Cosio, T.; Campione, E.; Novelli, G.; Giardina, E.; et al. Overview of the molecular determinants contributing to the expression of Psoriasis and Psoriatic Arthritis phenotypes. J. Cell. Mol. Med. 2020, 24, 13554–13563. [Google Scholar] [CrossRef]
- Takeshita, J.; Grewal, S.; Langan, S.M.; Mehta, N.N.; Ogdie, A.; Van Voorhees, A.S.; Gelfand, J.M. Psoriasis and comorbid diseases. J. Am. Acad. Dermatol. 2017, 76, 393–403. [Google Scholar] [CrossRef]
- TAKAHASHI, H.; IIZUKA, H. Psoriasis and metabolic syndrome. J. Dermatol. 2012, 39, 212–218. [Google Scholar] [CrossRef]
- Afonina, I.S.; Van Nuffel, E.; Beyaert, R. Immune responses and therapeutic options in psoriasis. Cell. Mol. Life Sci. 2021, 1–9. [Google Scholar]
- O’Rielly, D.D.; Rahman, P. Genetic, Epigenetic and Pharmacogenetic Aspects of Psoriasis and Psoriatic Arthritis. Rheum. Dis. Clin. North Am. 2015, 41, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Prinz, J.C. Melanocytes: Target Cells of an HLA-C*06:02–Restricted Autoimmune Response in Psoriasis. J. Investig. Dermatol. 2017, 137, 2053–2058. [Google Scholar] [CrossRef] [PubMed]
- Lowes, M.A.; Suárez-Fariñas, M.; Krueger, J.G. Immunology of Psoriasis. Annu. Rev. Immunol. 2014, 32, 227–255. [Google Scholar] [CrossRef] [PubMed]
- Elder, J.T. PSORS1: Linking Genetics and Immunology. J. Investig. Dermatol. 2006, 126, 1205–1206. [Google Scholar] [CrossRef] [PubMed]
- Nair, R.P.; Stuart, P.E.; Nistor, I.; Hiremagalore, R.; Chia, N.V.C.; Jenisch, S.; Weichenthal, M.; Abecasis, G.R.; Lim, H.W.; Christophers, E.; et al. Sequence and Haplotype Analysis Supports HLA-C as the Psoriasis Susceptibility 1 Gene. Am. J. Hum. Genet. 2006, 78, 827–851. [Google Scholar] [CrossRef]
- Arakawa, A.; Siewert, K.; Stöhr, J.; Besgen, P.; Kim, S.-M.; Rühl, G.; Nickel, J.; Vollmer, S.; Thomas, P.; Krebs, S.; et al. Melanocyte antigen triggers autoimmunity in human psoriasis. J. Exp. Med. 2015, 212, 2203–2212. [Google Scholar] [CrossRef]
- Capon, F. The Genetic Basis of Psoriasis. Int. J. Mol. Sci. 2017, 18, 2526. [Google Scholar] [CrossRef]
- Jordan, C.T.; Cao, L.; Roberson, E.D.O.; Pierson, K.C.; Yang, C.-F.; Joyce, C.E.; Ryan, C.; Duan, S.; Helms, C.A.; Liu, Y.; et al. PSORS2 Is Due to Mutations in CARD14. Am. J. Hum. Genet. 2012, 90, 784–795. [Google Scholar] [CrossRef]
- Jordan, C.T.; Cao, L.; Roberson, E.D.O.; Duan, S.; Helms, C.A.; Nair, R.P.; Duffin, K.C.; Stuart, P.E.; Goldgar, D.; Hayashi, G.; et al. Rare and Common Variants in CARD14, Encoding an Epidermal Regulator of NF-kappaB, in Psoriasis. Am. J. Hum. Genet. 2012, 90, 796–808. [Google Scholar] [CrossRef]
- Zhang, P.; Su, Y.; Zhao, M.; Huang, W.; Lu, Q. Abnormal histone modifications in PBMCs from patients with psoriasis vulgaris. Eur. J. Dermatology 2011, 21, 552–557. [Google Scholar] [CrossRef]
- Ovejero-Benito, M.C.; Reolid, A.; Sánchez-Jiménez, P.; Saiz-Rodríguez, M.; Muñoz-Aceituno, E.; Llamas-Velasco, M.; Martín-Vilchez, S.; Cabaleiro, T.; Román, M.; Ochoa, D.; et al. Histone modifications associated with biological drug response in moderate-to-severe psoriasis. Exp. Dermatol. 2018, 27, 1361–1371. [Google Scholar] [CrossRef]
