Mechanistic Modelling Identifies and Addresses the Risks of Empiric Concentration-Guided Sorafenib Dosing
Abstract
1. Introduction
2. Results
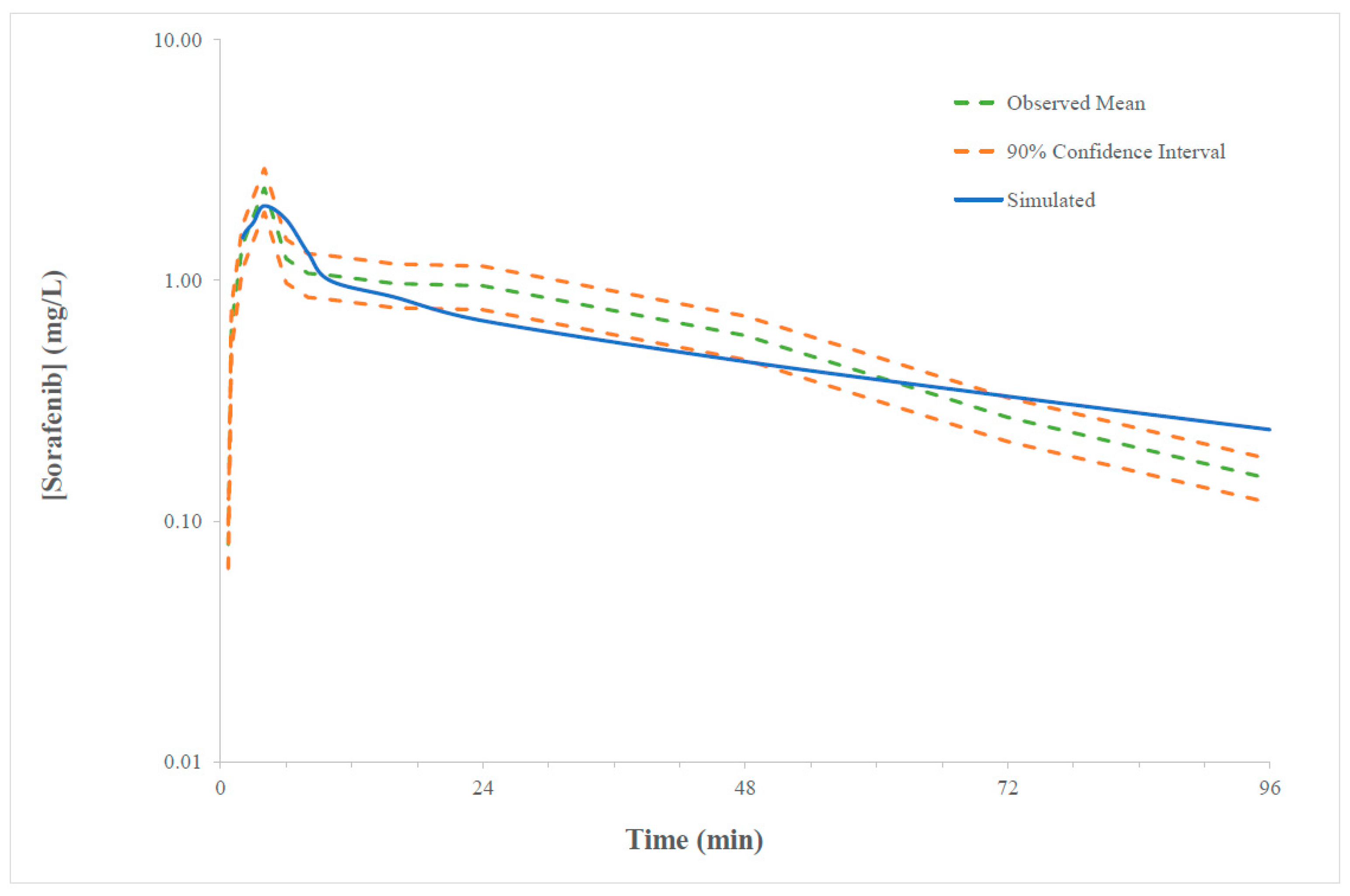
2.1. Verification of the Sorafenib PBPK Compound Model
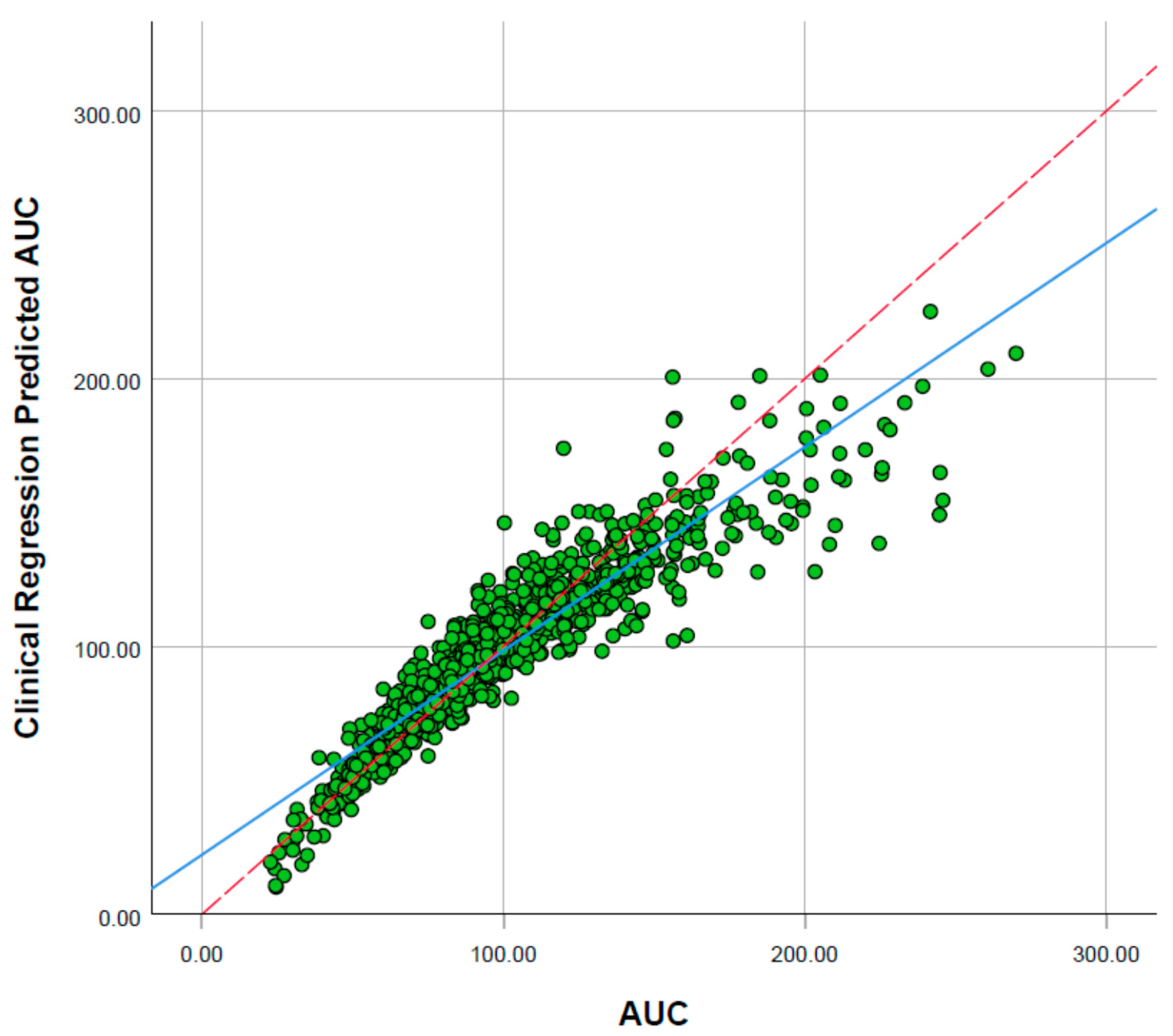
2.2. Sorafenib Exposure in Cancer Patient
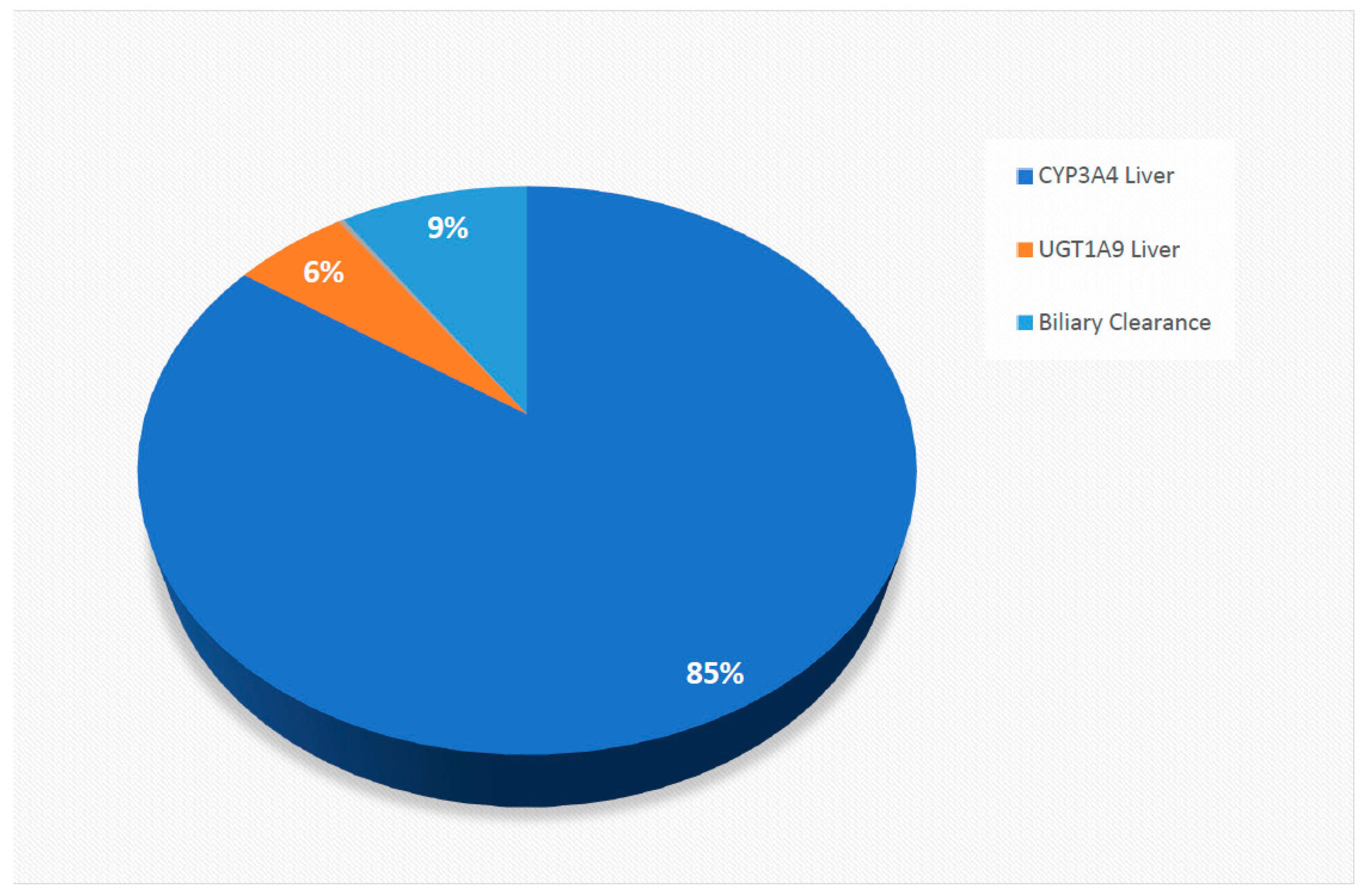
2.3. Physiological and Molecular Characteristics Driving Variability in Sorafenib Exposure
2.4. Impact of Dose Individualisation
3. Discussion
4. Materials and Methods
4.1. Development and Verification of the Sorafenib PBPK Model Structural Model
4.2. Development of the Sorafenib Compound Model
4.3. Population Model
4.4. Simulated Trial Designs
4.5. Validation of the Sorafenib Compound Model
4.6. Physiological and Molecular Characteristics Driving Variability in Sorafenib Exposure
4.7. Impact of Dose Individualisation
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Vogel, A.; Cervantes, A.; Chau, I.; Daniele, B.; Llovet, J.M.; Meyer, T.; Nault, J.C.; Neumann, U.; Ricke, J.; Sangro, B.; et al. Hepatocellular carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018, 29 (Suppl. 4), 238–255. [Google Scholar] [CrossRef]
- Wang, H.-T.; Xia, M. A meta-analysis of efficacy and safety of sorafenib versus other targeted agents for metastatic renal cell carcinoma. Medicine 2019, 98, e13779. [Google Scholar] [CrossRef]
- Strumberg, D.; Clark, J.W.; Awada, A.; Moore, M.J.; Richly, H.; Hendlisz, A.; Hirte, H.W.; Eder, J.P.; Lenz, H.-J.; Schwartz, B. Safety, Pharmacokinetics, and Preliminary Antitumor Activity of Sorafenib: A Review of Four Phase I Trials in Patients with Advanced Refractory Solid Tumors. Oncologist 2007, 12, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Awada, A.; Hendlisz, A.; Gil, T.; Bartholomeus, S.; Mano, M.; de Valeriola, D.; Strumberg, D.; Brendel, E.; Haase, C.G.; Schwartz, B.; et al. Phase I safety and pharmacokinetics of BAY 43-9006 administered for 21 days on/7 days off in patients with advanced, refractory solid tumours. Br. J. Cancer 2005, 92, 1855–1861. [Google Scholar] [CrossRef]
