Quorum Sensing Inhibitors to Quench P. aeruginosa Pathogenicity
Abstract
:1. Introduction
2. QS-Controlled Pathogenicity of P. aeruginosa
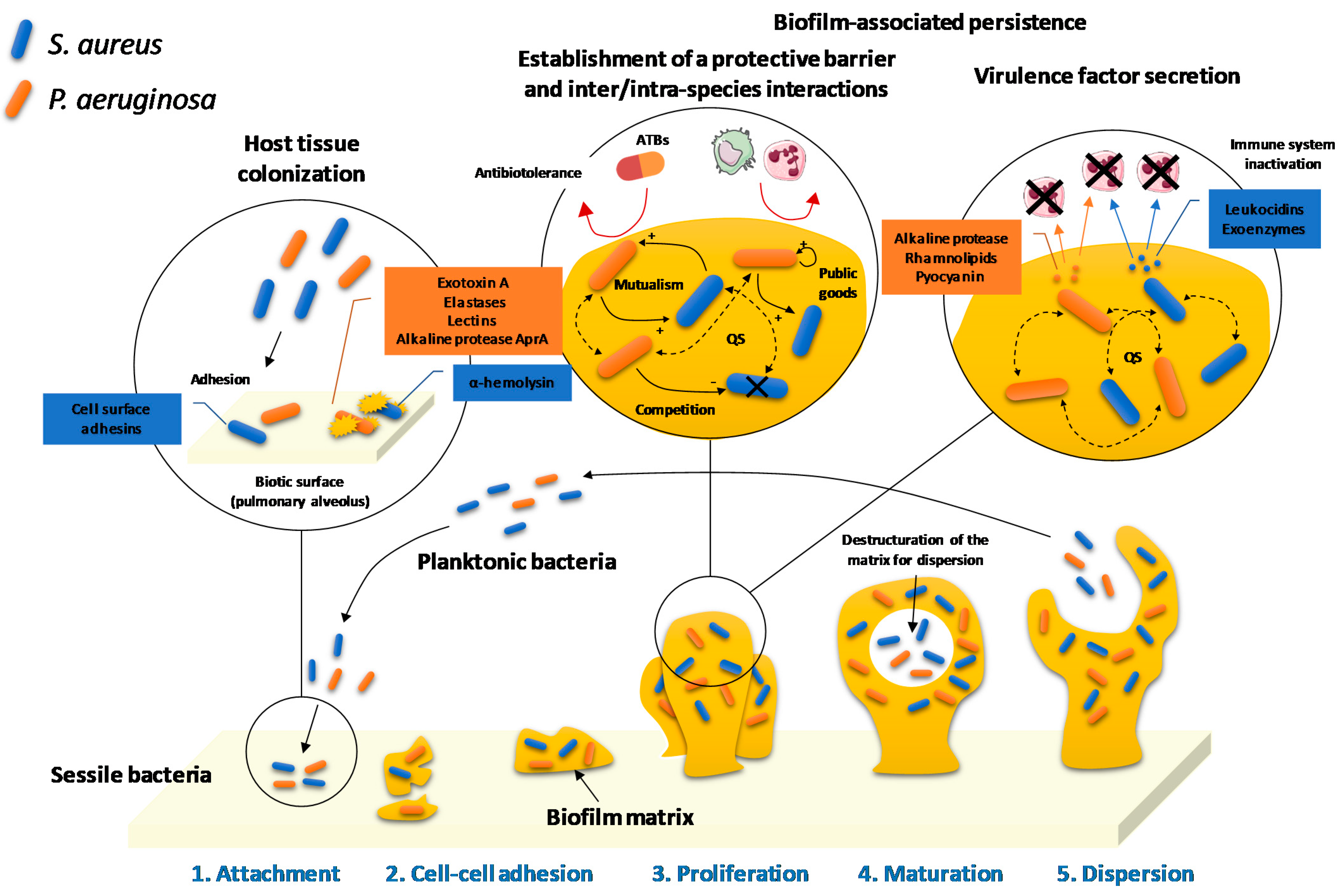
2.1. Biofilm Development
2.2. Virulence Factor Secretion
2.2.1. Proteolytic Enzymes
2.2.2. Exotoxins
2.2.3. Siderophores
2.2.4. Other Pro-Infectious Molecules
Rhamnolipids
Hydrogen Cyanide
Lectins
2.3. Host Tissue Colonization-Promoting Microbial Interactions
2.3.1. Biofilm-Associated Collaboration
2.3.2. Intra- and Inter-Species Competition
Indirect Competition
Direct Competition
Bacteriocins
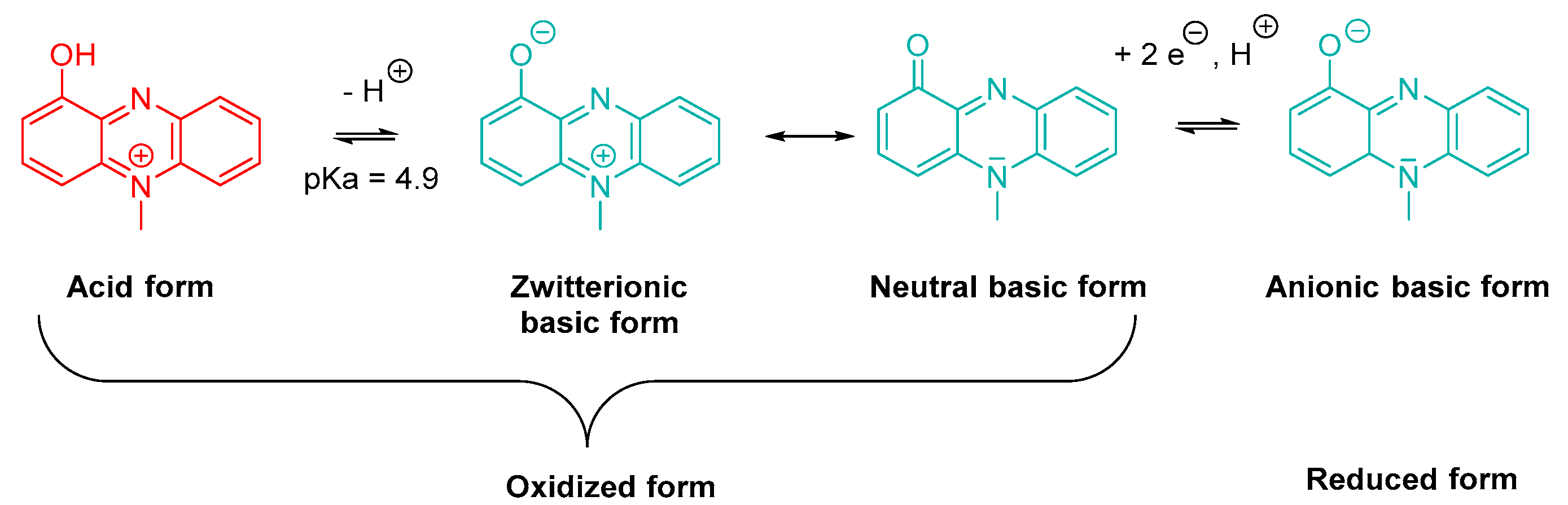
Pro-Oxidative Toxins
3. QS Molecular Pathways in P. aeruginosa
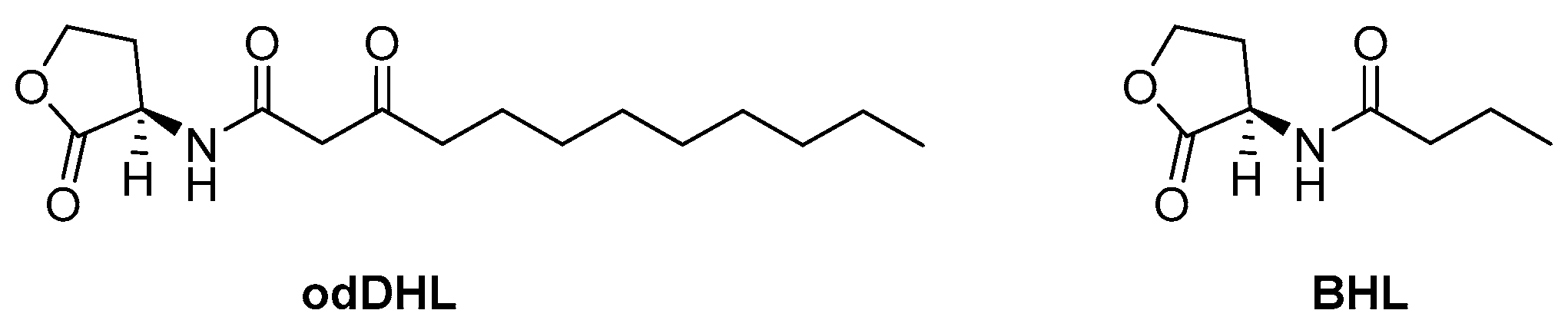
3.1. Las and Rhl Systems
3.2. Pqs System
4. P. aeruginosa QS-Quenching Anti-Virulence Agents
4.1. LasR-Targeting Pan-QSIs
4.1.1. AHL Autoinducer Analog QSIs
4.1.2. AHL Autoinducer Non-Analog QSIs
4.2. RhlR-Modulating QSIs
4.3. PqsR and AQ Synthase-Targeting QSIs
4.3.1. PqsR Inhibitors
AQ Autoinducer Analog QSIs
AQ Autoinducer Non-Analog QSIs
4.3.2. AQ Synthase Inhibitors
PqsA Inhibitors
PqsD Inhibitors
4.4. Multi-Target QSIs
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ABC | ATP binding cassette |
| ACP | Acyl carrier protein |
| AIs | Autoinducers |
| AIPs | Autoinducing peptides |
| AHL | N-acyl-homoserine lactone |
| AQs | 2-alkyl-4(1H)-quinolones |
| AQNOs | 2-alkyl-4-quinolone N-oxides |
| ATB | Antibiotic |
| AVAs | Anti-virulence agents |
| BHL | N-butanoyl-L-homoserine lactone |
| CA | Chorismic acid |
| CIO | Cyanide-insensitive terminal oxidase |
| DHQ | 2,4-dihydroxyquinoline |
| ECDC | European Centre for Disease Prevention and Control |
| eDNA | Extracellular DNA |
| EPS | Exopolysaccharides |
| ETA | Exotoxin A |
| EU/EEA | European Union/European Economic Area |
| HCL | HomoCysteine Lactone |
| HHQ | 2-heptyl-4(1H)-quinolone |
| HQNO | 2-heptyl-4-quinolone N-oxide |
| HSL | HomoSerine Lactone |
| IgA | Immunoglobulin A |
| IgG | Immunoglobulin G |
| LPS | Lipopolysaccharide |
| MDR | Multidrug-resistant |
| MvfR | Multiple virulence-factor regulator |
| NQNO | 2-nonyl-4-quinolone N-oxide |
| odDHL | N-(3-oxododecanoyl)-L-homoserine lactone |
| OMV | Outer membrane vesicles |
| PCA | Phenazine-1-carboxylic acid |
| Pe | Phosphatidylethanolamine |
| PG | Phosphatidylglycerol |
| PIV | Type IV proteases |
| PLLs | Phosphotriesterase-like lactonases |
| PQS | Pseudomonas quinolone signal |
| Quorum Quenching | |
| QS | Quorum Sensing |
| QSIs | Quorum Sensing inhibitors |
| SOD | Superoxide dismutase |
| sPLA2-IIA | Soluble Type IIA phospholipase A2 |
References
- Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 3 December 2021).
- European Center for Disease Prevention and Control. Antimicrobial Resistance in the EU/EEA (EARS-Net)—Annual Epidemiological Report for 2019; ECDC: Stockholm, Sweden, 2020. [Google Scholar]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Kapoor, G.; Saigal, S.; Elongavan, A. Action and Resistance Mechanisms of Antibiotics: A Guide for Clinicians. J. Anaesthesiol. Clin. Pharmacol. 2017, 33, 300–305. [Google Scholar] [CrossRef]
- Li, X.-Z.; Plésiat, P.; Nikaido, H. The Challenge of Efflux-Mediated Antibiotic Resistance in Gram-Negative Bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef] [Green Version]
- Levine, C.; Hiasa, H.; Marians, K.J. DNA Gyrase and Topoisomerase IV: Biochemical Activities, Physiological Roles during Chromosome Replication, and Drug Sensitivities. Biochim. Biophys. Acta BBA Gene Struct. Expr. 1998, 1400, 29–43. [Google Scholar] [CrossRef]
- Castillo-Juárez, I.; Maeda, T.; Mandujano-Tinoco, E.A.; Tomás, M.; Pérez-Eretza, B.; García-Contreras, S.J.; Wood, T.K.; García-Contreras, R. Role of Quorum Sensing in Bacterial Infections. World J. Clin. Cases 2015, 3, 575. [Google Scholar] [CrossRef]
- Khalifa, A.B.H.; Moissenet, D.; Vu Thien, H.; Khedher, M. Virulence factors in Pseudomonas aeruginosa: Mechanisms and modes of regulation. Ann. Biol. Clin. 2011, 69, 393–403. [Google Scholar] [CrossRef]
- Shaw, E.; Wuest, W.M. Virulence Attenuating Combination Therapy: A Potential Multi-Target Synergy Approach to Treat Pseudomonas aeruginosa Infections in Cystic Fibrosis Patients. RSC Med. Chem. 2020, 11, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Jacobs, H.M.; Matwichuk, M.; Wong, C.; Wozniak, D.J.; Parsek, M.R. The Versatile Pseudomonas aeruginosa Biofilm Matrix Protein CdrA Promotes Aggregation through Different Extracellular Exopolysaccharide Interactions. J. Bacteriol. 2020, 202, e00216-20. [Google Scholar] [CrossRef] [PubMed]