- Nedoszytko, B.; Szczerkowska-Dobosz, A.; Stawczyk-Macieja, M.; Owczarczyk-Saczonek, A.; Reich, A.; Bartosińska, J.; Batycka-Baran, A.; Czajkowski, R.; Dobrucki, I.; Dobrucki, L.; et al. Pathogenesis of psoriasis in the “omic” era. Part II. Genetic, genomic and epigenetic changes in psoriasis. Adv. Dermatology Allergol. 2020, 37, 283–298. [Google Scholar] [CrossRef] [PubMed]
- Gervin, K.; Vigeland, M.D.; Mattingsdal, M.; Hammerø, M.; Nygård, H.; Olsen, A.O.; Brandt, I.; Harris, J.R.; Undlien, D.E.; Lyle, R. DNA Methylation and Gene Expression Changes in Monozygotic Twins Discordant for Psoriasis: Identification of Epigenetically Dysregulated Genes. PLoS Genet. 2012, 8, e1002454. [Google Scholar] [CrossRef] [PubMed]
- Brandt, D.; Sergon, M.; Abraham, S.; Mäbert, K.; Hedrich, C.M. TCR + CD3 + CD4 − CD8 − effector T cells in psoriasis. Clin. Immunol. 2017, 181, 51–59. [Google Scholar] [CrossRef]
- Chandra, A.; Senapati, S.; Roy, S.; Chatterjee, G.; Chatterjee, R. Epigenome-wide DNA methylation regulates cardinal pathological features of psoriasis. Clin. Epigenetics 2018, 10, 108. [Google Scholar] [CrossRef] [PubMed]
- Roberson, E.D.O.; Liu, Y.; Ryan, C.; Joyce, C.E.; Duan, S.; Cao, L.; Martin, A.; Liao, W.; Menter, A.; Bowcock, A.M. A Subset of Methylated CpG Sites Differentiate Psoriatic from Normal Skin. J. Investig. Dermatol. 2012, 132, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Ekman, A.-K.; Bivik Eding, C.; Enerbäck, C. Genome-Wide DNA Methylation Profiling Identifies Differential Methylation in Uninvolved Psoriatic Epidermis. J. Investig. Dermatol. 2018, 138, 1088–1093. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Nylander, E.; Coates, P.J.; Fahraeus, R.; Nylander, K. Correlation between Reversal of DNA Methylation and Clinical Symptoms in Psoriatic Epidermis Following Narrow-Band UVB Phototherapy. J. Investig. Dermatol. 2015, 135, 2077–2083. [Google Scholar] [CrossRef]
- Ghasemi, N.; Razavi, S.; Nikzad, E. Multiple Sclerosis: Pathogenesis, Symptoms, Diagnoses and Cell-Based Therapy. Cell J. 2017, 19, 4867. [Google Scholar]
- Lassmann, H. Multiple Sclerosis Pathology. Cold Spring Harb. Perspect. Med. 2018, 8, a028936. [Google Scholar] [CrossRef] [PubMed]
- Tafti, D.; Ehsan, M.; Xixis, K.L. Multiple Sclerosis. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2021. [Google Scholar]
- Compston, A.; Coles, A. Multiple sclerosis. Lancet 2008, 372, 1502–1517. [Google Scholar] [CrossRef]
- Celius, E.G.; Thompson, H.; Pontaga, M.; Langdon, D.; Laroni, A.; Potra, S.; Bharadia, T.; Yeandle, D.; Shanahan, J.; van Galen, P.; et al. Disease Progression in Multiple Sclerosis: A Literature Review Exploring Patient Perspectives. Patient Prefer. Adherence 2021, 15, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Jennum, P.; Wanscher, B.; Frederiksen, J.; Kjellberg, J. The socioeconomic consequences of multiple sclerosis: A controlled national study. Eur. Neuropsychopharmacol. 2012, 22, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Sattler, A.; Wagner, U.; Rossol, M.; Sieper, J.; Wu, P.; Krause, A.; Schmidt, W.A.; Radmer, S.; Kohler, S.; Romagnani, C.; et al. Cytokine-induced human IFN-γ–secreting effector-memory Th cells in chronic autoimmune inflammation. Blood 2009, 113, 1948–1956. [Google Scholar] [CrossRef]