- Moore, M.; Hirte, H.W.; Siu, L.; Oza, A.; Hotte, S.J.; Petrenciuc, O.; Cihon, F.; Lathia, C.; Schwartz, B. Phase I study to determine the safety and pharmacokinetics of the novel Raf kinase and VEGFR inhibitor BAY 43-9006, administered for 28 days on/7 days off in patients with advanced, refractory solid tumors. Ann. Oncol. 2005, 16, 1688–1694. [Google Scholar] [CrossRef]
- Jain, L.; Woo, S.; Gardner, E.R.; Dahut, W.L.; Kohn, E.C.; Kummar, S.; Mould, D.R.; Giaccone, G.; Yarchoan, R.; Venitz, J.; et al. Population pharmacokinetic analysis of sorafenib in patients with solid tumours. Br. J. Clin. Pharmacol. 2011, 72, 294–305. [Google Scholar] [CrossRef]
- Sharma, M.; Holmes, H.M.; Mehta, H.B.; Chen, H.; Aparasu, R.R.; Shih, Y.-C.T.; Giordano, S.H.; Johnson, M.L. The concomitant use of tyrosine kinase inhibitors and proton pump inhibitors: Prevalence, predictors, and impact on survival and discontinuation of therapy in older adults with cancer. Cancer 2019, 125, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Ruanglertboon, W.; Sorich, M.J.; Logan, J.M.; Rowland, A.; Hopkins, A.M. The effect of proton pump inhibitors on survival outcomes in advanced hepatocellular carcinoma treated with sorafenib. J. Cancer Res. Clin. Oncol. 2020, 146, 2693–2697. [Google Scholar] [CrossRef] [PubMed]
- Lalani, A.A.; McKay, R.R.; Lin, X.; Simantov, R.; Kaymakcalan, M.D.; Choueiri, T.K. Proton Pump Inhibitors and Survival Outcomes in Patients With Metastatic Renal Cell Carcinoma. Clin. Genitourin. Cancer 2017, 15, 724–732. [Google Scholar] [CrossRef] [PubMed]
- Au, T.H.; Bailey, E.B.; Patel, S.B.; Tantravahi, S.K.; Agarwal, N.; Stenehjem, D.D. Effect of concomitant proton pump inhibitor (PPI) on effectiveness of tyrosine kinase inhibitor (TKI) in patients with metastatic renal cell carcinoma (mRCC). J. Clin. Oncol. 2016, 34 (Suppl. 2), 608. [Google Scholar] [CrossRef]
- Fukudo, M.; Ito, T.; Mizuno, T.; Shinsako, K.; Hatano, E.; Uemoto, S.; Kamba, T.; Yamasaki, T.; Ogawa, O.; Seno, H.; et al. Exposure-toxicity relationship of sorafenib in Japanese patients with renal cell carcinoma and hepatocellular carcinoma. Clin. Pharmacokinet. 2014, 53, 185–196. [Google Scholar] [CrossRef] [PubMed]