- Devaraj, A.; Buzzo, J.R.; Mashburn-Warren, L.; Gloag, E.S.; Novotny, L.A.; Stoodley, P.; Bakaletz, L.O.; Goodman, S.D. The Extracellular DNA Lattice of Bacterial Biofilms Is Structurally Related to Holliday Junction Recombination Intermediates. Proc. Natl. Acad. Sci. USA 2019, 116, 25068–25077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santajit, S.; Indrawattana, N. Mechanisms of Antimicrobial Resistance in ESKAPE Pathogens. BioMed Res. Int. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Nadell, C.D.; Drescher, K.; Foster, K.R. Spatial Structure, Cooperation and Competition in Biofilms. Nat. Rev. Microbiol. 2016, 14, 589–600. [Google Scholar] [CrossRef] [PubMed]
- Stewart, P.S.; Costerton, J.W. Antibiotic Resistance of Bacteria in Biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Alhede, M.; Bjarnsholt, T.; Givskov, M.; Alhede, M. Pseudomonas aeruginosa Biofilms. In Advances in Applied Microbiology; Elsevier: Amsterdam, The Netherlands, 2014; Volume 86, pp. 1–40. ISBN 978-0-12-800262-9. [Google Scholar]
- Jefferson, K.K. What Drives Bacteria to Produce a Biofilm? FEMS Microbiol. Lett. 2004, 236, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.-F.; Purmal, K.; Chin, S.; Chan, X.-Y.; Koh, C.-L.; Sam, C.-K.; Chan, K.-G. N-Acyl Homoserine Lactone Production by Klebsiella pneumoniae Isolated from Human Tongue Surface. Sensors 2012, 12, 3472–3483. [Google Scholar] [CrossRef] [Green Version]
- Abe, K.; Nomura, N.; Suzuki, S. Biofilms: Hot Spots of Horizontal Gene Transfer (HGT) in Aquatic Environments, with a Focus on a New HGT Mechanism. FEMS Microbiol. Ecol. 2020, 96, fiaa031. [Google Scholar] [CrossRef]
- Jyot, J.; Balloy, V.; Jouvion, G.; Verma, A.; Touqui, L.; Huerre, M.; Chignard, M.; Ramphal, R. Type II Secretion System of Pseudomonas aeruginosa: In Vivo Evidence of a Significant Role in Death Due to Lung Infection. J. Infect. Dis. 2011, 203, 1369–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsumoto, K. Role of Bacterial Proteases in Pseudomonal and Serratial keratitis. Biol. Chem. 2004, 385. [Google Scholar] [CrossRef]
- Laarman, A.J.; Bardoel, B.W.; Ruyken, M.; Fernie, J.; Milder, F.J.; van Strijp, J.A.G.; Rooijakkers, S.H.M. Pseudomonas aeruginosa Alkaline Protease Blocks Complement Activation via the Classical and Lectin Pathways. J. Immunol. 2012, 188, 386–393. [Google Scholar] [CrossRef] [Green Version]
- Iiyama, K.; Takahashi, E.; Lee, J.M.; Mon, H.; Morishita, M.; Kusakabe, T.; Yasunaga-Aoki, C. Alkaline Protease Contributes to Pyocyanin Production in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef]
- Conibear, T.C.R.; Willcox, M.D.P.; Flanagan, J.L.; Zhu, H. Characterization of Protease IV Expression in Pseudomonas aeruginosa Clinical Isolates. J. Med. Microbiol. 2012, 61, 180–190. [Google Scholar] [CrossRef]
- Fitzgerald, D. Receptor-Mediated Internalization of Pseudomonas Toxin by Mouse Fibroblasts. Cell 1980, 21, 867–873. [Google Scholar] [CrossRef]
- Yousefi-Avarvand, A.; Khashei, R.; Ebrahim-Saraie, H.S.; Emami, A.; Zomorodian, K.; Motamedifar, M. The Frequency of Exotoxin A and Exoenzymes S and U Genes Among Clinical Isolates of Pseudomonas aeruginosa in Shiraz, Iran. Int. J. Mol. Cell. Med. 2015, 4, 167–173. [Google Scholar] [PubMed]
- Hall, S.; McDermott, C.; Anoopkumar-Dukie, S.; McFarland, A.; Forbes, A.; Perkins, A.; Davey, A.; Chess-Williams, R.; Kiefel, M.; Arora, D.; et al. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins 2016, 8, 236. [Google Scholar] [CrossRef]
- Gonçalves, T.; Vasconcelos, U. Colour Me Blue: The History and the Biotechnological Potential of Pyocyanin. Molecules 2021, 26, 927. [Google Scholar] [CrossRef] [PubMed]
- Nadal Jimenez, P.; Koch, G.; Thompson, J.A.; Xavier, K.B.; Cool, R.H.; Quax, W.J. The Multiple Signaling Systems Regulating Virulence in Pseudomonas aeruginosa. Microbiol. Mol. Biol. Rev. 2012, 76, 46–65. [Google Scholar] [CrossRef] [Green Version]
- Ran, H.; Hassett, D.J.; Lau, G.W. Human Targets of Pseudomonas aeruginosa Pyocyanin. Proc. Natl. Acad. Sci. USA 2003, 100, 14315–14320. [Google Scholar] [CrossRef] [Green Version]
- Allen, L.; Dockrell, D.H.; Pattery, T.; Lee, D.G.; Cornelis, P.; Hellewell, P.G.; Whyte, M.K.B. Pyocyanin Production by Pseudomonas aeruginosa Induces Neutrophil Apoptosis and Impairs Neutrophil-Mediated Host Defenses In Vivo. J. Immunol. 2005, 174, 3643–3649. [Google Scholar] [CrossRef] [Green Version]