- Young, H.A.; Hardy, K.J. Role of interferon-gamma in immune cell regulation. J. Leukoc. Biol. 1995, 58, 373–381. [Google Scholar] [CrossRef]
- Cao, Y.; Goods, B.A.; Raddassi, K.; Nepom, G.T.; Kwok, W.W.; Love, J.C.; Hafler, D.A. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci. Transl. Med. 2015, 7, ra74–ra287. [Google Scholar] [CrossRef]
- Takeshita, Y.; Ransohoff, R.M. Inflammatory cell trafficking across the blood-brain barrier: Chemokine regulation and in vitro models. Immunol. Rev. 2012, 248, 228–239. [Google Scholar] [CrossRef]
- Correale, J.; Farez, M.F. The Role of Astrocytes in Multiple Sclerosis Progression. Front. Neurol. 2015, 6, 217. [Google Scholar] [CrossRef]
- Celarain, N.; Tomas-Roig, J. Aberrant DNA methylation profile exacerbates inflammation and neurodegeneration in multiple sclerosis patients. J. Neuroinflammation 2020, 17, 21. [Google Scholar] [CrossRef] [PubMed]
- Denic, A.; Wootla, B.; Rodriguez, M. CD8 + T cells in multiple sclerosis. Expert Opin. Ther. Targets 2013, 17, 1053–1066. [Google Scholar] [CrossRef]
- Jelcic, I.; Al Nimer, F.; Wang, J.; Lentsch, V.; Planas, R.; Jelcic, I.; Madjovski, A.; Ruhrmann, S.; Faigle, W.; Frauenknecht, K.; et al. Memory B Cells Activate Brain-Homing, Autoreactive CD4+ T Cells in Multiple Sclerosis. Cell 2018, 175, 85–100.e23. [Google Scholar] [CrossRef] [PubMed]
- Krumbholz, M.; Derfuss, T.; Hohlfeld, R.; Meinl, E. B cells and antibodies in multiple sclerosis pathogenesis and therapy. Nat. Rev. Neurol. 2012, 8, 613–623. [Google Scholar] [CrossRef]
- Sawcer, S.; Franklin, R.J.M.; Ban, M. Multiple sclerosis genetics. Lancet Neurol. 2014, 13, 700–709. [Google Scholar] [CrossRef]
- Gourraud, P.-A.; Harbo, H.F.; Hauser, S.L.; Baranzini, S.E. The genetics of multiple sclerosis: An up-to-date review. Immunol. Rev. 2012, 248, 87–103. [Google Scholar] [CrossRef]
- Lill, C.M. Recent Advances and Future Challenges in the Genetics of Multiple Sclerosis. Front. Neurol. 2014, 5, 130. [Google Scholar] [CrossRef] [PubMed]
- Mo, X.-B.; Lei, S.-F.; Qian, Q.-Y.; Guo, Y.-F.; Zhang, Y.-H.; Zhang, H. Integrative analysis revealed potential causal genetic and epigenetic factors for multiple sclerosis. J. Neurol. 2019, 266, 2699–2709. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Nie, P.; Peng, D.; He, Z.; Liu, M.; Xie, Y.; Miao, Y.; Zuo, Z.; Ren, J. m6AVar: A database of functional variants involved in m6A modification. Nucleic Acids Res. 2018, 46, D139–D145. [Google Scholar] [CrossRef] [PubMed]
- da Costa, T.P.; El-Cheikh, M.C.; Carneiro, K. Epigenetic Therapy as a Putative Molecular Target to Modulate B Cell Biology and Behavior in the Context of Immunological Disorders. J. Immunol. Res. 2020, 2020, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Jamebozorgi, K.; Rostami, D.; Pormasoumi, H.; Taghizadeh, E.; Barreto, G.E.; Sahebkar, A. Epigenetic aspects of multiple sclerosis and future therapeutic options. Int. J. Neurosci. 2021, 131, 56–64. [Google Scholar] [CrossRef]