- Ruanglertboon, W.; Sorich, M.J.; Rowland, A.; Hopkins, A.M. Effect of early adverse events resulting in sorafenib dose adjustments on survival outcomes of advanced hepatocellular carcinoma patients. Int. J. Clin. Oncol. 2020, 25, 1672–1677. [Google Scholar] [CrossRef] [PubMed]
- de Wit, D.; Guchelaar, H.J.; den Hartigh, J.; Gelderblom, H.; van Erp, N.P. Individualized dosing of tyrosine kinase inhibitors: Are we there yet? Drug Discov. Today 2015, 20, 18–36. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Schoell, A.; Groenland, S.L.; Scherf-Clavel, O.; van Dyk, M.; Huisinga, W.; Michelet, R.; Jaehde, U.; Steeghs, N.; Huitema, A.D.R.; Kloft, C. Therapeutic drug monitoring of oral targeted antineoplastic drugs. Eur. J. Clin. Pharmacol. 2020, 29, iv238–iv255. [Google Scholar] [CrossRef]
- Semrad, T.J.; Gandara, D.R.; Lara, P.N., Jr. Enhancing the clinical activity of sorafenib through dose escalation: Rationale and current experience. Ther. Adv. Med. Oncol. 2011, 3, 95–100. [Google Scholar] [CrossRef]
- Escudier, B.; Szczylik, C.; Hutson, T.E.; Demkow, T.; Staehler, M.; Rolland, F.; Negrier, S.; Laferriere, N.; Scheuring, U.J.; Cella, D.; et al. Randomized phase II trial of first-line treatment with sorafenib versus interferon Alfa-2a in patients with metastatic renal cell carcinoma. J. Clin. Oncol. 2009, 27, 1280–1289. [Google Scholar] [CrossRef] [PubMed]
- Verheijen, R.B.; Yu, H.; Schellens, J.H.M.; Beijnen, J.H.; Steeghs, N.; Huitema, A.D.R. Practical Recommendations for Therapeutic Drug Monitoring of Kinase Inhibitors in Oncology. Clin. Pharmacol. Ther. 2017, 102, 765–776. [Google Scholar] [CrossRef]
- Rowland, A.; van Dyk, M.; Mangoni, A.A.; Miners, J.O.; McKinnon, R.A.; Wiese, M.D.; Rowland, A.; Kichenadasse, G.; Gurney, H.; Sorich, M.J. Kinase inhibitor pharmacokinetics: Comprehensive summary and roadmap for addressing inter-individual variability in exposure. Expert Opin. Drug Metab. Toxicol. 2017, 13, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Darwich, A.; Ogungbenro, K.; Hatley, O.; Rostami-Hodjegan, A. Role of pharmacokinetic modeling and simulation in precision dosing of anticancer drugs. Transl. Cancer Res. 2017. [Google Scholar] [CrossRef]
- Hopkins, A.M.; Menz, B.D.; Wiese, M.D.; Kichenadasse, G.; Gurney, H.; McKinnon, R.A.; Rowland, A.; Sorich, M.J. Nuances to precision dosing strategies of targeted cancer medicines. Pharmacol. Res. Perspect. 2020, 8, e00625. [Google Scholar] [CrossRef]
