Appraisal of Bioactive Compounds of Betel Fruit as Antimalarial Agents by Targeting Plasmepsin 1 and 2: A Computational Approach
Abstract
1. Introduction
2. Results
2.1. GC-MS Analysis
2.2. ADME Analysis
2.3. Toxicity Analysis
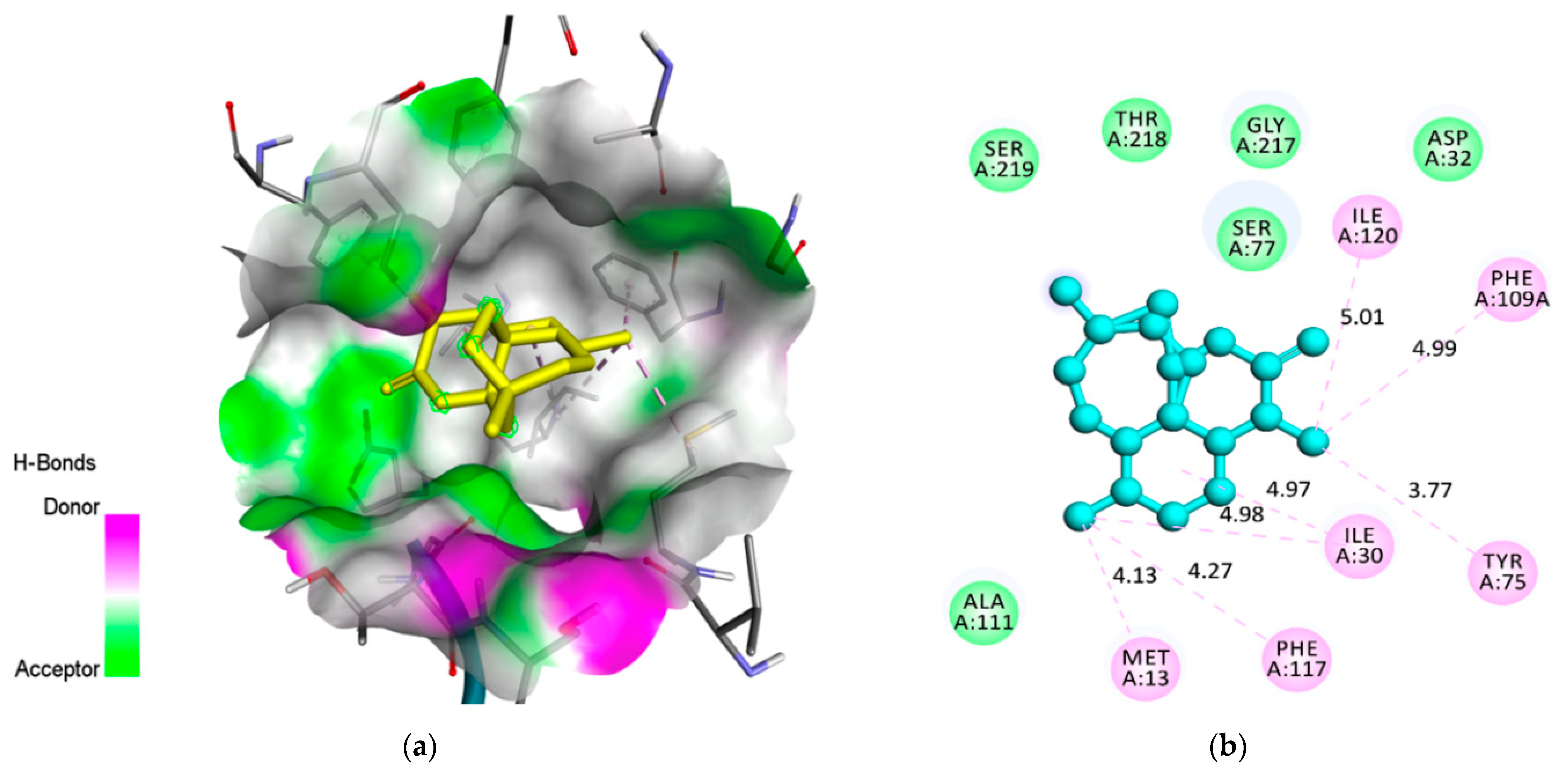
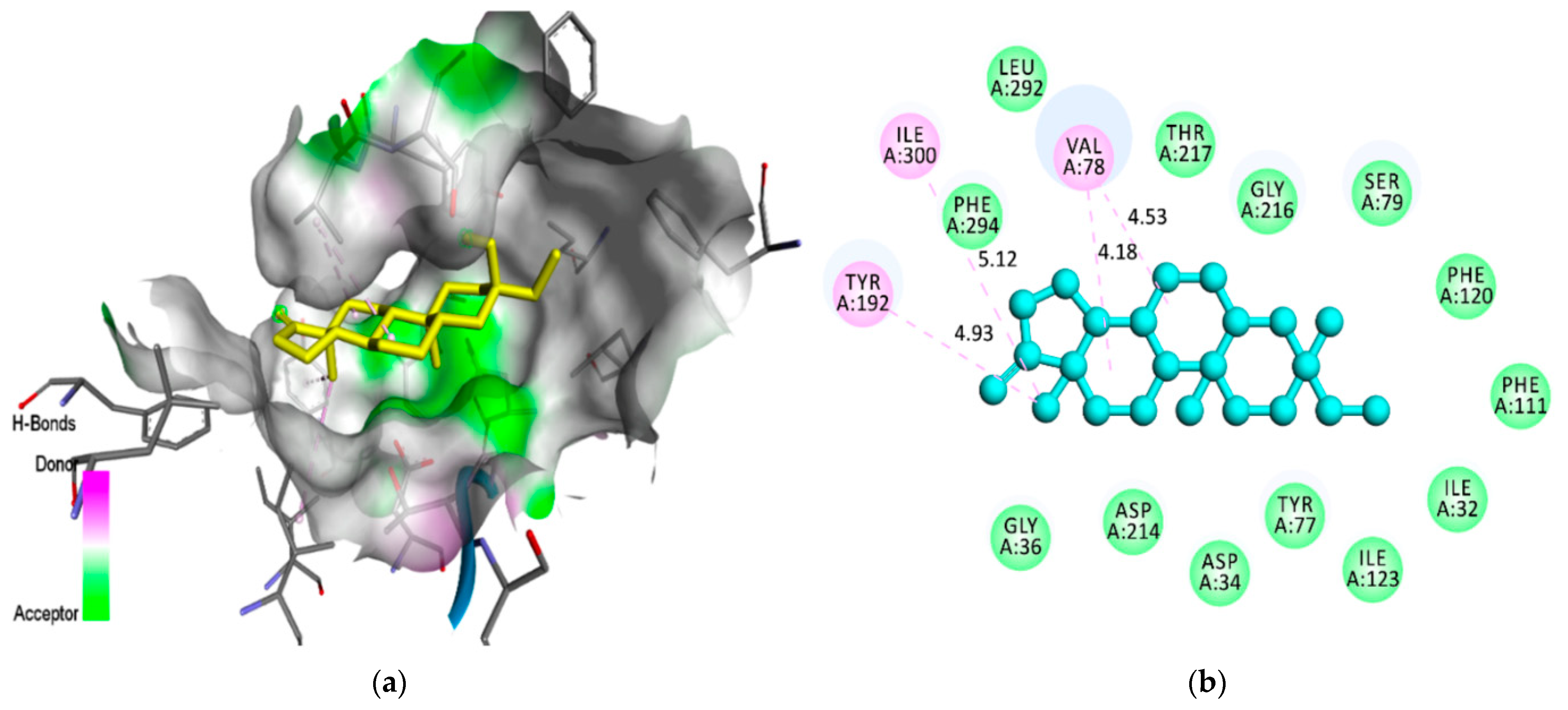
2.4. Molecular Docking Analysis
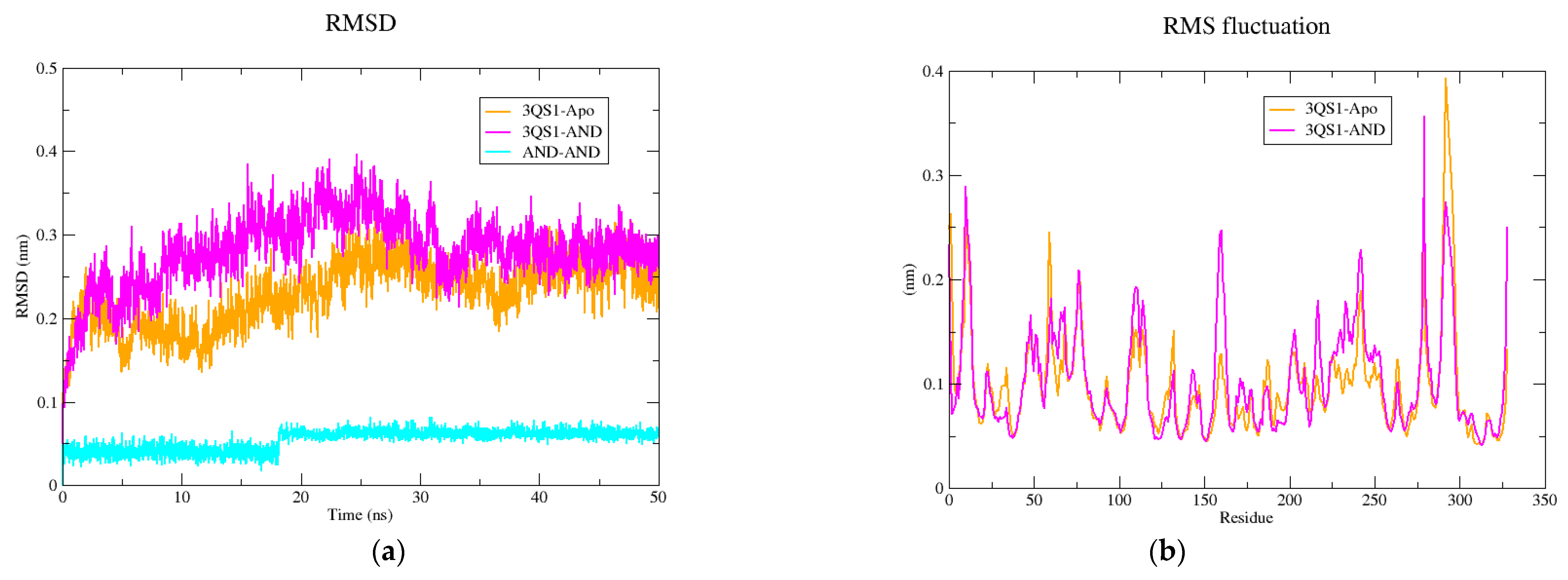
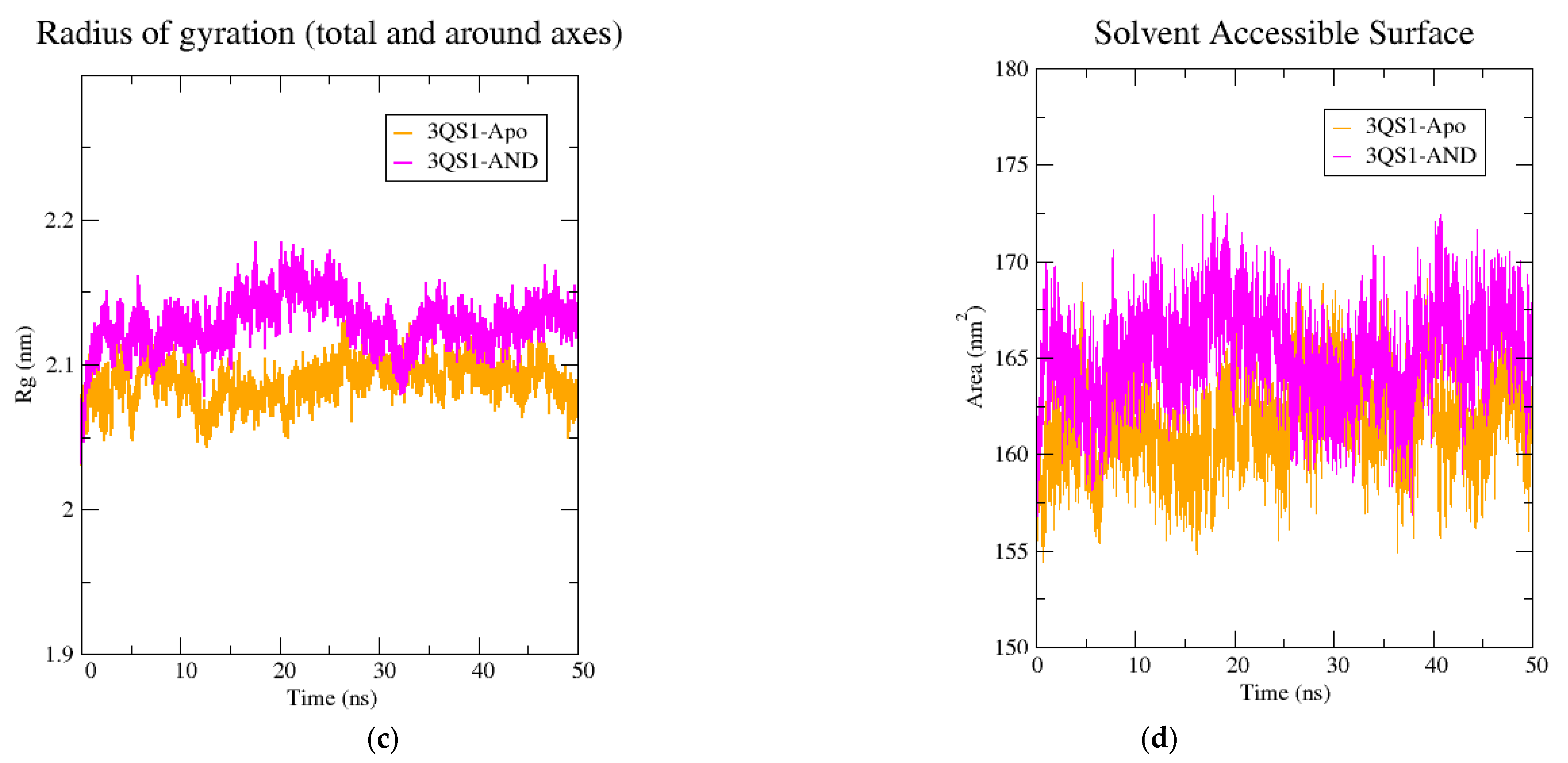
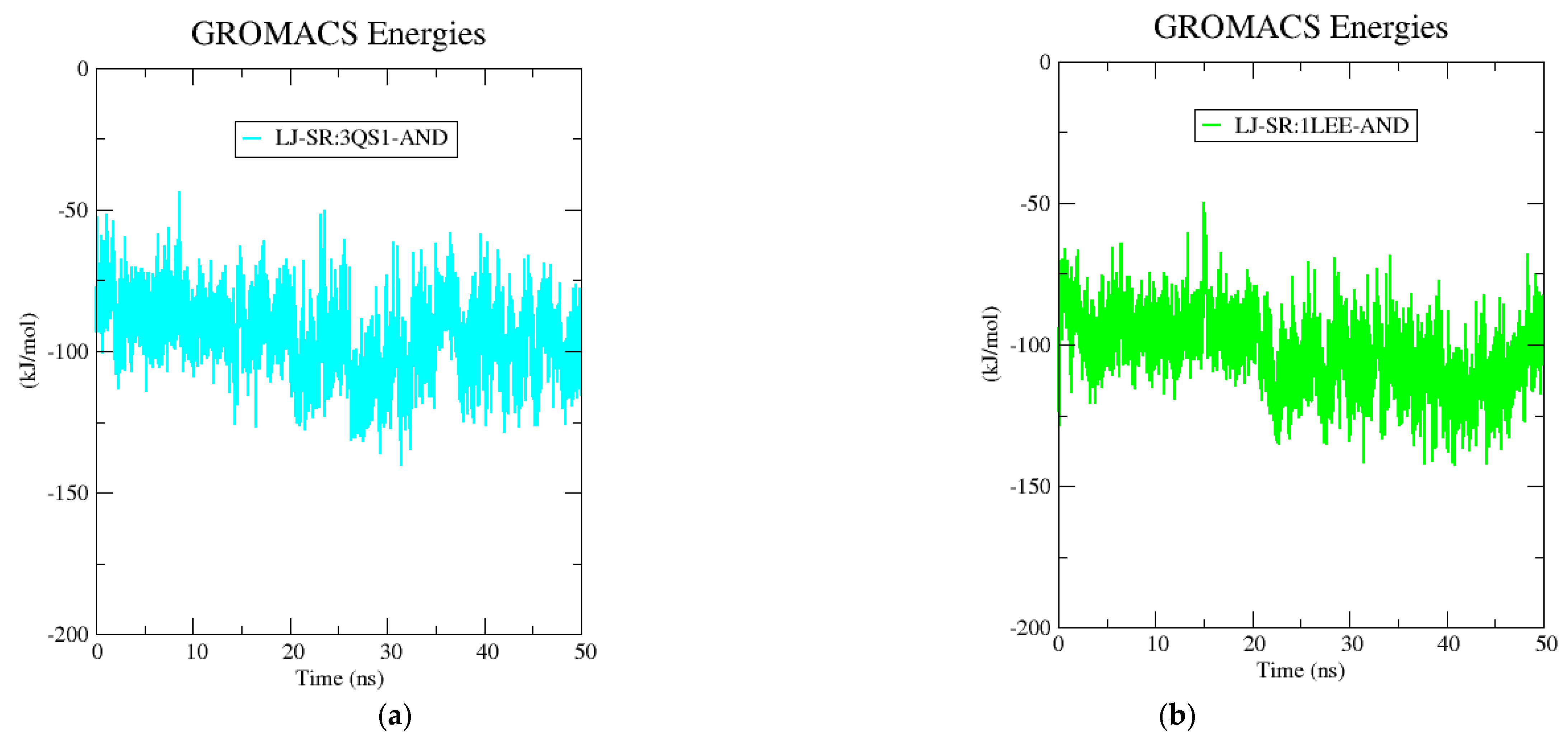
2.5. Molecular Dynamics Simulation
3. Discussion
4. Materials and Methods
4.1. Plant Collection
4.2. Sample Preparation
4.3. Gas Chromatography-Mass Spectrometer (GC-MS) Analysis
4.4. In Silico ADMET Analysis
4.5. Computational Molecular Docking Analysis
4.5.1. Preparation of the Receptors
4.5.2. Preparation of the Ligands
4.5.3. Molecular Docking
4.6. Molecular Dynamics Simulation
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nugroho, Y. Aktivitas antimalaria (in vivo) kombinasi buah sirih (Piper betle L.), daun miyana (Plectranthus scutellarioides (L.) R. Br.), madu dan kuning telur pada mencit yang diinfeksi Plasmodium berghei. Bul. Penelit. Kesehat. 2011, 39, 129–137. [Google Scholar]
- Antinori, S.; Galimberti, L.; Milazzo, L.; Corbellino, M. Biology of human malaria plasmodia including Plasmodium knowlesi. Mediterr. J. Hematol. Infect. Dis. 2012, 4, e2012013. [Google Scholar] [CrossRef]
- Harijanto, P.N.; Nugroho, A.; Gunawan, C.A. Malaria dari Molekuler ke Klinis; Penerbit Buku Kedokteran EGC: Jakarta, Indonesia, 2009. [Google Scholar]
- Antony, H.A.; Parija, S.C. Antimalarial drug resistance: An overview. Trop. Parasitol. 2016, 6, 30–41. [Google Scholar] [CrossRef]
- Conrad, M.D.; Rosenthal, P.J. Antimalarial drug resistance in Africa: The calm before the storm? Lancet. Infect. Dis. 2019, 19, e338–e351. [Google Scholar] [CrossRef]
- Pribadi, W.; Muljono, R. Resistensi parasit malaria terhadap obat malaria. In Parasitologi Kedokteran; Gandahusada, S., Ilahude, H., Pribadi, W., Eds.; Gaya Baru: Jakarta, Indonesia, 2004; pp. 197–198. [Google Scholar]
- Gulati, M.; Narula, A.; Vishnu, R.; Katyal, G.; Negi, A.; Ajaz, I.; Narula, K.; Chauhan, G.; Kant, R.; Lumb, V.; et al. Plasmepsin II as a Potential Drug Target for Resistant Malaria. DU J. Undergrad. Res. Innov. 2015, 1, 85–95. [Google Scholar]
- Soh, B.Y.; Song, H.O.; Lee, Y.; Lee, J.; Kaewintajuk, K.; Lee, B.; Choi, Y.Y.; Cho, J.H.; Choi, S.; Park, H. Identification of active Plasmodium falciparum calpain to establish screening system for Pf-calpain-based drug development. Malar. J. 2013, 12, 47. [Google Scholar] [CrossRef] [PubMed]
- Cheuka, P.M.; Dziwornu, G.; Okombo, J.; Chibale, K. Plasmepsin inhibitors in antimalarial drug discovery: Medicinal chemistry and target validation (2000 to present). J. Med. Chem. 2020, 63, 4445–4467. [Google Scholar] [CrossRef]