- Tóth, D.M.; Ocskó, T.; Balog, A.; Markovics, A.; Mikecz, K.; Kovács, L.; Jolly, M.; Bukiej, A.A.; Ruthberg, A.D.; Vida, A.; et al. Amelioration of Autoimmune Arthritis in Mice Treated with the DNA Methyltransferase Inhibitor 5′-Azacytidine. Arthritis Rheumatol. 2019, 71, 1265–1275. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Wu, A.; Tesmer, L.; Ray, D.; Yousif, N.; Richardson, B. Demethylation of CD40LG on the Inactive X in T Cells from Women with Lupus. J. Immunol. 2007, 179, 6352–6358. [Google Scholar] [CrossRef] [PubMed]
- Petralia, M.C.; Mazzon, E.; Basile, M.S.; Cutuli, M.; Di Marco, R.; Scandurra, F.; Saraceno, A.; Fagone, P.; Nicoletti, F.; Mangano, K. Effects of Treatment with the Hypomethylating Agent 5-aza-2′-deoxycytidine in Murine Type II Collagen-Induced Arthritis. Pharmaceuticals 2019, 12, 174. [Google Scholar] [CrossRef] [PubMed]
- Mangano, K.; Fagone, P.; Bendtzen, K.; Meroni, P.L.; Quattrocchi, C.; Mammana, S.; Di Rosa, M.; Malaguarnera, L.; Coco, M.; Magro, G.; et al. Hypomethylating Agent 5-Aza-2′-deoxycytidine (DAC) Ameliorates Multiple Sclerosis in Mouse Models. J. Cell. Physiol. 2014, 229, 1918–1925. [Google Scholar] [CrossRef] [PubMed]
- Morinobu, A.; Tanaka, S.; Nishimura, K.; Takahashi, S.; Kageyama, G.; Miura, Y.; Kurosaka, M.; Saegusa, J.; Kumagai, S. Expression and Functions of Immediate Early Response Gene X-1 (IEX-1) in Rheumatoid Arthritis Synovial Fibroblasts. PLoS ONE 2016, 11, e0164350. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Zhang, B. Trichostatin A, an Inhibitor of Histone Deacetylase, Inhibits the Viability and Invasiveness of Hypoxic Rheumatoid Arthritis Fibroblast-Like Synoviocytes via PI3K/Akt Signaling. J. Biochem. Mol. Toxicol. 2016, 30, 163–169. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, A.; Sharma, M.; Prabhakar, B.; Holterman, M.; Jayaraman, S. Amelioration of progressive autoimmune encephalomyelitis by epigenetic regulation involves selective repression of mature neutrophils during the preclinical phase. Exp. Neurol. 2018, 304, 14–20. [Google Scholar] [CrossRef]
- Jayaraman, A.; Soni, A.; Prabhakar, B.S.; Holterman, M.; Jayaraman, S. The epigenetic drug Trichostatin A ameliorates experimental autoimmune encephalomyelitis via T cell tolerance induction and impaired influx of T cells into the spinal cord. Neurobiol. Dis. 2017, 108, 1–12. [Google Scholar] [CrossRef]
- Jorn Bovenschen, H.; van de Kerkhof, P.C.; van Erp, P.E.; Woestenenk, R.; Joosten, I.; Koenen, H.J.P.M. Foxp3+ Regulatory T Cells of Psoriasis Patients Easily Differentiate into IL-17A-Producing Cells and Are Found in Lesional Skin. J. Investig. Dermatol. 2011, 131, 1853–1860. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Zhao, D.; Kirschbaum, M.; Zhang, C.; Lin, C.-L.; Todorov, I.; Kandeel, F.; Forman, S.; Zeng, D. HDAC inhibitor reduces cytokine storm and facilitates induction of chimerism that reverses lupus in anti-CD3 conditioning regimen. Proc. Natl. Acad. Sci. USA 2008, 105, 4796–4801. [Google Scholar] [CrossRef] [PubMed]
- Elkashef, S.M.; Lin, A.-P.; Myers, J.; Sill, H.; Jiang, D.; Dahia, P.L.M.; Aguiar, R.C.T. IDH Mutation, Competitive Inhibition of FTO, and RNA Methylation. Cancer Cell 2017, 31, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Su, R.; Dong, L.; Li, C.; Nachtergaele, S.; Wunderlich, M.; Qing, Y.; Deng, X.; Wang, Y.; Weng, X.; Hu, C.; et al. R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m6A/MYC/CEBPA Signaling. Cell 2018, 172, 90–105.e23. [Google Scholar] [CrossRef]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691.e10. [Google Scholar] [CrossRef]