- Kang, J.S.; Lee, M.H. Overview of therapeutic drug monitoring. Korean J. Intern. Med. 2009, 24, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Thomson, A. TDM—A Multidisciplinary Approach. Ther. Drug Monit. 1997, 19, 490. [Google Scholar] [CrossRef]
- Sánchez-Hernández, J.G.; Rebollo, N.; Martin-Suarez, A.; Calvo, M.V.; Muñoz, F. A 3-year prospective study of a multidisciplinary early proactive therapeutic drug monitoring programme of infliximab treatments in inflammatory bowel disease. Br. J. Clin. Pharmacol. 2020, 86, 1165–1175. [Google Scholar] [CrossRef] [PubMed]
- Sorich, M.J.; Mutlib, F.; van Dyk, M.; Hopkins, A.M.; Polasek, T.M.; Marshall, J.C.; Rodrigues, A.D.; Rowland, A. Use of Physiologically Based Pharmacokinetic Modeling to Identify Physiological and Molecular Characteristics Driving Variability in Axitinib Exposure: A Fresh Approach to Precision Dosing in Oncology. J. Clin. Pharmacol. 2019, 59, 872–879. [Google Scholar] [CrossRef] [PubMed]
- Rowland, A.; van Dyk, M.; Hopkins, A.M.; Mounzer, R.; Polasek, T.M.; Rostami-Hodjegan, A.; Sorich, M.J. Physiologically Based Pharmacokinetic Modeling to Identify Physiological and Molecular Characteristics Driving Variability in Drug Exposure. Clin. Pharmacol. Ther. 2018, 104, 1219–1228. [Google Scholar] [CrossRef] [PubMed]
- Polasek, T.M.; Rostami-Hodjegan, A. Virtual Twins: Understanding the Data Required for Model-Informed Precision Dosing. Clin. Pharmacol. Ther. 2020, 107, 742–745. [Google Scholar] [CrossRef]
- Kluwe, F.; Michelet, R.; Mueller-Schoell, A.; Maier, C.; Klopp-Schulze, L.; van Dyk, M.; Mikus, G.; Huisinga, W.; Kloft, C. Perspectives on Model-Informed Precision Dosing in the Digital Health Era: Challenges, Opportunities, and Recommendations. Clin. Pharmacol. Ther. 2020, 109, 29–36. [Google Scholar] [CrossRef]
- Polasek, T.M.; Polak, S.; Doogue, M.P.; Rostami-Hodjegan, A.; Miners, J.O. Assessment of inter-individual variability in predicted phenytoin clearance. Eur. J. Clin. Pharmacol. 2009, 65, 1203–1210. [Google Scholar] [CrossRef]