- Favuzza, P.; de Lera Ruiz, M.; Thompson, J.K.; Triglia, T.; Ngo, A.; Steel, R.W.J.; Vavrek, M.; Christensen, J.; Healer, J.; Boyce, C.; et al. Dual Plasmepsin-Targeting Antimalarial Agents Disrupt Multiple Stages of the Malaria Parasite Life Cycle. Cell Host Microbe 2020, 27, 642–658.e12. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Ziegler, R.; Hamann, A. Inhibition of phosphoenolpyruvate carboxykinase gene expression by metformin in cultured hepatocytes. Chin. Med. J. 2002, 115, 1843–1848. [Google Scholar]
- Ismail, H.M.; Barton, V.; Phanchana, M.; Charoensutthivarakul, S.; Wong, M.H.L.; Hemingway, J.; Biagini, G.A.; O’Neill, P.M.; Ward, S.A. Artemisinin activity-based probes identify multiple molecular targets within the asexual stage of the malaria parasites Plasmodium falciparum 3D7. Proc. Natl. Acad. Sci. USA 2016, 113, 2080–2085. [Google Scholar] [CrossRef]
- Nerdy. In silico docking roselle (Hibiscus sabdariffa L.) calyces flavonoids as antimalarial against plasmepsin 1 and plasmepsin 2. Asian J. Pharm. Clin. Res. 2017, 10, 183–186. [Google Scholar] [CrossRef][Green Version]
- Rakib, A.; Nain, Z.; Islam, M.A.; Sami, S.A.; Mahmud, S.; Islam, A.; Ahmed, S.; Siddiqui, A.B.F.; Babu, S.M.O.F.; Hossain, P.; et al. A molecular modelling approach for identifying antiviral selenium-containing heterocyclic compounds that inhibit the main protease of SARS-CoV-2: An in silico investigation. Brief. Bioinform. 2021, 22, 1476–1498. [Google Scholar] [CrossRef] [PubMed]
- Venkateswarlu, K.; Devanna, N. Pharmacological evaluations (analgesic activity) of ‘Piper Betel’. Int. J. Pharmamedix India 2014, 2, 688–693. [Google Scholar]
- Rekha, V.P.B.; Kollipara, M.; Srinivasa Gupta, B.R.S.S.; Bharath, Y.; Pulicherla, K.K. A Review on Piper betle L.: Nature’s promising medicinal reservoir. Am. J. Ethnomed. 2014, 1, 276–289. [Google Scholar]
- Chakraborty, D.; Shah, B. Antimicrobial, anti-oxidative and anti-hemolytic activity of Piper betel leaf extracts. Int. J. Pharm. Pharm. Sci. 2011, 3, 192–199. [Google Scholar]
- Makatamba, V.; Fatimawali; Rundengan, G. Analisis senyawa tannin dan aktifitas antibakteri fraksi buah sirih (Piper betle L.) terhadap Streptococcus mutans. J. MIPA 2020, 9, 75–80. [Google Scholar] [CrossRef]
- Pratiwi, N.P.R.K.; Muderawan, I.W. Analisis kandungan kimia ekstrak daun sirih hijau (Piper betle) dengan GC-MS. EJournal Univ. Pendidik. Ganesha 2016, 304–310. [Google Scholar]
- Tjandra, R.; Fatimawali; Datu, O. Analisis senyawa alkaloid dan uji daya hambat ekstrak buah sirih (Piper betle L.) terhadap bakteri Staphylococcus epidermidis. eBiomedik 2020, 8, 165–171. [Google Scholar] [CrossRef]
- Parwata, I.O.A.; Santi, S.R.; Sulaksana, I.; Alit Widiarthini, I. Aktivitas larvasida minyak atsiri pada daun sirih (Piper betle linn) terhadap larva nyamuk Aedes aegypti. J. Kim. 2012, 5, 88–93. [Google Scholar]