- Moayedi, A.; Nowroozi, J.; Sepahi, A.A. Cytotoxic Effect of Pyocyanin on Human Pancreatic Cancer Cell Line (Panc-1). Iran. J. Basic Med. Sci. 2018. [Google Scholar] [CrossRef]
- Rada, B.; Leto, T.L. Pyocyanin Effects on Respiratory Epithelium: Relevance in Pseudomonas aeruginosa Airway Infections. Trends Microbiol. 2013, 21, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Cox, C.D. Role of Pyocyanin in the Acquisition of Iron from Transferrin. Infect. Immun. 1986, 52, 263–270. [Google Scholar] [CrossRef] [Green Version]
- Cornelis, P. Iron Uptake and Metabolism in Pseudomonads. Appl. Microbiol. Biotechnol. 2010, 86, 1637–1645. [Google Scholar] [CrossRef]
- Brandel, J.; Humbert, N.; Elhabiri, M.; Schalk, I.J.; Mislin, G.L.A.; Albrecht-Gary, A.-M. Pyochelin, a Siderophore of Pseudomonas aeruginosa: Physicochemical Characterization of the Iron(III), Copper(II) and Zinc(II) Complexes. Dalton Trans. 2012, 41, 2820. [Google Scholar] [CrossRef] [PubMed]
- Meyer, J.-M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, I.A. Pyoverdin Is Essential for Virulence of Pseudomonas aeruginosa. Infect Immun. 1996, 64, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cezard, C.; Farvacques, N.; Sonnet, P. Chemistry and Biology of Pyoverdines, Pseudomonas Primary Siderophores. Curr. Med. Chem. 2014, 22, 165–186. [Google Scholar] [CrossRef]
- Braud, A.; Hoegy, F.; Jezequel, K.; Lebeau, T.; Schalk, I.J. New Insights into the Metal Specificity of the Pseudomonas aeruginosa Pyoverdine-Iron Uptake Pathway. Environ. Microbiol. 2009, 11, 1079–1091. [Google Scholar] [CrossRef]
- Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. Impact of Siderophore Production on Pseudomonas aeruginosa Infections in Immunosuppressed Mice. Infect. Immun. 2000, 68, 1834–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, D.; Kirienko, D.R.; Webster, P.; Fisher, A.L.; Kirienko, N.V. Pyoverdine, a Siderophore from Pseudomonas aeruginosa, Translocates into C. Elegans, Removes Iron, and Activates a Distinct Host Response. Virulence 2018, 9, 804–817. [Google Scholar] [CrossRef] [Green Version]
- McClure, C.D.; Schiller, N.L. Inhibition of Macrophage Phagocytosis by Pseudomonas aeruginosa Rhamnolipids In Vitro and In Vivo. Curr. Microbiol. 1996, 33, 109–117. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Asfahl, K.L.; Li, N.; Sun, F.; Xiao, J.; Shen, D.; Dandekar, A.A.; Wang, M. Conditional Quorum-Sensing Induction of a Cyanide-Insensitive Terminal Oxidase Stabilizes Cooperating Populations of Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 4999. [Google Scholar] [CrossRef]
- Williams, H.D.; Zlosnik, J.E.A.; Ryall, B. Oxygen, Cyanide and Energy Generation in the Cystic Fibrosis Pathogen Pseudomonas aeruginosa. In Advances in Microbial Physiology; Elsevier: Amsterdam, The Netherlands, 2006; Volume 52, pp. 1–71. ISBN 978-0-12-027752-0. [Google Scholar]
- Lenney, W.; Gilchrist, F.J. Pseudomonas aeruginosa and Cyanide Production. Eur. Respir. J. 2011, 37, 482–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zlosnik, J.E.A.; Tavankar, G.R.; Bundy, J.G.; Mossialos, D.; O′Toole, R.; Williams, H.D. Investigation of the Physiological Relationship between the Cyanide-Insensitive Oxidase and Cyanide Production in Pseudomonas aeruginosa. Microbiology 2006, 152, 1407–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grishin, A.V.; Krivozubov, M.S.; Karyagina, A.S.; Gintsburg, A.L. Pseudomonas aeruginosa Lectins as Targets for Novel Antibacterials. Acta Nat. 2015, 7, 29–41. [Google Scholar] [CrossRef] [Green Version]
- Heilmann, S.; Krishna, S.; Kerr, B. Why Do Bacteria Regulate Public Goods by Quorum Sensing?—How the Shapes of Cost and Benefit Functions Determine the Form of Optimal Regulation. Front. Microbiol. 2015, 6, 767. [Google Scholar] [CrossRef]
- Rendueles, O.; Ghigo, J.-M. Mechanisms of Competition in Biofilm Communities. Microbiol. Spectr. 2015, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornelis, P.; Dingemans, J. Pseudomonas aeruginosa Adapts Its Iron Uptake Strategies in Function of the Type of Infections. Front. Cell. Infect. Microbiol. 2013, 3, 75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rendueles, O.; Ghigo, J.-M. Multi-Species Biofilms: How to Avoid Unfriendly Neighbors. FEMS Microbiol. Rev. 2012, 36, 972–989. [Google Scholar] [CrossRef]