- Chetty, M.; Cain, T.; Wedagedera, J.; Rostami-Hodjegan, A.; Jamei, M. Application of Physiologically Based Pharmacokinetic (PBPK) Modeling Within a Bayesian Framework to Identify Poor Metabolizers of Efavirenz (PM), Using a Test Dose of Efavirenz. Front. Pharmacol. 2018, 9, 247. [Google Scholar] [CrossRef]
- Tsamandouras, N.; Rostami-Hodjegan, A.; Aarons, L. Combining the ‘bottom up’ and ‘top down’ approaches in pharmacokinetic modelling: Fitting PBPK models to observed clinical data. Br. J. Clin. Pharmacol. 2015, 79, 48–55. [Google Scholar] [CrossRef]
- Tylutki, Z.; Polak, S.; Wiśniowska, B. Top-down, Bottom-up and Middle-out Strategies for Drug Cardiac Safety Assessment via Modeling and Simulations. Curr. Pharmacol. Rep. 2016, 2, 171–177. [Google Scholar] [CrossRef]
- Polasek, T.M.; Tucker, G.T.; Sorich, M.J.; Wiese, M.D.; Mohan, T.; Rostami-Hodjegan, A.; Korprasertthaworn, P.; Perera, V.; Rowland, A. Prediction of olanzapine exposure in individual patients using physiologically based pharmacokinetic modelling and simulation. Br. J. Clin. Pharmacol. 2018, 84, 462–476. [Google Scholar] [CrossRef]
- Van Dyk, M.; Rowland, A. PBPK modeling as an approach to evaluate the effect of covariates and drug-drug interactions on variability in EGFR kinase inhibitor exposure. Transl. Cancer Res. 2017, 6, S1600–S1615. [Google Scholar] [CrossRef]
- Minami, H.; Kawada, K.; Ebi, H.; Kitagawa, K.; Kim, Y.-I.; Araki, K.; Mukai, H.; Tahara, M.; Nakajima, H.; Nakajima, K. Phase I and pharmacokinetic study of sorafenib, an oral multikinase inhibitor, in Japanese patients with advanced refractory solid tumors. Cancer Sci. 2008, 99, 1492–1498. [Google Scholar] [CrossRef] [PubMed]
- Shebley, M.; Sandhu, P.; Riedmaier, A.E.; Jamei, M.; Narayanan, R.; Patel, A.; Peters, S.A.; Reddy, V.P.; Zheng, M.; de Zwart, L.; et al. Physiologically Based Pharmacokinetic Model Qualification and Reporting Procedures for Regulatory Submissions: A Consortium Perspective. Clin. Pharmacol. Ther. 2018, 104, 88–110. [Google Scholar] [CrossRef]
- Basu, S.; Lien, Y.T.; Vozmediano, V.; Schlender, J.-F.; Eissing, T.; Schmidt, S.; Niederalt, C. Physiologically Based Pharmacokinetic Modeling of Monoclonal Antibodies in Pediatric Populations Using PK-Sim. Front. Pharmacol. 2020, 11, 868. [Google Scholar] [CrossRef] [PubMed]