- Nagori, K.; Singh, M.K.; Alexander, A.; Kumar, T.; Dewangan, D.; Badwaik, H.; Tripathi, D.K. Piper betle L.: A review on its ethnobotany, phytochemistry, pharmacological profile and profiling by new hyphenated technique DART-MS (Direct Analysis in Real Time Mass Spectrometry). J. Pharm. Res. 2011, 4, 2991–2997. [Google Scholar]
- Mohottalage, S.; Tabacchi, R.; Guerin, P.M. Components from Sri Lankan Piper betle L. leaf oil and their analogues showing toxicity against the housefly, Musca domestica. Flavour Fragr. J. 2007, 22, 130–138. [Google Scholar] [CrossRef]
- Walker, D.K. The use of pharmacokinetic and pharmacodynamic data in the assessment of drug safety in early drug development. Br. J. Clin. Pharmacol. 2004, 58, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Lipinski, C.A. Lead- and drug-like compounds: The rule-of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Mahmud, S.; Mita, M.A.; Biswas, S.; Paul, G.K.; Promi, M.M.; Afrose, S.; Hasan, R.; Shimu, S.S.; Zaman, S.; Uddin, S.; et al. Molecular docking and dynamics study to explore phytochemical ligand molecules against the main protease of SARS-CoV-2 from extensive phytochemical datasets. Expert Rev. Clin. Pharmacol. 2021, 14, 1305–1315. [Google Scholar] [CrossRef]
- Singh, S.; Gupta, A.K.; Verma, A. Molecular Properties and Bioactivity Score of The Aloe vera Antioxidant Compounds—In Order to Lead Fnding. Res. J. Pharm. Biol. Chem. Sci. 2013, 4, 876–881. [Google Scholar]
- Lei, T.; Li, Y.; Song, Y.; Li, D.; Sun, H.; Hou, T. ADMET evaluation in drug discovery: 15. Accurate prediction of rat oral acute toxicity using relevance vector machine and consensus modeling. J. Cheminform. 2016, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, P.; Begluitti, G.; Tincher, T.; Wheeler, J.; Mumtaz, M. Prediction of Acute Mammalian Toxicity Using QSAR Methods: A Case Study of Sulfur Mustard and Its Breakdown Products. Molecules 2012, 17, 8982–9001. [Google Scholar] [CrossRef]
- Anderson, S.E.; Shane, H.L. Investigative immunotoxicology. Methods Mol. Biol. 2018, 1803, 27–46. [Google Scholar] [CrossRef] [PubMed]
- Moura, P.A.; Dame, J.B.; Fidock, D.A. Role of Plasmodium falciparum digestive vacuole plasmepsins in the specificity and antimalarial mode of action of cysteine and aspartic protease inhibitors. Antimicrob. Agents Chemother. 2009, 53, 4968–4978. [Google Scholar] [CrossRef]
- Sargsyan, K.; Grauffel, C.; Lim, C. How molecular size impacts RMSD applications in molecular dynamics simulations. J. Chem. Theory Comput. 2017, 13, 1518–1524. [Google Scholar] [CrossRef]
- Ladokun, O.A.; Abiola, A.; Okikiola, D.; Ayodeji, F. GC-MS and Molecular Docking Studies of Hunteria umbellata Methanolic Extract as a Potent Anti-Diabetic. Inform. Med. Unlocked 2018, 13, 1–8. [Google Scholar] [CrossRef]