- Czajkowski, R.; Jafra, S. Quenching of Acyl-Homoserine Lactone-Dependent Quorum Sensing by Enzymatic Disruption of Signal Molecules. Acta Biochim. Pol. 2009, 56, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Qazi, S.; Middleton, B.; Muharram, S.H.; Cockayne, A.; Hill, P.; O’Shea, P.; Chhabra, S.R.; Camara, M.; Williams, P. N-Acylhomoserine Lactones Antagonize Virulence Gene Expression and Quorum Sensing in Staphylococcus aureus. Infect. Immun. 2006, 74, 910–919. [Google Scholar] [CrossRef] [Green Version]
- Khan, B.A.; Yeh, A.J.; Cheung, G.Y.; Otto, M. Investigational Therapies Targeting Quorum-Sensing for the Treatment of Staphylococcus aureus Infections. Expert Opin. Investig. Drugs 2015, 24, 689–704. [Google Scholar] [CrossRef] [PubMed]
- Davies, D.G.; Marques, C.N.H. A Fatty Acid Messenger Is Responsible for Inducing Dispersion in Microbial Biofilms. J. Bacteriol. 2009, 191, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Pernet, E.; Guillemot, L.; Burgel, P.-R.; Martin, C.; Lambeau, G.; Sermet-Gaudelus, I.; Sands, D.; Leduc, D.; Morand, P.C.; Jeammet, L.; et al. Pseudomonas aeruginosa Eradicates Staphylococcus aureus by Manipulating the Host Immunity. Nat. Commun. 2014, 5, 5105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pickard, J.M.; Zeng, M.Y.; Caruso, R.; Núñez, G. Gut Microbiota: Role in Pathogen Colonization, Immune Responses, and Inflammatory Disease. Immunol. Rev. 2017, 279, 70–89. [Google Scholar] [CrossRef] [PubMed]
- Majeed, H.; Gillor, O.; Kerr, B.; Riley, M.A. Competitive Interactions in Escherichia coli Populations: The Role of Bacteriocins. ISME J. 2011, 5, 71–81. [Google Scholar] [CrossRef] [Green Version]
- Gillor, O.; Etzion, A.; Riley, M.A. The Dual Role of Bacteriocins as Anti- and Probiotics. Appl. Microbiol. Biotechnol. 2008, 81, 591–606. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Baron, S.S.; Rowe, J.J. Antibiotic Action of Pyocyanin. Antimicrob. Agents Chemother. 1981, 20, 814–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, S.S.; Terranova, G.; Rowe, J.J. Molecular Mechanism of the Antimicrobial Action of Pyocyanin. Curr. Microbiol. 1989, 18, 223–230. [Google Scholar] [CrossRef]
- Hazan, R.; Que, Y.A.; Maura, D.; Strobel, B.; Majcherczyk, P.A.; Hopper, L.R.; Wilbur, D.J.; Hreha, T.N.; Barquera, B.; Rahme, L.G. Auto Poisoning of the Respiratory Chain by a Quorum-Sensing-Regulated Molecule Favors Biofilm Formation and Antibiotic Tolerance. Curr. Biol. 2016, 26, 195–206. [Google Scholar] [CrossRef] [Green Version]
- Hoffman, L.R.; Deziel, E.; D’Argenio, D.A.; Lepine, F.; Emerson, J.; McNamara, S.; Gibson, R.L.; Ramsey, B.W.; Miller, S.I. Selection for Staphylococcus Aureus Small-Colony Variants Due to Growth in the Presence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2006, 103, 19890–19895. [Google Scholar] [CrossRef] [Green Version]
- Filkins, L.M.; Graber, J.A.; Olson, D.G.; Dolben, E.L.; Lynd, L.R.; Bhuju, S.; O’Toole, G.A. Coculture of Staphylococcus aureus with Pseudomonas aeruginosa Drives S. aureus towards Fermentative Metabolism and Reduced Viability in a Cystic Fibrosis Model. J. Bacteriol. 2015, 197, 2252–2264. [Google Scholar] [CrossRef] [Green Version]
- Novick, R.P.; Geisinger, E. Quorum Sensing in Staphylococci. Annu. Rev. Genet. 2008, 42, 541–564. [Google Scholar] [CrossRef]
- Sufrin, J.; Finckbeiner, S.; Oliver, C. Marine-Derived Metabolites of S-Adenosylmethionine as Templates for New Anti-Infectives. Mar. Drugs 2009, 7, 401–434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soukarieh, F.; Williams, P.; Stocks, M.J.; Cámara, M. Pseudomonas aeruginosa Quorum Sensing Systems as Drug Discovery Targets: Current Position and Future Perspectives. J. Med. Chem. 2018, 61, 10385–10402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, T.T.; Sullivan, S.A.; Cusick, J.K.; Schweizer, H.P. β-Ketoacyl Acyl Carrier Protein Reductase (FabG) Activity of the Fatty Acid Biosynthetic Pathway is a Determining Factor of 3-Oxo-Homoserine Lactone Acyl Chain Lengths. Microbiology 2002, 148, 3849–3856. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, A.; Wood, T.K. Connecting Quorum Sensing, c-Di-GMP, Pel Polysaccharide, and Biofilm Formation in Pseudomonas aeruginosa through Tyrosine Phosphatase TpbA (PA3885). PLoS Pathog. 2009, 5, e1000483. [Google Scholar] [CrossRef] [PubMed]
- Muimhneacháin, E.Ó.; Reen, F.J.; O’Gara, F.; McGlacken, G.P. Analogues of Pseudomonas aeruginosa Signalling Molecules to Tackle Infections. Org. Biomol. Chem. 2018, 16, 169–179. [Google Scholar] [CrossRef]
- Yan, S.; Wu, G. Can Biofilm Be Reversed Through Quorum Sensing in Pseudomonas aeruginosa? Front. Microbiol. 2019, 10, 1582. [Google Scholar] [CrossRef] [Green Version]