- Hennig, S.; Holthouse, F.; Staatz, C.E. Comparing dosage adjustment methods for once-daily tobramycin in paediatric and adolescent patients with cystic fibrosis. Clin. Pharmacokinet. 2015, 54, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; He, C.-Y.; Yin, N.-G.; Liu, F.; Jia, Y.-T.; Liu, Y. A population pharmacokinetic model for individualised dosage regimens of vancomycin in Chinese neonates and young infants. Oncotarget 2017, 8, 105211–105221. [Google Scholar] [CrossRef]
- Rowland, A.; Ruanglertboon, W.; van Dyk, M.; Wijayakumara, D.; Wood, L.; Meech, R.; Mackenzie, P.; Rodrigues, A.; Marshall, J.; Sorich, M. Plasma Extracellular Nanovesicle (Exosome) derived biomarkers for ADME pathways: A novel approach to characterise variability in drug exposure. Br. J. Clin. Pharmacol. 2019, 85, 216–226. [Google Scholar] [CrossRef]
- Kumar, S.; Sinha, N.; Gerth, K.A.; Rahman, M.A.; Yallapu, M.M.; Midde, N.M. Specific packaging and circulation of cytochromes P450, especially 2E1 isozyme, in human plasma exosomes and their implications in cellular communications. Biochem. Biophys. Res. Commun. 2017, 491, 675–680. [Google Scholar] [CrossRef]
- Achour, B.; Al-Majdoub, Z.M.; Grybos-Gajniak, A.; Lea, K.; Kilford, P.; Zhang, M.; Knight, D.; Barber, J.; Schageman, J.; Rostami-Hodjegan, A. Liquid Biopsy Enables Quantification of the Abundance and Interindividual Variability of Hepatic Enzymes and Transporters. Clin. Pharmacol. Ther. 2021, 109, 222. [Google Scholar] [CrossRef] [PubMed]
- Rowland Yeo, K.; Jamei, M.; Yang, J.; Tucker, G.T.; Rostami-Hodjegan, A. Physiologically based mechanistic modelling to predict complex drug-drug interactions involving simultaneous competitive and time-dependent enzyme inhibition by parent compound and its metabolite in both liver and gut-the effect of diltiazem on the time-course of exposure to triazolam. Eur. J. Pharm. Sci. 2010, 39, 298–309. [Google Scholar] [PubMed]
- European Medicines Agency. Nexavar: European Public Assessment Reports (EPAR)-Scientific Discussion. 3 March 2007. Available online: http://www.ema.europa.eu/docs/en_GB/document_library/EPAR-Scientific_Discussion/human/000690/WC500027707.pdf (accessed on 20 February 2021).