- Rakib, A.; Paul, A.; Chy, M.N.U.; Sami, S.A.; Baral, S.K.; Majumder, M.; Tareq, A.M.; Amin, M.N.; Shahriar, A.; Zia Uddin, M.; et al. Biochemical and computational approach of selected phytocompounds from Tinospora crispa in the management of COVID-19. Molecules 2020, 25, 3936. [Google Scholar] [CrossRef]
- Salmaso, V.; Moro, S. Bridging molecular docking to molecular dynamics in exploring ligand-protein recognition process: An overview. Front. Pharmacol. 2018, 9, 923. [Google Scholar] [CrossRef]
- Binder, K.; Horbach, J.; Kob, W.; Wolfgang, P.; Fathollah, V. Molecular dynamics simulations. J. Phys. Condens. Matter 2004, 16, S429–S453. [Google Scholar] [CrossRef]
- Jha, P.K.; Sknepnek, R.; Guerrero-García, G.I.; De La Cruz, M.O. A graphics processing unit implementation of coulomb interaction in molecular dynamics. J. Chem. Theory Comput. 2010, 6, 3058–3065. [Google Scholar] [CrossRef]
- Semiromi, D.T.; Azimian, A.R. Molecular dynamics simulation of nonodroplets with the modified Lennard-Jones potential function. Heat Mass Transf. Stoffuebertragung 2010, 47, 579–588. [Google Scholar] [CrossRef]
- Gellatly, N.; Sewell, F. Regulatory acceptance of in silico approaches for the safety assessment of cosmetic-related substances. Comput. Toxicol. 2019, 11, 82–89. [Google Scholar] [CrossRef]
- Soni, A.; Pandey, K.; Ray, P.; Jayaram, B. Genomes to hits in silico—A country path today, a highway romorrow: A case study of Chikungunya. Curr. Pharm. Des. 2013, 19, 4687–4700. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, P.; Eckert, A.O.; Schrey, A.K.; Preissner, R. ProTox-II: A webserver for the prediction of toxicity of chemicals. Nucleic Acids Res. 2018, 46, W257–W263. [Google Scholar] [CrossRef] [PubMed]
- Tallei, T.E.; Tumilaar, S.G.; Niode, N.J.; Fatimawali; Kepel, B.J.; Idroes, R.; Effendi, Y.; Sakib, S.A.; Emran, T.B. Potential of plant bioactive compounds as SARS-CoV-2 main protease (Mpro) and spike (S) glycoprotein inhibitors: A molecular docking study. Scientifica 2020, 2020, 6307457. [Google Scholar] [CrossRef]
- Morris, G.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Tumilaar, S.G.; Fatimawali, F.; Jane, N.N.; Yunus, E.; Rinaldi, I.; Akroman, A.A.; Ahmed, R.; Bin, E.T.; Ekawati, T.T. The potential of leaf extract of Pangium edule Reinw as HIV-1 protease inhibitor: A computational biology approach. J. Appl. Pharm. Sci. 2020, 11, 101–110. [Google Scholar] [CrossRef]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminform. 2011, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Sailah, I.; Tumilaar, S.G.; Lombogia, L.T.; Celik, I.; Tallei, T.E. Molecular docking and dynamics simulations study of selected phytoconstituents of “Pangi” (Pangium edule Reinw) leaf as anti-SARS-CoV-2. Philipp. J. 2021, 150, 925–937. [Google Scholar]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1, 19–25. [Google Scholar] [CrossRef]