- Pesci, E.C.; Pearson, J.P.; Seed, P.C.; Iglewski, B.H. Regulation of Las and Rhl Quorum Sensing in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 3127–3132. [Google Scholar] [CrossRef] [Green Version]
- Turkina, M.V.; Vikström, E. Bacteria-Host Crosstalk: Sensing of the Quorum in the Context of Pseudomonas aeruginosa Infections. J. Innate Immun. 2019, 11, 263–279. [Google Scholar] [CrossRef]
- García-Reyes, S.; Soberón-Chávez, G.; Cocotl-Yanez, M. The Third Quorum-Sensing System of Pseudomonas aeruginosa: Pseudomonas Quinolone Signal and the Enigmatic PqsE Protein. J. Med. Microbiol. 2020, 69, 25–34. [Google Scholar] [CrossRef]
- Lin, J.; Cheng, J.; Wang, Y.; Shen, X. The Pseudomonas Quinolone Signal (PQS): Not Just for Quorum Sensing Anymore. Front. Cell. Infect. Microbiol. 2018, 8, 230. [Google Scholar] [CrossRef] [PubMed]
- Brouwer, S.; Pustelny, C.; Ritter, C.; Klinkert, B.; Narberhaus, F.; Haussler, S. The PqsR and RhlR Transcriptional Regulators Determine the Level of Pseudomonas Quinolone Signal Synthesis in Pseudomonas aeruginosa by Producing Two Different PqsABCDE MRNA Isoforms. J. Bacteriol. 2014, 196, 4163–4171. [Google Scholar] [CrossRef] [Green Version]
- Williams, P.; Cámara, M. Quorum Sensing and Environmental Adaptation in Pseudomonas aeruginosa: A Tale of Regulatory Networks and Multifunctional Signal Molecules. Curr. Opin. Microbiol. 2009, 12, 182–191. [Google Scholar] [CrossRef]
- Rémy, B.; Mion, S.; Plener, L.; Elias, M.; Chabrière, E.; Daudé, D. Interference in Bacterial Quorum Sensing: A Biopharmaceutical Perspective. Front. Pharmacol. 2018, 9, 203. [Google Scholar] [CrossRef]
- Haque, S.; Ahmad, F.; Dar, S.A.; Jawed, A.; Mandal, R.K.; Wahid, M.; Lohani, M.; Khan, S.; Singh, V.; Akhter, N. Developments in Strategies for Quorum Sensing Virulence Factor Inhibition to Combat Bacterial Drug Resistance. Microb. Pathog. 2018, 121, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Geske, G.D.; Wezeman, R.J.; Siegel, A.P.; Blackwell, H.E. Small Molecule Inhibitors of Bacterial Quorum Sensing and Biofilm Formation. J. Am. Chem. Soc. 2005, 127, 12762–12763. [Google Scholar] [CrossRef] [PubMed]
- Ishida, T.; Ikeda, T.; Takiguchi, N.; Kuroda, A.; Ohtake, H.; Kato, J. Inhibition of Quorum Sensing in Pseudomonas aeruginosa by N-Acyl Cyclopentylamides. Appl. Environ. Microbiol. 2007, 73, 3183–3188. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.M.; Bu, Y.; Suga, H. Library Screening for Synthetic Agonists and Antagonists of a Pseudomonas aeruginosa Autoinducer. Chem. Biol. 2003, 10, 563–571. [Google Scholar] [CrossRef] [Green Version]
- Smith, K.M.; Bu, Y.; Suga, H. Induction and Inhibition of Pseudomonas aeruginosa Quorum Sensing by Synthetic Autoinducer Analogs. Chem. Biol. 2003, 10, 81–89. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.D.; Rossi, F.M.; Welsh, M.A.; Nyffeler, K.E.; Blackwell, H.E. A Comparative Analysis of Synthetic Quorum Sensing Modulators in Pseudomonas aeruginosa: New Insights into Mechanism, Active Efflux Susceptibility, Phenotypic Response, and Next-Generation Ligand Design. J. Am. Chem. Soc. 2015, 137, 14626–14639. [Google Scholar] [CrossRef]
- Hodgkinson, J.T.; Galloway, W.R.J.D.; Wright, M.; Mati, I.K.; Nicholson, R.L.; Welch, M.; Spring, D.R. Design, Synthesis and Biological Evaluation of Non-Natural Modulators of Quorum Sensing in Pseudomonas aeruginosa. Org. Biomol. Chem. 2012, 10, 6032. [Google Scholar] [CrossRef]
- Amara, N.; Gregor, R.; Rayo, J.; Dandela, R.; Daniel, E.; Liubin, N.; Willems, H.M.E.; Ben-Zvi, A.; Krom, B.P.; Meijler, M.M. Fine-Tuning Covalent Inhibition of Bacterial Quorum Sensing. ChemBioChem 2016, 17, 825–835. [Google Scholar] [CrossRef]
- Geske, G.D.; O’Neill, J.C.; Miller, D.M.; Mattmann, M.E.; Blackwell, H.E. Modulation of Bacterial Quorum Sensing with Synthetic Ligands: Systematic Evaluation of N-Acylated Homoserine Lactones in Multiple Species and New Insights into Their Mechanisms of Action. J. Am. Chem. Soc. 2007, 129, 13613–13625. [Google Scholar] [CrossRef] [Green Version]
- Amara, N.; Mashiach, R.; Amar, D.; Krief, P.; Spieser, S.A.H.; Bottomley, M.J.; Aharoni, A.; Meijler, M.M. Covalent Inhibition of Bacterial Quorum Sensing. J. Am. Chem. Soc. 2009, 131, 10610–10619. [Google Scholar] [CrossRef]
- Liu, H.; Gong, Q.; Luo, C.; Liang, Y.; Kong, X.; Wu, C.; Feng, P.; Wang, Q.; Zhang, H.; Wireko, M.A. Synthesis and Biological Evaluation of Novel L-Homoserine Lactone Analogs as Quorum Sensing Inhibitors of Pseudomonas aeruginosa. Chem. Pharm. Bull. 2019, 67, 1088–1098. [Google Scholar] [CrossRef]