- Liu, B.; Crewe, H.K.; Ozdemir, M.; Yeo, K.R.; Tucker, G.; Rostami-Hodjegan, A. The absorption kinetics of ketoconazole plays a major role in explaining the reported variability in the level of interaction with midazolam: Interplay between formulation and inhibition of gut wall and liver metabolism. Biopharm. Drug Dispos. 2017, 38, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Schwenger, E.; Reddy, V.P.; Moorthy, G.; Sharma, P.; Tomkinson, H.; Masson, E.; Vishwanathan, K. Harnessing Meta-analysis to Refine an Oncology Patient Population for Physiology-Based Pharmacokinetic Modeling of Drugs. Clin. Pharmacol. Ther. 2018, 103, 271–280. [Google Scholar] [CrossRef]
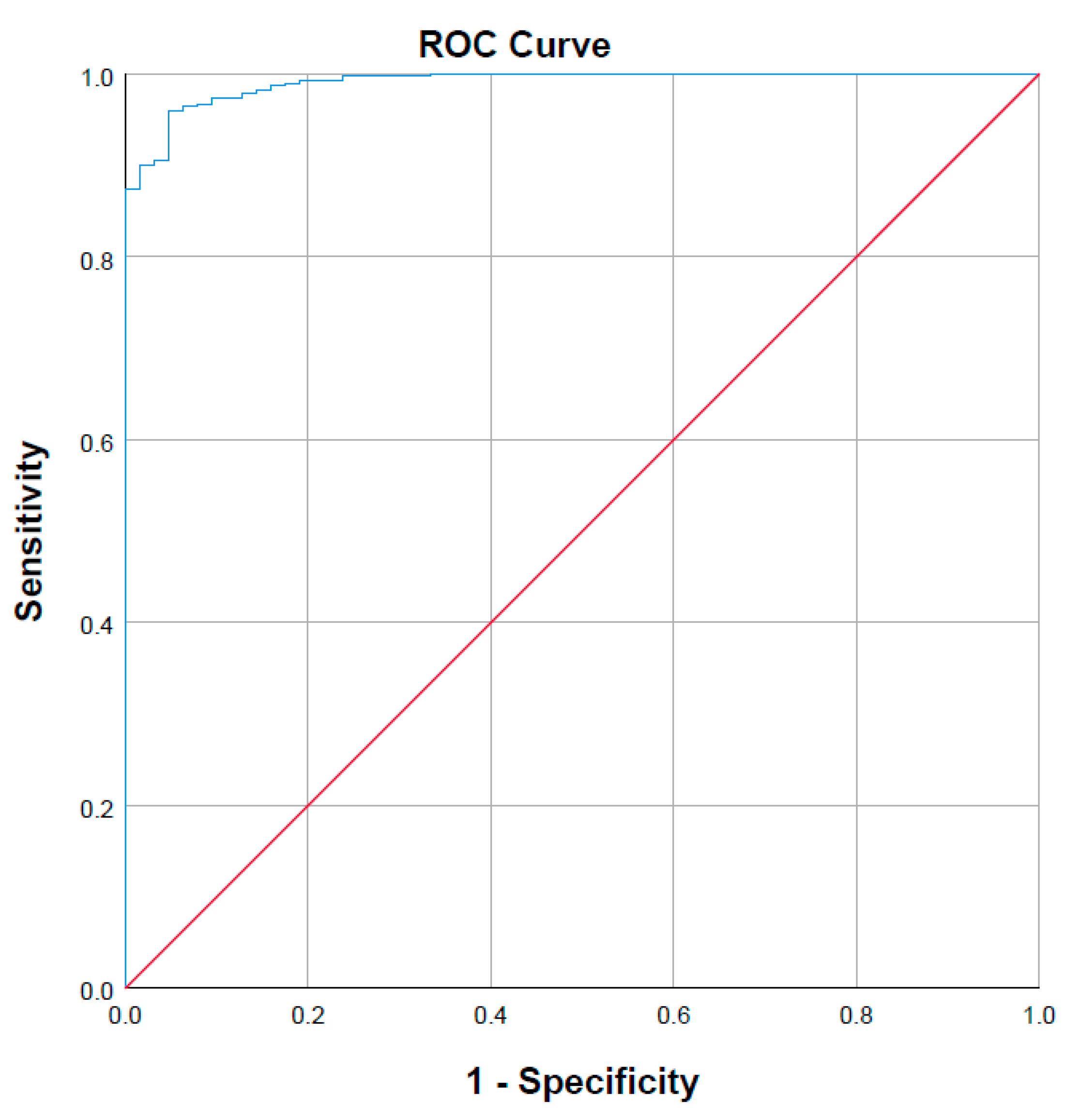
| Model | R2 | Std. Error of the Estimate | R2 Change | AUC ROC | AUC ROC Change |
|---|---|---|---|---|---|
| a | 0.631 | 0.24141 | 0.631 | 0.953 | 0.953 |
| b | 0.781 | 0.18614 | 0.150 | 0.981 | 0.028 |
| c | 0.868 | 0.14458 | 0.087 | 0.990 | 0.009 |
| d | 0.873 | 0.14156 | 0.006 | 0.991 | 0.001 |
| e | 0.883 | 0.13619 | 0.010 | 0.991 | - |
| f | 0.883 | 0.13595 | 0.001 | 0.991 | - |
| Predicted Therapeutic Cmax | Percentage Correct | |||
|---|---|---|---|---|
| Sub-Therapeutic | Therapeutic | |||
| Observed Therapeutic Cmax | Sub-therapeutic | 60 (true negative) | 3 (false negative) | 95.2 |
| Therapeutic | 47 (false positive) | 890 (true positive) | 95.0 | |
| Dosing Protocol | Day 14 | Day 28 | ||||
|---|---|---|---|---|---|---|
| <4.78 µg/mL | 4.78 to 5.78 µg/mL | >5.78 µg/mL | <4.78 µg/mL | 4.78 to 5.78 µg/mL | >5.78 µg/mL | |
| Flat dosing | 62 | 116 | 322 | 62 | 116 | 322 |
| Concentration-guided dosing | 62 | 116 | 322 | 5 | 130 | 365 |
| Concentration-guided dosing with MIDS | 34 | 135 | 331 | 5 | 164 | 336 |
| Parameter | Value | Source |