- Celik, I.; Yadav, R.; Duzgun, Z.; Albogami, S.; El-Shehawi, A.M.; Fatimawali; Idroes, R.; Tallei, T.E.; Emran, T.B. Interactions of the receptor binding domain of SARS-CoV-2 variants with hACE2: Insights from molecular docking analysis and molecular dynamic simulation. Biology 2021, 10, 880. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Mackerell, A.D. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. J. Comput. Chem. 2013, 34, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
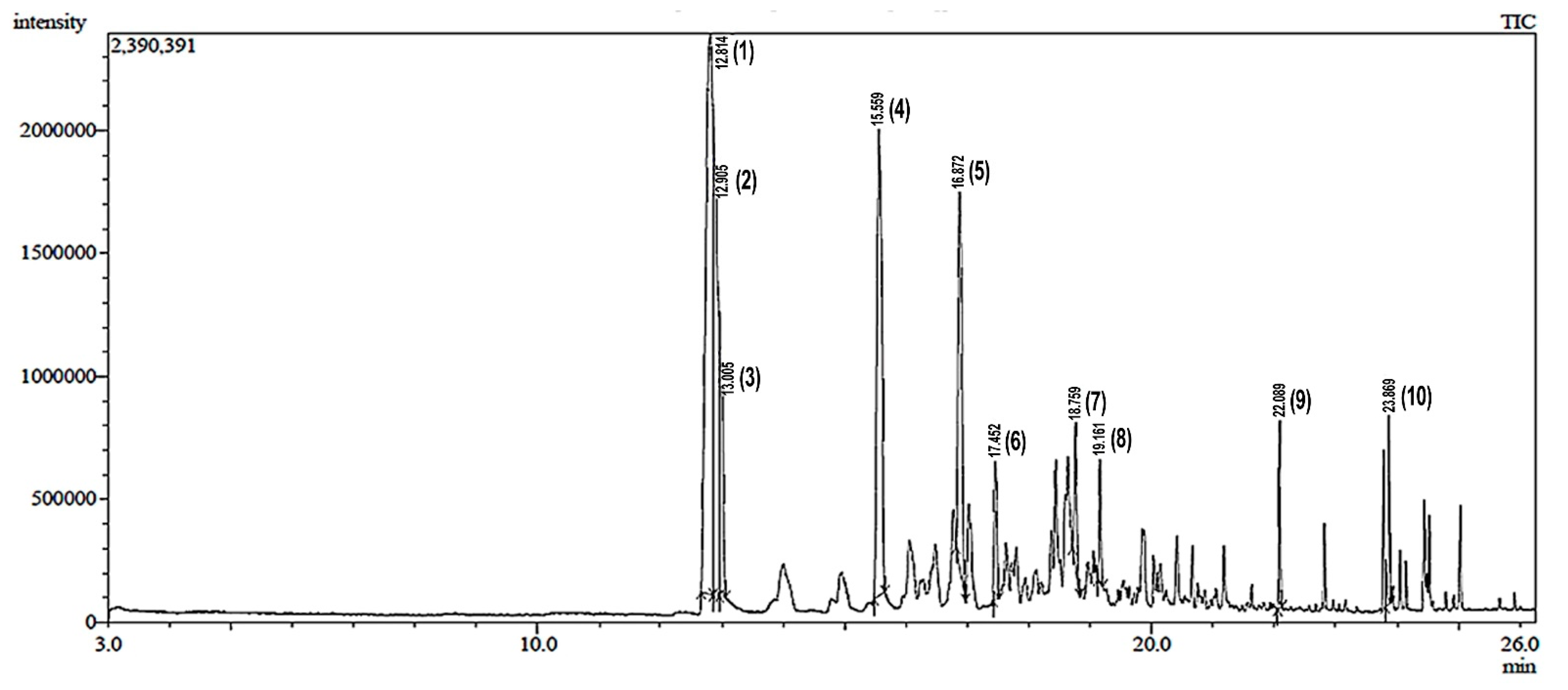
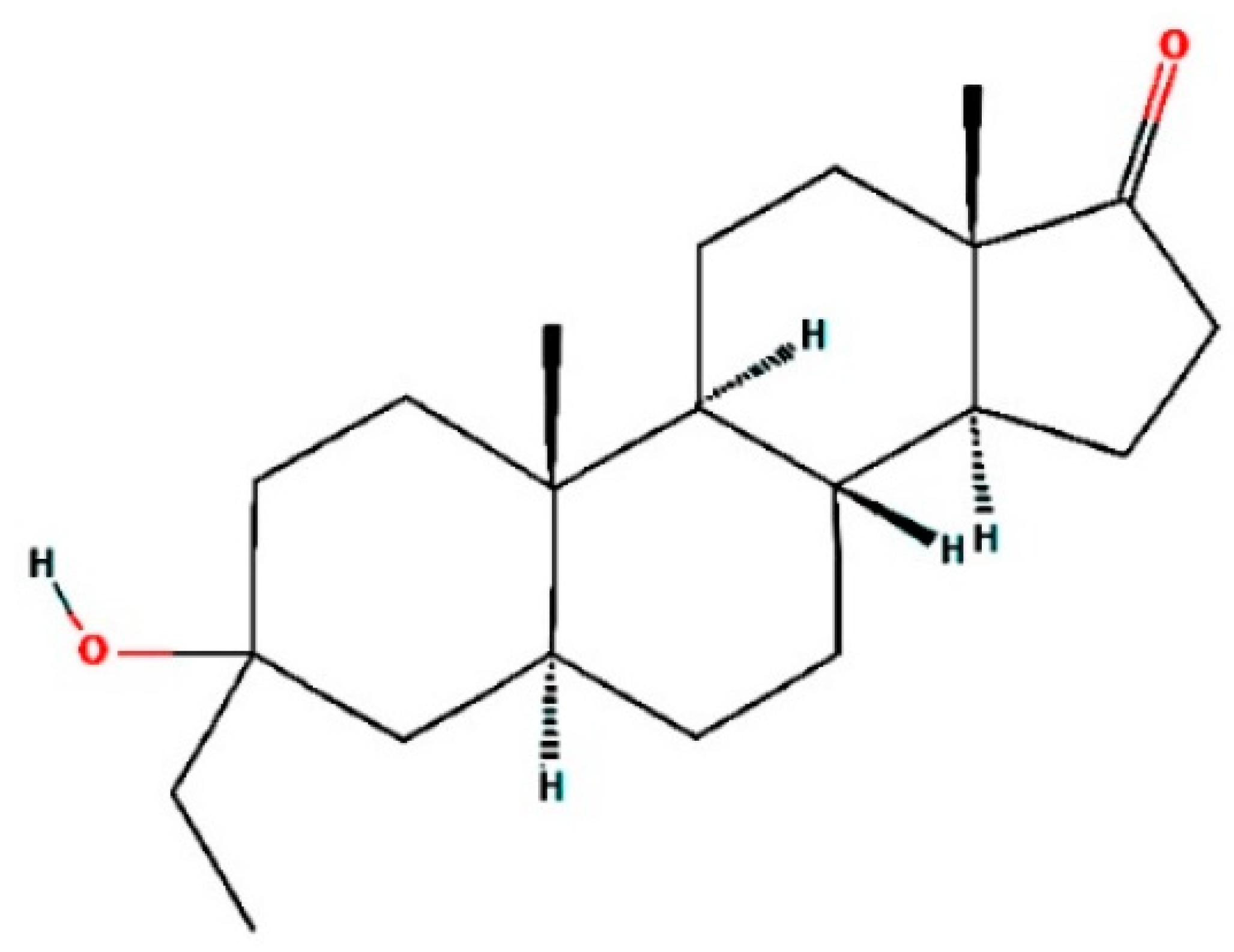


| Peak | Retention Time (min) | Probable Compound Name #Hit1 | Probable Compound Name #Hit2 | Probable Compound Name #Hit3 | Retention Area (%) |
|---|---|---|---|---|---|
| 1 | 12.814 | Phenol, 2-methoxy-3-(2-propenyl)- | Phenol, 2-methoxy-4-(2-propenyl)- | Phenol, 2-methoxy-4-(2-propenyl)- | 32.22 |
| 2 | 12.905 | 4-Nitroisopropylbenzene | 4-Nitroisopropylbenzene | 3-Nitroisopropylbenzene | 16.99 |
| 3 | 13.005 | Guaiacol, 3-allyl- | p-Eugenol | p-Eugenol | 7.10 |
| 4 | 15.559 | Benzoic acid, 2,4-dimethyl- | Benzoic acid, 2,4-dimethyl- | Benzoic acid, 2,6-dimethyl- | 18.86 |
| 5 | 16.872 | Delta-Cadinene | delta-Cadinene | delta-Cadinene | 11.85 |
| 6 | 17.452 | Nerolidol | Nerolidol b (cis or trans) | d-Nerolidol | 3.04 |
| 7 | 18.759 | alpha-Cadinol | Epiglobulol | Torreyol | 2.84 |
| 8 | 19.161 | Androstan-17-one, 3-ethyl-3-hydroxy-, (5 alpha)- | Longipinocarveol, trans- | Neoclovenoxid-alcohol | 1.95 |
| 9 | 22.089 | Hexadecanoic acid, methyl ester | Hexadecanoic acid, methyl ester | Hexadecanoic acid, methyl ester | 2.37 |
| 10 | 23.869 | 9-Octadecenoic acid, methyl ester | 9-Octadecenoic acid (Z)-, methyl ester | 9-Octadecenoic acid (Z)-, methyl ester | 2.78 |
| Ligand Properties | PubChem ID | Mol. Weight < 500 g/mol | No. H-Bond Donors < 5 | No. H-Bond Acceptors < 10 | Log p < 5 | No. of Violation |
|---|---|---|---|---|---|---|
| Androstan-17-one, ethyl-3-hydroxy-, (5 alpha)- | 14681481 | 318.50 | 1 | 2 | 4.4 | 0 |
| Torreyol | 11990360 | 222.37 | 1 | 1 | 3.3 | 0 |
| Delta-cadinene | 12306054 | 204.35 | 0 | 0 | 3.8 | 0 |
| Epiglobulol | 11858788 | 222.37 | 1 | 1 | 3.7 | 0 |
| Longipinocarveol, trans- | 534645 | 220.35 | 1 | 1 | 3.8 | 0 |
| Alpha-Cadinol | 6431302 | 223.37 | 3 | 5 | 3.78 | 0 |
| Neoclovenoxid-alcohol | 16211877 | 220.35 | 1 | 6 | 3.22 | 1 |
| 9-Octadecenoic acid, methyl ester | 5280590 | 34.06 | 1 | 1 | 0.57 | 0 |
| d-Nerolidol | 5356544 | 194.31 | 1 | 1 | 3.54 | 0 |
| Nerolidol | 5284507 | 222.37 | 1 | 1 | 4.19 | 0 |
| Benzoic acid, 2,4-dimethyl- | 11897 | 150 | 1 | 2 | 2 | 0 |
| Nerolidol b (cis or trans) | 131753171 | 233.26 | 1 | 3 | 4.5 | 0 |
| Eugenol | 3314 | 164 | 1 | 2 | 2.2 | 0 |
| 3-Nitroisopropylbenzene | 591251 | 165.19 | 0 | 2 | 2.07 | 0 |