- McInnis, C.E.; Blackwell, H.E. Thiolactone Modulators of Quorum Sensing Revealed through Library Design and Screening. Bioorg. Med. Chem. 2011, 19, 4820–4828. [Google Scholar] [CrossRef] [Green Version]
- Jiang, K.; Yan, X.; Yu, J.; Xiao, Z.; Wu, H.; Zhao, M.; Yue, Y.; Zhou, X.; Xiao, J.; Lin, F. Design, Synthesis, and Biological Evaluation of 3-Amino-2-Oxazolidinone Derivatives as Potent Quorum-Sensing Inhibitors of Pseudomonas aeruginosa PAO1. Eur. J. Med. Chem. 2020, 194, 112252. [Google Scholar] [CrossRef]
- Hentzer, M.; Riedel, K.; Rasmussen, T.B.; Heydorn, A.; Andersen, J.B.; Parsek, M.R.; Rice, S.A.; Eberl, L.; Molin, S.; Høiby, N.; et al. Inhibition of Quorum Sensing in Pseudomonas aeruginosa Biofilm Bacteria by a Halogenated Furanone Compound. Microbiology 2002, 148, 87–102. [Google Scholar] [CrossRef] [Green Version]
- Wu, H. Synthetic Furanones Inhibit Quorum-Sensing and Enhance Bacterial Clearance in Pseudomonas aeruginosa Lung Infection in Mice. J. Antimicrob. Chemother. 2004, 53, 1054–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.; Wang, P.-C.; Ma, H.-M.; Chen, S.-Y.; Fu, Y.-H.; Liu, Y.-Y.; Wang, X.; Yu, G.-C.; Huang, T.; Hibbs, D.E.; et al. Design, Synthesis and Evaluation of Halogenated Furanone Derivatives as Quorum Sensing Inhibitors in Pseudomonas aeruginosa. Eur. J. Pharm. Sci. 2019, 140, 105058. [Google Scholar] [CrossRef]
- Muh, U.; Schuster, M.; Heim, R.; Singh, A.; Olson, E.R.; Greenberg, E.P. Novel Pseudomonas aeruginosa Quorum-Sensing Inhibitors Identified in an Ultra-High-Throughput Screen. Antimicrob. Agents Chemother. 2006, 50, 3674–3679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muh, U.; Hare, B.J.; Duerkop, B.A.; Schuster, M.; Hanzelka, B.L.; Heim, R.; Olson, E.R.; Greenberg, E.P. A Structurally Unrelated Mimic of a Pseudomonas aeruginosa Acyl-Homoserine Lactone Quorum-Sensing Signal. Proc. Natl. Acad. Sci. USA 2006, 103, 16948–16952. [Google Scholar] [CrossRef] [Green Version]
- Shah, M.D.; Kharkar, P.S.; Sahu, N.U.; Peerzada, Z.; Desai, K.B. Potassium 2-Methoxy-4-Vinylphenolate: A Novel Hit Exhibiting Quorum-Sensing Inhibition in Pseudomonas aeruginosa via LasIR/RhlIR Circuitry. RSC Adv. 2019, 9, 40228–40239. [Google Scholar] [CrossRef] [Green Version]
- Srinivasarao, S.; Nandikolla, A.; Nizalapur, S.; Yu, T.T.; Pulya, S.; Ghosh, B.; Murugesan, S.; Kumar, N.; Sekhar, K.V.G.C. Design, Synthesis and Biological Evaluation of 1,2,3-Triazole Based 2-Aminobenzimidazoles as Novel Inhibitors of LasR Dependent Quorum Sensing in Pseudomonas aeruginosa. RSC Adv. 2019, 9, 29273–29292. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Moustafa, D.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The RhlR Quorum-Sensing Receptor Controls Pseudomonas aeruginosa Pathogenesis and Biofilm Development Independently of Its Canonical Homoserine Lactone Autoinducer. PLOS Pathog. 2017, 13, e1006504. [Google Scholar] [CrossRef]
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A Quorum-Sensing Inhibitor Blocks Pseudomonas aeruginosa Virulence and Biofilm Formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986. [Google Scholar] [CrossRef] [Green Version]
- Eibergen, N.R.; Moore, J.D.; Mattmann, M.E.; Blackwell, H.E. Potent and Selective Modulation of the RhlR Quorum Sensing Receptor by Using Non-Native Ligands: An Emerging Target for Virulence Control in Pseudomonas aeruginosa. ChemBioChem 2015, 16, 2348–2356. [Google Scholar] [CrossRef] [Green Version]
- Caiazza, N.C.; Shanks, R.M.Q.; O’Toole, G.A. Rhamnolipids Modulate Swarming Motility Patterns of Pseudomonas aeruginosa. J. Bacteriol. 2005, 187, 7351–7361. [Google Scholar] [CrossRef] [Green Version]
- Boles, B.R.; Thoendel, M.; Singh, P.K. Rhamnolipids Mediate Detachment of Pseudomonas aeruginosa from Biofilms: Rhamnolipid-Mediated Biofilm Detachment. Mol. Microbiol. 2005, 57, 1210–1223. [Google Scholar] [CrossRef]
- Zulianello, L.; Canard, C.; Köhler, T.; Caille, D.; Lacroix, J.-S.; Meda, P. Rhamnolipids Are Virulence Factors That Promote Early Infiltration of Primary Human Airway Epithelia by Pseudomonas aeruginosa. Infect. Immun. 2006, 74, 3134–3147. [Google Scholar] [CrossRef] [Green Version]
- Qiu, M.-N.; Wang, F.; Chen, S.-Y.; Wang, P.-C.; Fu, Y.-H.; Liu, Y.-Y.; Wang, X.; Wang, F.-B.; Wang, C.; Yang, H.-W.; et al. Novel 2,8-Bit Derivatives of Quinolines Attenuate Pseudomonas aeruginosa Virulence and Biofilm Formation. Bioorg. Med. Chem. Lett. 2019, 29, 749–754. [Google Scholar] [CrossRef]