|---|---|---|
| Physicochemical properties Molecular weight Log Po:w Hydrogen bond donor Species | 464.82 g/mol 4.54 3 Base | [43] [43] |
| Protein binding B/P fup | 0.55 0.0048 | [43] [43] |
| Absorption (ADAM model) fa ka (L/h) | 0.99 1.75 | Predicted Predicted |
| Permeability Peff, man (10−4 cm/s) Caco-2 (10−6 cm/s) | 4.01 24.1 | Predicted |
| Formulation Solid formulation | Immediate release | [43] |
| In vivo pharmacokinetic properties (full PBPK model) Prediction model Kp scalar | 1 0.7 | Predicted |
| CYP metabolism: ISEF adjusted recombinant enzyme kinetics (CLint; μL/min/pmol) CYP3A4 | 2.6 | [18] |
| UGT metabolism: ISEF adjusted recombinant enzyme kinetics (CLint; μL/min/mg) UGT1A9 | 20.1 | [18] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruanglertboon, W.; Sorich, M.J.; Hopkins, A.M.; Rowland, A. Mechanistic Modelling Identifies and Addresses the Risks of Empiric Concentration-Guided Sorafenib Dosing. Pharmaceuticals 2021, 14, 389. https://doi.org/10.3390/ph14050389
Ruanglertboon W, Sorich MJ, Hopkins AM, Rowland A. Mechanistic Modelling Identifies and Addresses the Risks of Empiric Concentration-Guided Sorafenib Dosing. Pharmaceuticals. 2021; 14(5):389. https://doi.org/10.3390/ph14050389
Chicago/Turabian StyleRuanglertboon, Warit, Michael J. Sorich, Ashley M. Hopkins, and Andrew Rowland. 2021. "Mechanistic Modelling Identifies and Addresses the Risks of Empiric Concentration-Guided Sorafenib Dosing" Pharmaceuticals 14, no. 5: 389. https://doi.org/10.3390/ph14050389
APA StyleRuanglertboon, W., Sorich, M. J., Hopkins, A. M., & Rowland, A. (2021). Mechanistic Modelling Identifies and Addresses the Risks of Empiric Concentration-Guided Sorafenib Dosing. Pharmaceuticals, 14(5), 389. https://doi.org/10.3390/ph14050389