| 4-Nitroisopropylbenzene | 15749 | 165 | 0 | 2 | 2.12 | 0 |
| Benzoic acid, 2,6-dimethyl- | 12439 | 150 | 1 | 2 | 2.3 | 0 |
| Phenol, 2-methoxy-3-(2-propenyl)- | 596373 | 125 | 1 | 2 | 2.98 | 0 |
| Phenol, 2-methoxy-4-(2-propenyl)- | 3313 | 125 | 1 | 3 | 2.9 | 0 |
| Hexadecanoic acid, methyl ester | 8181 | 270 | 0 | 2 | 5.6 | 0 |
| Guaicoal | 460 | 312 | 5 | 6 | 0.05 | 0 |
| Artemisinin (control) | 68827 | 282.33 | 0 | 5 | 2.8 | 0 |
| Compounds | LD50 (mg/kg) | Predicted Toxicity Class | Hepatotoxicity (Prediction/ Probability) | Carcinogenicity (Prediction/ Probability) | Immuno- Toxicity (Prediction/ Probability) | Mutagenicity (Prediction/ Probability) | Cytotoxicity (Prediction/ Probability) |
|---|---|---|---|---|---|---|---|
| Androstan-17-one, ethyl-3-hydroxy-, (5-alpha)- | 3000 | 5 | −/0.52 | −/0.78 | +/0.79 | −/0.96 | −/0.82 |
| Torreyol | 2830 | 5 | −/0.82 | −/0.66 | +/0.69 | −/0.91 | −/0.87 |
| Delta-cadinene | 4390 | 5 | −/0.82 | −/0.75 | −/0.68 | −/0.68 | −/0.69 |
| Epiglobulol | 2000 | 4 | −/0.77 | −/0.69 | −/0.87 | −/0.75 | −/0.89 |
| Longipinocarveol, trans- | 5000 | 5 | −/0.89 | −/0.64 | +/0.62 | −/0.92 | −/0.96 |
| Alpha-Cadinol | 2830 | 5 | −/0.82 | −/0.66 | +/0.69 | −/0.91 | −/0.87 |
| Neoclovenoxid-alcohol | 2000 | 4 | −/0.77 | −/0.75 | −/0.94 | −/0.75 | −/0.86 |
| 9-Octadecenoic acid, methyl ester | 3000 | 5 | −/0.59 | −/0.56 | −/0.96 | −/0.98 | −/0.70 |
| d-Nerolidol | 5000 | 5 | −/0.81 | −/0.65 | −/0.99 | −/0.91 | −/0.81 |
| Nerolidol | 5000 | 5 | −/0.81 | −/0.65 | −/0.99 | −/0.91 | −/0.81 |
| Benzoic acid, 2,4-dimethyl- | 3200 | 5 | +/0.52 | −/0.72 | −/0.99 | −/0.97 | −/0.88 |
| Nerolidol b (cis or trans) | 5000 | 6 | −/0.75 | −/0.66 | −/0.99 | −/0.92 | −/0.79 |
| Eugenol | 1930 | 4 | −/0.67 | −/0.73 | −/0.83 | −/0.97 | −/0.90 |
| 3-Nitroisopropylbenzene | 430 | 4 | −/0.51 | −/0.52 | −/0.86 | −/0.57 | −/0.79 |
| 4-Nitroisopropylbenzene | 1000 | 4 | −/0.51 | −/0.52 | −/0.96 | −/0.57 | −/0.79 |
| Benzoic acid, 2,6-dimethyl- | 4480 | 5 | +/0.52 | −/0.72 | −/0.99 | −/0.97 | −/0.88 |
| Phenol, 2-methoxy-3-(2-propenyl)- | 1230 | 4 | −/0.68 | −/0.72 | −/0.70 | −/0.84 | −/0.86 |
| Phenol, 2-methoxy-4-(2-propenyl)- | 916 | 4 | −/0.74 | −/0.62 | −/0.70 | −/0.84 | −/0.86 |
| Hexadecanoic acid, methyl ester | 5000 | 5 | −/0.58 | −/0.55 | −/0.90 | −/0.83 | −/0.70 |
| Guaicoal | 520 | 4 | −/0.72 | +/0.56 | −/0.85 | −/0.99 | −/0.81 |
| Artemisinin | 4228 | 5 | −/0.72 | −/0.63 | +/0.70 | −/0.63 | −/0.97 |
| Ligand Properties | Binding Free Energy (kcal/mol) | |
|---|---|---|
| 1LEE (Plasmepsin 2) | 3QS1 (Plasmepsin 1) | |
| Androstan-17-one, ethyl-3-hydroxy-, (5-alpha)- | −8.0 | −9.1 |
| Torreyol | −6.6 | −6.4 |
| Delta-cadinene | −6.4 | −6.3 |
| Epiglobulol | −6.4 | −6.3 |
| Longipinocarveol, trans- | −6.1 | −7.1 |
| Alpha-Cadinol | −6.0 | −6.1 |
| Neoclovenoxid-alcohol | −6.0 | −6.0 |