- Lu, C.; Kirsch, B.; Zimmer, C.; de Jong, J.C.; Henn, C.; Maurer, C.K.; Müsken, M.; Häussler, S.; Steinbach, A.; Hartmann, R.W. Discovery of Antagonists of PqsR, a Key Player in 2-Alkyl-4-Quinolone-Dependent Quorum Sensing in Pseudomonas aeruginosa. Chem. Biol. 2012, 19, 381–390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamal, A.A.M.; Petrera, L.; Eberhard, J.; Hartmann, R.W. Structure–Functionality Relationship and Pharmacological Profiles of Pseudomonas aeruginosa Alkylquinolone Quorum Sensing Modulators. Org. Biomol. Chem. 2017, 15, 4620–4630. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Maurer, C.K.; Kirsch, B.; Steinbach, A.; Hartmann, R.W. Overcoming the Unexpected Functional Inversion of a PqsR Antagonist in Pseudomonas aeruginosa: An In Vivo Potent Antivirulence Agent Targeting Pqs Quorum Sensing. Angew. Chem. Int. Ed. 2014, 53, 1109–1112. [Google Scholar] [CrossRef]
- Ilangovan, A.; Fletcher, M.; Rampioni, G.; Pustelny, C.; Rumbaugh, K.; Heeb, S.; Cámara, M.; Truman, A.; Chhabra, S.R.; Emsley, J.; et al. Structural Basis for Native Agonist and Synthetic Inhibitor Recognition by the Pseudomonas aeruginosa Quorum Sensing Regulator PqsR (MvfR). PLoS Pathog. 2013, 9, e1003508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grossman, S.; Soukarieh, F.; Richardson, W.; Liu, R.; Mashabi, A.; Emsley, J.; Williams, P.; Cámara, M.; Stocks, M.J. Novel Quinazolinone Inhibitors of the Pseudomonas aeruginosa Quorum Sensing Transcriptional Regulator PqsR. Eur. J. Med. Chem. 2020, 208, 112778. [Google Scholar] [CrossRef] [PubMed]
- Soukarieh, F.; Mashabi, A.; Richardson, W.; Oton, E.V.; Romero, M.; Roberston, S.N.; Grossman, S.; Sou, T.; Liu, R.; Halliday, N.; et al. Design and Evaluation of New Quinazolin-4(3H)-One Derived PqsR Antagonists as Quorum Sensing Quenchers in Pseudomonas aeruginosa. ACS Infect. Dis. 2021, 7, 2666–2685. [Google Scholar] [CrossRef] [PubMed]
- Aleksić, I.; Šegan, S.; Andrić, F.; Zlatović, M.; Moric, I.; Opsenica, D.M.; Senerovic, L. Long-Chain 4-Aminoquinolines as Quorum Sensing Inhibitors in Serratia marcescens and Pseudomonas aeruginosa. ACS Chem. Biol. 2017, 12, 1425–1434. [Google Scholar] [CrossRef] [Green Version]
- Aleksic, I.; Jeremic, J.; Milivojevic, D.; Ilic-Tomic, T.; Šegan, S.; Zlatović, M.; Opsenica, D.M.; Senerovic, L. N-Benzyl Derivatives of Long-Chained 4-Amino-7-Chloro-Quinolines as Inhibitors of Pyocyanin Production in Pseudomonas aeruginosa. ACS Chem. Biol. 2019, 14, 2800–2809. [Google Scholar] [CrossRef]
- Soukarieh, F.; Oton, E.V.; Dubern, J.-F.; Gomes, J.; Halliday, N.; de Pilar Crespo, M.; Ramírez-Prada, J.; Insuasty, B.; Abonia, R.; Quiroga, J.; et al. In Silico and in Vitro-Guided Identification of Inhibitors of Alkylquinolone-Dependent Quorum Sensing in Pseudomonas aeruginosa. Molecules 2018, 23, 257. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.S.A.; Empting, M.; Hamed, M.; Hartmann, R.W. Preparation of Five- and Six-Membered Nitrogen Heterocycles as PQSR Inverse Agonists for the Treatment and Prevention of Bacterial Infections. WO 2020/007938 A1, 3 July 2019. [Google Scholar]
- Zender, M.; Witzgall, F.; Kiefer, A.; Kirsch, B.; Maurer, C.K.; Kany, A.M.; Xu, N.; Schmelz, S.; Börger, C.; Blankenfeldt, W.; et al. Flexible Fragment Growing Boosts Potency of Quorum-Sensing Inhibitors against Pseudomonas aeruginosa Virulence. ChemMedChem 2020, 15, 188–194. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.-B.; Liu, J.; Huang, Z.-X.; Yu, J.-H.; Xu, X.-F.; Sun, P.-H.; Lin, J.; Chen, W.-M. Design, Synthesis and Biological Evaluation of 2-Substituted 3-Hydroxy-6-Methyl-4H-Pyran-4-One Derivatives as Pseudomonas aeruginosa Biofilm Inhibitors. Eur. J. Med. Chem. 2018, 158, 753–766. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Hou, J.-S.; Li, Y.-B.; Miao, Z.-Y.; Sun, P.-H.; Lin, J.; Chen, W.-M. Novel 2-Substituted 3-Hydroxy-1,6-Dimethylpyridin-4(1H)-Ones as Dual-Acting Biofilm Inhibitors of Pseudomonas aeruginosa. J. Med. Chem. 2020, 63, 10921–10945. [Google Scholar] [CrossRef]
- Starkey, M.; Lepine, F.; Maura, D.; Bandyopadhaya, A.; Lesic, B.; He, J.; Kitao, T.; Righi, V.; Milot, S.; Tzika, A.; et al. Identification of Anti-Virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity. PLoS Pathog. 2014, 10, e1004321. [Google Scholar] [CrossRef]