- Huang, Y.; Yan, J.; Li, Q.; Li, J.; Gong, S.; Zhou, H.; Gan, J.; Jiang, H.; Jia, G.-F.; Luo, C.; et al. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015, 43, 373–384. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhou, B.; Liu, W.; Zhang, M.; Shen, Z.; Han, Z.; Jiang, Q.; Yang, Q.; Song, C.; Wang, R.; et al. Identification of A Novel Small-Molecule Binding Site of the Fat Mass and Obesity Associated Protein (FTO). J. Med. Chem. 2015, 58, 7341–7348. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 2020, 38, 79–96.e11. [Google Scholar] [CrossRef]
- Singh, B.; Kinne, H.E.; Milligan, R.D.; Washburn, L.J.; Olsen, M.; Lucci, A. Important Role of FTO in the Survival of Rare Panresistant Triple-Negative Inflammatory Breast Cancer Cells Facing a Severe Metabolic Challenge. PLoS ONE 2016, 11, e0159072. [Google Scholar] [CrossRef]
- Islam, M.S.; Leissing, T.M.; Chowdhury, R.; Hopkinson, R.J.; Schofield, C.J. 2-Oxoglutarate-Dependent Oxygenases. Annu. Rev. Biochem. 2018, 87, 585–620. [Google Scholar] [CrossRef] [PubMed]
- Rose, N.R.; McDonough, M.A.; King, O.N.F.; Kawamura, A.; Schofield, C.J. Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev. 2011, 40, 4364. [Google Scholar] [CrossRef]
- Yang, S.; Wei, J.; Cui, Y.-H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. m6A mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Chan, M.W.Y.; Chang, C.-B.; Tung, C.-H.; Sun, J.; Suen, J.-L.; Wu, S.-F. Low-Dose 5-Aza-2′-deoxycytidine Pretreatment Inhibits Experimental Autoimmune Encephalomyelitis by Induction of Regulatory T Cells. Mol. Med. 2014, 20, 248–256. [Google Scholar] [CrossRef]
- Börner, C.; Martella, E.; Höllt, V.; Kraus, J. Regulation of Opioid and Cannabinoid Receptor Genes in Human Neuroblastoma and T Cells by the Epigenetic Modifiers Trichostatin A and 5-Aza-2′-Deoxycytidine. Neuroimmunomodulation 2012, 19, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Camelo, S.; Iglesias, A.H.; Hwang, D.; Due, B.; Ryu, H.; Smith, K.; Gray, S.G.; Imitola, J.; Duran, G.; Assaf, B.; et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin A ameliorates experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2005, 164, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Ge, Z.; Da, Y.; Xue, Z.; Zhang, K.; Zhuang, H.; Peng, M.; Li, Y.; Li, W.; Simard, A.; Hao, J.; et al. Vorinostat, a histone deacetylase inhibitor, suppresses dendritic cell function and ameliorates experimental autoimmune encephalomyelitis. Exp. Neurol. 2013, 241, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.W.; Song, J.W.; Ha, N.; Choi, Y.I.; Kim, S. CKD-506, a novel HDAC6-selective inhibitor, improves renal outcomes and survival in a mouse model of systemic lupus erythematosus. Sci. Rep. 2018, 8, 17297. [Google Scholar] [CrossRef]