| 9-Octadecenoic acid, methyl ester | −5.9 | −5.8 |
| d- Nerolidol | −5.8 | −6.1 |
| Nerolidol | −5.8 | −6.1 |
| Benzoic acid, 2,4-dimethyl- | −5.6 | −5.6 |
| Nerolidol b (cis or trans) | −5.4 | −5.6 |
| Eugenol | −5.4 | −5.5 |
| 3-Nitroisopropylbenzene | −5.3 | −6.0 |
| 4-Nitroisopropylbenzene | −5.2 | −5.8 |
| Benzoic acid, 2,6-dimethyl- | −5.0 | −5.1 |
| Phenol, 2-methoxy-3-(2-propenyl)- | −5.0 | −5.3 |
| Phenol, 2-methoxy-4-(2-propenyl)- | −4,9 | −5.0 |
| Hexadecanoic acid, methyl ester | −4.9 | −4.9 |
| Guaicoal | −4.5 | −4.7 |
| Artemisinin (control) | −6.7 | −7.7 |
| Receptor Name | Binding Affinity (kcal/mol) | No. H-Bond | Interacting Residues | Distance (Å) | Category | Type of Interaction |
|---|---|---|---|---|---|---|
| Plasmepsin 1 (3QS1) | −9.1 | 1 | Ser(A77) | 2.74 | H-Bond | Conventional |
| Tyr(A75) | 3.83 | Hydrophobic | Pi-Sigma | |||
| Met(A13) | 4.92 | Hydrophobic | Alkyl | |||
| Ile(A30) | 3.89 | Hydrophobic | Alkyl | |||
| Phe(A117) | - | Electrostatic | Van der Waals | |||
| Ile(A120) | - | Electrostatic | Van der Waals | |||
| Phe(A109) | - | Electrostatic | Van der Waals | |||
| Val(A76) | - | Electrostatic | Van der Waals | |||
| Asp(A32) | - | Electrostatic | Van der Waals | |||
| Thr(A218) | - | Electrostatic | Van der Waals | |||
| Gly(A217) | - | Electrostatic | Van der Waals | |||
| Plasmepsin 2 (1LEE) | −8 | 0 | Ile(A300) | 5.12 | Hydrophobic | Pi-Alkyl/Alkyl |
| Val(A78) | 4.18 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Val(A78) | 4.53 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Tyr(A192) | 4.93 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Gly(A36) | - | Electrostatic | Van der Waals | |||
| Asp(A214) | - | Electrostatic | Van der Waals | |||
| Asp(A34) | - | Electrostatic | Van der Waals | |||
| Tyr(A77) | - | Electrostatic | Van der Waals | |||
| Ile(A123) | - | Electrostatic | Van der Waals | |||
| Ile(A32) | - | Electrostatic | Van der Waals | |||
| Phe(A111) | - | Electrostatic | Van der Waals | |||
| Phe(A120) | - | Electrostatic | Van der Waals | |||
| Ser(A79) | - | Electrostatic | Van der Waals | |||
| Gly(A216) | - | Electrostatic | Van der Waals | |||
| Thr(A217) | - | Electrostatic | Van der Waals | |||
| Leu(A292) | - | Electrostatic | Van der Waals | |||
| Phe(A294) | - | Electrostatic | Van der Waals |
| Receptor Name | Binding Affinity (kcal/mol) | No. H-Bond | Interacting Residues | Distance (Å) | Category | Type of Interaction |
|---|---|---|---|---|---|---|
| Plasmepsin 1 (3QS1) | −7.7 | 0 | Ile(A120) | 5.01 | Hydrophobic | Pi-Alkyl/Alkyl |
| Phe(A109) | 4.99 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Tyr(A75) | 3.77 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Ile(A30) | 4.97 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Ile(A30) | 4.98 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Phe(A117) | 4.27 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Met(A13) | 4.13 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Ala(A111) | - | Electrostatic | Van der Waals | |||