- Ren, J.; Catalina, M.D.; Eden, K.; Liao, X.; Read, K.A.; Luo, X.; McMillan, R.P.; Hulver, M.W.; Jarpe, M.; Bachali, P.; et al. Selective Histone Deacetylase 6 Inhibition Normalizes B Cell Activation and Germinal Center Formation in a Model of Systemic Lupus Erythematosus. Front. Immunol. 2019, 10, 2512. [Google Scholar] [CrossRef]
- Du, C.; Liu, C.; Kang, J.; Zhao, G.; Ye, Z.; Huang, S.; Li, Z.; Wu, Z.; Pei, G. MicroRNA miR-326 regulates TH-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 2009, 10, 1252–1259. [Google Scholar] [CrossRef]
| Method Name | Abbreviation | Method Principle | Sensitivity | Ref. |
|---|---|---|---|---|
| Dot blot technology | - | Semiquantitative antibody-dependent identification on a membrane | Medium | [54,55] |
| Immune-Northern blot | - | UV-crosslinking followed by detection with specific antibody on the nylon membrane | Medium | [57,58] |
| Methyl sensitivity of MazF RNA endonuclease | MAZTER-Seq | Restriction with endonuclease that cuts methylated but not unmethylated nucleotides | High | [72] |
| m6A-sensitive RNA-endoribonuclease-facilitated sequencing | m6A–REF-Seq | Restriction with endonuclease that cuts methylated but not unmethylated nucleotides | High | [73] |
| Site-specific cleavage and radioactive-labeling followed by ligation-assisted extraction and thin-layer chromatography | SCARLET | Restriction with endonuclease that cuts methylated but not unmethylated nucleotides | High | [64] |
| N6-methyladenosine sequencing | m6A-seq | Immunoprecipitation with m6A-specific antibody | Medium | [74] |
| Methylated RNA immunoprecipitation sequencing | MeRIP-Seq | Immunoprecipitation with m6A-specific antibody | Medium | [62] |
| m6A-level and isoform-characterization sequencing | m6A-LAIC-seq | Immunoprecipitation with m6A-specific antibody | Medium | [65] |
| Cross-linking immunoprecipitation | miCLIP, eCLIP, m6A-CLIP | Crosslinking and immunoprecipitation with m6A-specific antibody | High | [66,67,75] |
| FTO-assisted chemical labeling method | m6A-SEAL-Seq | FTO-assisted oxidation to hm6A for biotin labelling to immunoprecipitation | High | [76] |
| Autoimmune Disease | Cells/Tissues Studied | Analyzed Elements/Mechanisms | Observations | Ref. |
|---|---|---|---|---|
| SLE | PBMC | m6A enzymes Writers: METTL3, METTL14, WTAP Erasers: FTO and ALKBH5 Readers: YTHDF2 | Decreased expression of METTL14, ALKBH5 and YTHDF2 associated with clinical features of SLE patients | [134] |
| PBMC | m6A enzymes Writers: METTL3, METTL14, WTAP Erasers: FTO and ALKBH5 Readers: YTHDF2, Comparison between SLE, RA, HBV, TB patients | Decreased expression of ALKHB5 related to clinical symptoms; METTL3, WTAP and FTO as potential collaborators of autoreactivity in SLE | [135] | |
| RA | PBMC, m6AVar database | GWAS m6A-associated SNPs | Detection of m6A-SNPs within genes in immune cells and m6A regulators encoded genes | [156] |
| PBMC, LPS-stimulated THP-1 | m6A writer: METTL3 | LPS stimulation enhances expression and biological activity of METTL3, overexpression of METTL3 attenuates inflammation, | [157] | |
| PBMC | m6A enzymes Writers: METTL3, METTL14, WTAP Erasers: FTO and ALKBH5 Readers: YTHDF2 | global m6A content increase and ALKBH5, FTO, YTHDF2 decrease in RA | [158] | |
| MS | GEO database | Association between m6A-SNPs and gene expression | Identification of 13 m6A-SNPs, rs923829 in METTL21B and rs2288481 in DKKL1 gene and association with MS | [202] |
| Ps | Skin samples | m6A methylation pattern, DMR | Hypomethylated transcripts in psoriasis affected skin linked with Wnt signaling pathway | [160] |
| Drug | Mechanism of Action | Disease/Model of Study | Effect | Ref. |
|---|---|---|---|---|
| Azacytidine (5-azaC, AZA) | DNMT inhibitor | SLE/human isolated T cells (CD4+, CD8+) | Amelioration of SLE symptoms | [207] |
| MS/murine EAE model | Suppression of CNS inflammation | [226] | ||
| RA/murine model of proteoglycan-induced arthritis | Amelioration of autoimmune arthritis | [206] | ||
| Decitabine (5-aza-dC, DAC) | DNMT inhibitor | MS/murine EAE model | Improvement of disease course | [209,227] |
| RA/murine model of type II collagen induced arthritis | Amelioration of the clinical condition, diminished production of Th1 and Th17 proinflammatory cytokines | [208] | ||
| Trichostatin A (TSA) | Histone deacetylase inhibitor (Pan HDAC inhibitor) | MS/murine EAE model | Reduction of spinal cord inflammation, demyelination, neuronal and axonal loss and amelioration of disability | [228] |
| MS/murine EAE model | Amelioration of neurodegeneration, reduced number of neutrophils | [212] | ||
| MS/murine EAE model | Reduction of migration of T cells to the spinal cord and improved clinical outcome | [213] | ||
| RA/human RASFs | Proinflammatory cytokine suppression and induction of apoptosis | [210] | ||
| RA/human hypoxic RAFLS | Reduction of cell viability and increased apoptosis | [211] | ||
| Psoriasis/human CD4+ T cells | Prevention of Treg differentiation into Th17 cells | [214] | ||
| Vorinostat (SAHA) | Histone deacetylase inhibitor (Pan HDAC inhibitor) | MS/human moDCs, murine EAE model | Inhibition of moDCs function (activation, maturation, antigen presentation); amelioration of CNS inflammation and demyelination | [229] |
| CKD-506 | Selective histone deacetylase inhibitor | SLE/murine model of SLE | Decrease in the production of proinflammatory cytokines and improved renal outcomes | [230] |
| ACY-738 | Selective histone deacetylase inhibitor | SLE/murine model of SLE | Decrease in B cell activation signaling pathways and reduction of PC differentiation | [231] |
| miRNA sponges | miRNA depletion | MS/cell culture/murine EAE model | Reduced number of Th17 cells | [232] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wardowska, A. m6A RNA Methylation in Systemic Autoimmune Diseases—A New Target for Epigenetic-Based Therapy? Pharmaceuticals 2021, 14, 218. https://doi.org/10.3390/ph14030218
Wardowska A. m6A RNA Methylation in Systemic Autoimmune Diseases—A New Target for Epigenetic-Based Therapy? Pharmaceuticals. 2021; 14(3):218. https://doi.org/10.3390/ph14030218
Chicago/Turabian StyleWardowska, Anna. 2021. "m6A RNA Methylation in Systemic Autoimmune Diseases—A New Target for Epigenetic-Based Therapy?" Pharmaceuticals 14, no. 3: 218. https://doi.org/10.3390/ph14030218
APA StyleWardowska, A. (2021). m6A RNA Methylation in Systemic Autoimmune Diseases—A New Target for Epigenetic-Based Therapy? Pharmaceuticals, 14(3), 218. https://doi.org/10.3390/ph14030218