| Ser(A219) | - | Electrostatic | Van der Waals | |||
| Thr(A218) | - | Electrostatic | Van der Waals | |||
| Gly(A217) | - | Electrostatic | Van der Waals | |||
| Ser(A77) | - | Electrostatic | Van der Waals | |||
| Asp(A32) | - | Electrostatic | Van der Waals | |||
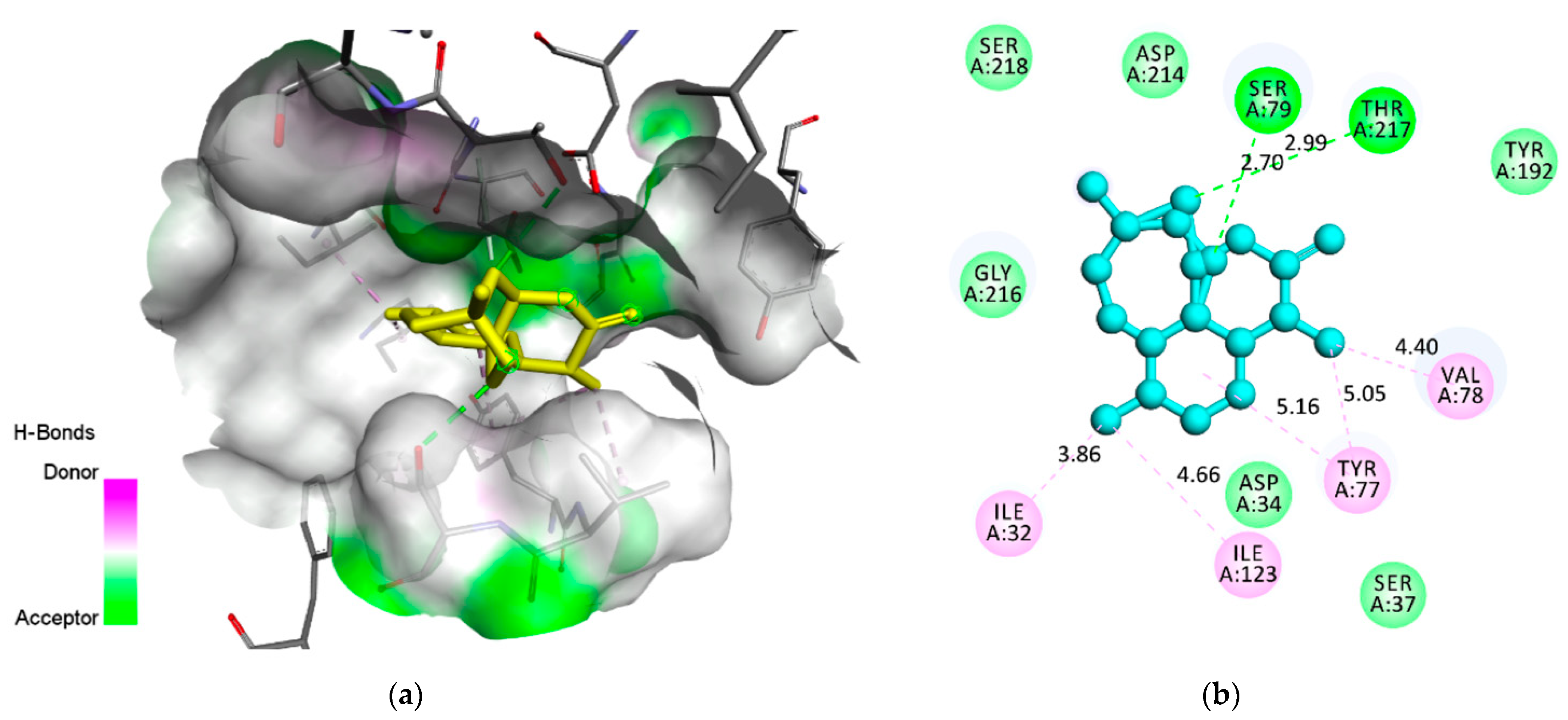
| Plasmepsin 2 (1LEE) | −6.7 | 2 | Ser(A79) | 2.70 | H-Bond | Conventional |
| Thr(A217) | 2.99 | H-Bond | Conventional | |||
| Val(A78) | 4.40 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Tyr(A77) | 5.05 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Tyr(A77) | 5.16 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Ile(A123) | 4.66 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Ile(A32) | 3.86 | Hydrophobic | Pi-Alkyl/Alkyl | |||
| Tyr(A192) | - | Electrostatic | Van der Waals | |||
| Ser(A37) | - | Electrostatic | Van der Waals | |||
| Asp(A34) | Electrostatic | Van der Waals | ||||
| Gly(A216) | - | Electrostatic | Van der Waals | |||
| Ser(A218) | - | Electrostatic | Van der Waals | |||
| Asp(A214) | - | Electrostatic | Van der Waals |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatimawali; Tallei, T.E.; Kepel, B.J.; Alorabi, M.; El-Shehawi, A.M.; Bodhi, W.; Tumilaar, S.G.; Celik, I.; Mostafa-Hedeab, G.; Mohamed, A.A.-R.; et al. Appraisal of Bioactive Compounds of Betel Fruit as Antimalarial Agents by Targeting Plasmepsin 1 and 2: A Computational Approach. Pharmaceuticals 2021, 14, 1285. https://doi.org/10.3390/ph14121285
Fatimawali, Tallei TE, Kepel BJ, Alorabi M, El-Shehawi AM, Bodhi W, Tumilaar SG, Celik I, Mostafa-Hedeab G, Mohamed AA-R, et al. Appraisal of Bioactive Compounds of Betel Fruit as Antimalarial Agents by Targeting Plasmepsin 1 and 2: A Computational Approach. Pharmaceuticals. 2021; 14(12):1285. https://doi.org/10.3390/ph14121285
Chicago/Turabian StyleFatimawali, Trina Ekawati Tallei, Billy Johnson Kepel, Mohammed Alorabi, Ahmed M. El-Shehawi, Widdhi Bodhi, Sefren Geiner Tumilaar, Ismail Celik, Gomaa Mostafa-Hedeab, Amany Abdel-Rahman Mohamed, and et al. 2021. "Appraisal of Bioactive Compounds of Betel Fruit as Antimalarial Agents by Targeting Plasmepsin 1 and 2: A Computational Approach" Pharmaceuticals 14, no. 12: 1285. https://doi.org/10.3390/ph14121285
APA StyleFatimawali, Tallei, T. E., Kepel, B. J., Alorabi, M., El-Shehawi, A. M., Bodhi, W., Tumilaar, S. G., Celik, I., Mostafa-Hedeab, G., Mohamed, A. A.-R., & Emran, T. B. (2021). Appraisal of Bioactive Compounds of Betel Fruit as Antimalarial Agents by Targeting Plasmepsin 1 and 2: A Computational Approach. Pharmaceuticals, 14(12), 1285. https://doi.org/10.3390/ph14121285