- Soukarieh, F.; Liu, R.; Romero, M.; Roberston, S.N.; Richardson, W.; Lucanto, S.; Oton, E.V.; Qudus, N.R.; Mashabi, A.; Grossman, S.; et al. Hit Identification of New Potent PqsR Antagonists as Inhibitors of Quorum Sensing in Planktonic and Biofilm Grown Pseudomonas aeruginosa. Front. Chem. 2020, 8, 204. [Google Scholar] [CrossRef] [PubMed]
- Zahler, R. Aryloxyacetylindoles and Analogs as Antibiotic Tolerance Inhibitors. WO 2016/112088 Al, 14 July 2016. [Google Scholar]
- Zender, M.; Klein, T.; Henn, C.; Kirsch, B.; Maurer, C.K.; Kail, D.; Ritter, C.; Dolezal, O.; Steinbach, A.; Hartmann, R.W. Discovery and Biophysical Characterization of 2-Amino-Oxadiazoles as Novel Antagonists of PqsR, an Important Regulator of Pseudomonas aeruginosa Virulence. J. Med. Chem. 2013, 56, 6761–6774. [Google Scholar] [CrossRef]
- Klein, T.; Henn, C.; de Jong, J.C.; Zimmer, C.; Kirsch, B.; Maurer, C.K.; Pistorius, D.; Müller, R.; Steinbach, A.; Hartmann, R.W. Identification of Small-Molecule Antagonists of the Pseudomonas aeruginosa Transcriptional Regulator PqsR: Biophysically Guided Hit Discovery and Optimization. ACS Chem. Biol. 2012, 7, 1496–1501. [Google Scholar] [CrossRef] [PubMed]
- Maura, D.; Rahme, L.G. Pharmacological Inhibition of the Pseudomonas aeruginosa MvfR Quorum-Sensing System Interferes with Biofilm Formation and Potentiates Antibiotic-Mediated Biofilm Disruption. Antimicrob. Agents Chemother. 2017, 61, e01362-17. [Google Scholar] [CrossRef] [Green Version]
- Coleman, J.P.; Hudson, L.L.; McKnight, S.L.; Farrow, J.M.; Calfee, M.W.; Lindsey, C.A.; Pesci, E.C. Pseudomonas aeruginosa PqsA Is an Anthranilate-Coenzyme A Ligase. J. Bacteriol. 2008, 190, 1247–1255. [Google Scholar] [CrossRef] [Green Version]
- Lesic, B.; Lépine, F.; Déziel, E.; Zhang, J.; Zhang, Q.; Padfield, K.; Castonguay, M.-H.; Milot, S.; Stachel, S.; Tzika, A.A.; et al. Inhibitors of Pathogen Intercellular Signals as Selective Anti-Infective Compounds. PLoS Pathog. 2007, 3, e126. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Sharma, I.; Pratihar, D.; Hudson, L.L.; Maura, D.; Guney, T.; Rahme, L.G.; Pesci, E.C.; Coleman, J.P.; Tan, D.S. Designed Small-Molecule Inhibitors of the Anthranilyl-CoA Synthetase PqsA Block Quinolone Biosynthesis in Pseudomonas aeruginosa. ACS Chem. Biol. 2016, 11, 3061–3067. [Google Scholar] [CrossRef]
- Storz, M.P.; Maurer, C.K.; Zimmer, C.; Wagner, N.; Brengel, C.; de Jong, J.C.; Lucas, S.; Müsken, M.; Häussler, S.; Steinbach, A.; et al. Validation of PqsD as an Anti-Biofilm Target in Pseudomonas aeruginosa by Development of Small-Molecule Inhibitors. J. Am. Chem. Soc. 2012, 134, 16143–16146. [Google Scholar] [CrossRef]
- Storz, M.P.; Allegretta, G.; Kirsch, B.; Empting, M.; Hartmann, R.W. From in Vitro to in Cellulo: Structure–Activity Relationship of (2-Nitrophenyl)Methanol Derivatives as Inhibitors of PqsD in Pseudomonas aeruginosa. Org. Biomol. Chem. 2014, 12, 6094–6104. [Google Scholar] [CrossRef] [Green Version]
- Allegretta, G.; Weidel, E.; Empting, M.; Hartmann, R.W. Catechol-Based Substrates of Chalcone Synthase as a Scaffold for Novel Inhibitors of PqsD. Eur. J. Med. Chem. 2015, 90, 351–359. [Google Scholar] [CrossRef]
- Allegretta, G.; Maurer, C.K.; Eberhard, J.; Maura, D.; Hartmann, R.W.; Rahme, L.; Empting, M. In-Depth Profiling of MvfR-Regulated Small Molecules in Pseudomonas aeruginosa after Quorum Sensing Inhibitor Treatment. Front. Microbiol. 2017, 8, 924. [Google Scholar] [CrossRef]
- Maura, D.; Drees, S.L.; Bandyopadhaya, A.; Kitao, T.; Negri, M.; Starkey, M.; Lesic, B.; Milot, S.; Déziel, E.; Zahler, R.; et al. Polypharmacology Approaches against the Pseudomonas aeruginosa MvfR Regulon and Their Application in Blocking Virulence and Antibiotic Tolerance. ACS Chem. Biol. 2017, 12, 1435–1443. [Google Scholar] [CrossRef]
- Thomann, A.; de Mello Martins, A.G.G.; Brengel, C.; Empting, M.; Hartmann, R.W. Application of Dual Inhibition Concept within Looped Autoregulatory Systems toward Antivirulence Agents against Pseudomonas aeruginosa Infections. ACS Chem. Biol. 2016, 11, 1279–1286. [Google Scholar] [CrossRef]
- Thomann, A.; Brengel, C.; Börger, C.; Kail, D.; Steinbach, A.; Empting, M.; Hartmann, R.W. Structure-Activity Relationships of 2-Sufonylpyrimidines as Quorum-Sensing Inhibitors to Tackle Biofilm Formation and EDNA Release of Pseudomonas aeruginosa. ChemMedChem 2016, 11, 2522–2533. [Google Scholar] [CrossRef]

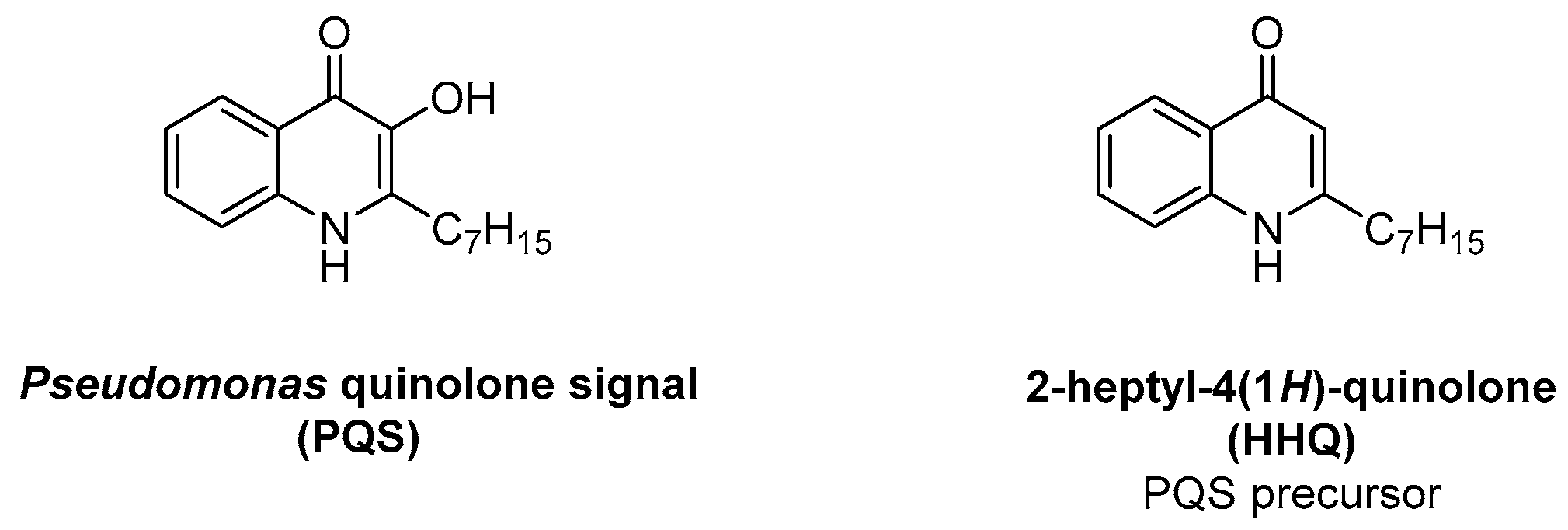

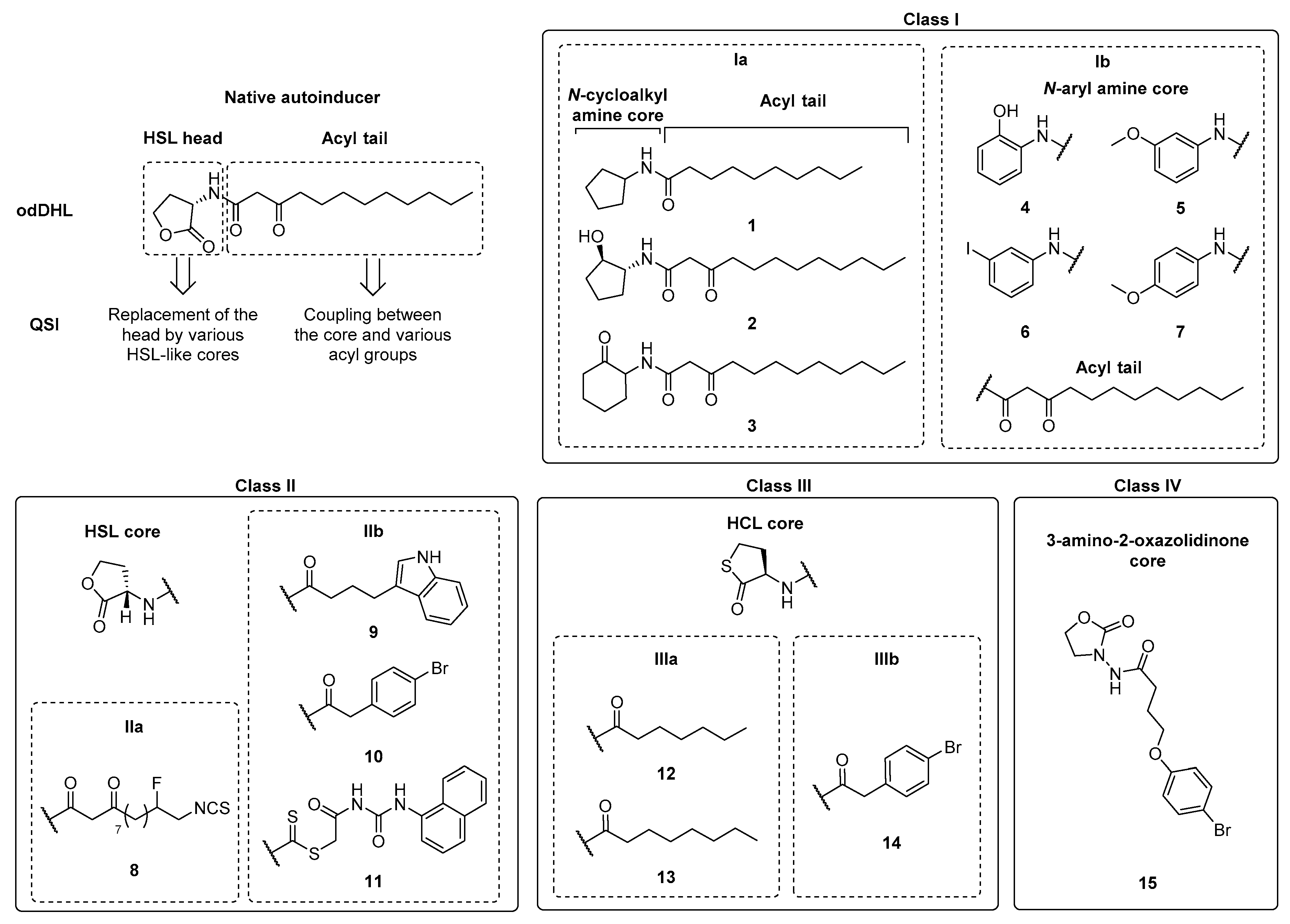
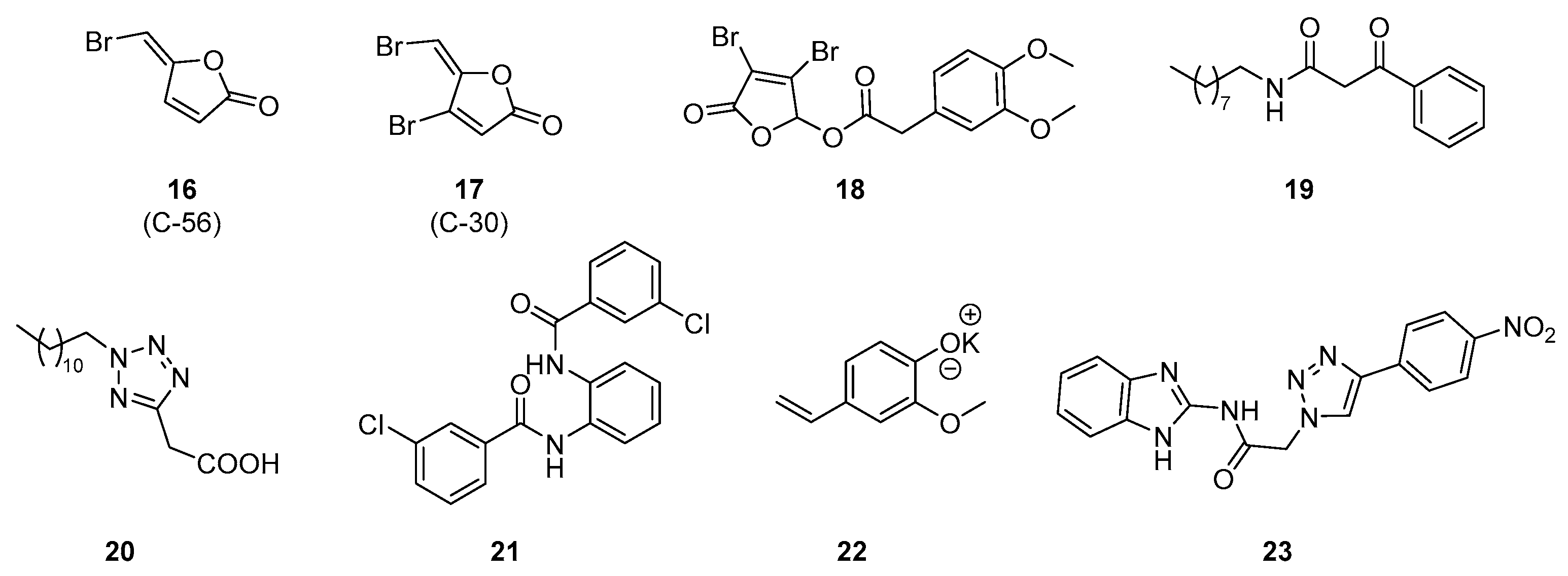
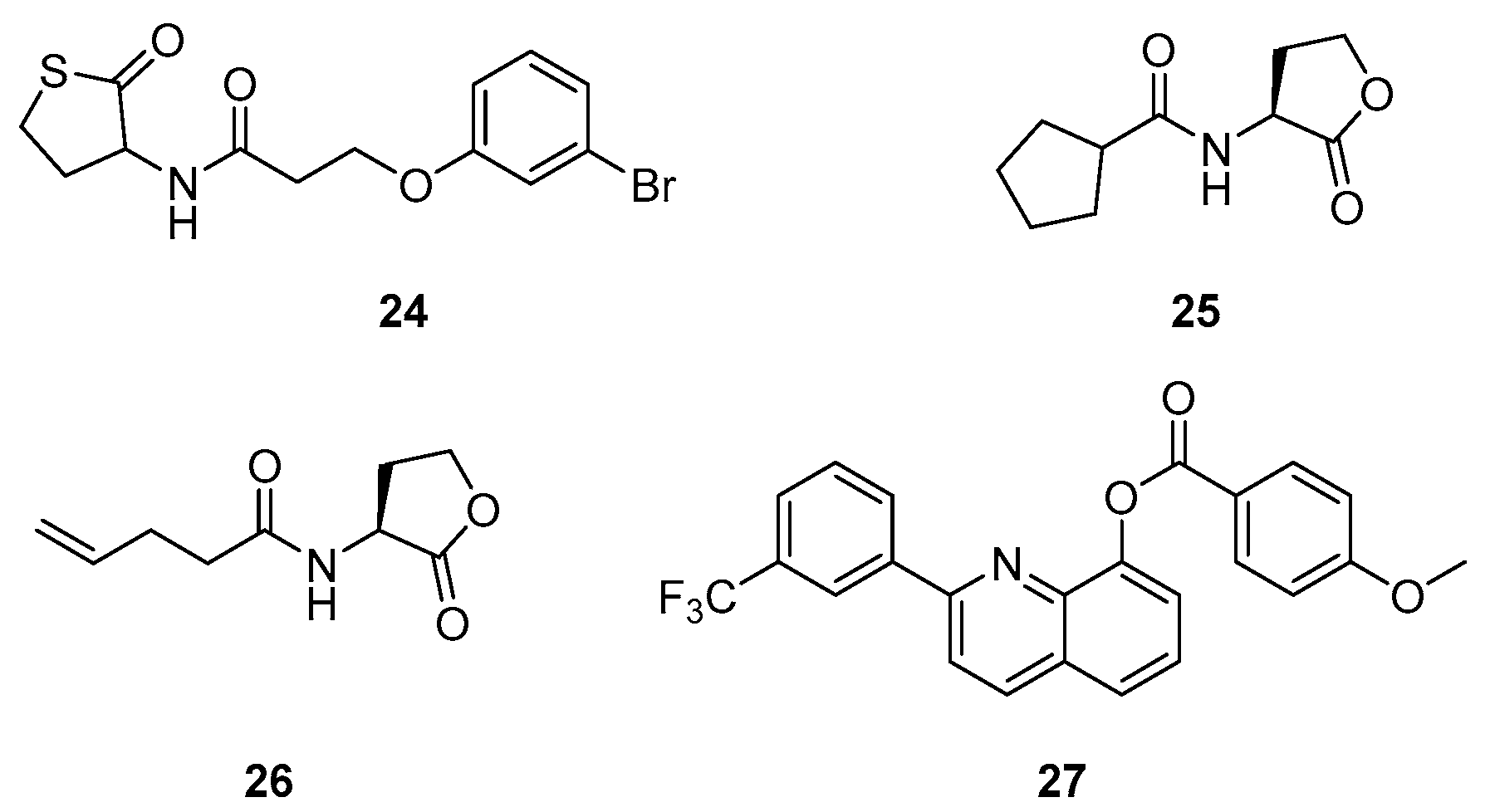
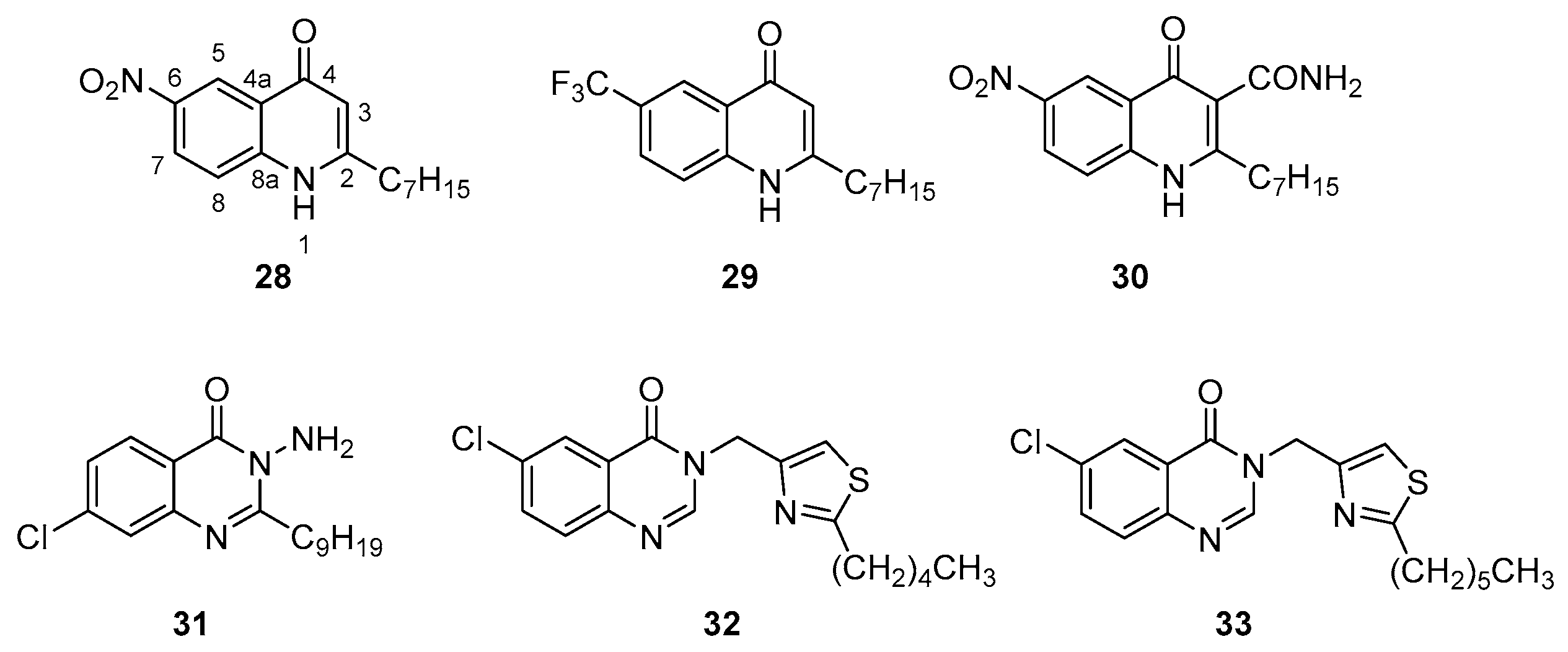
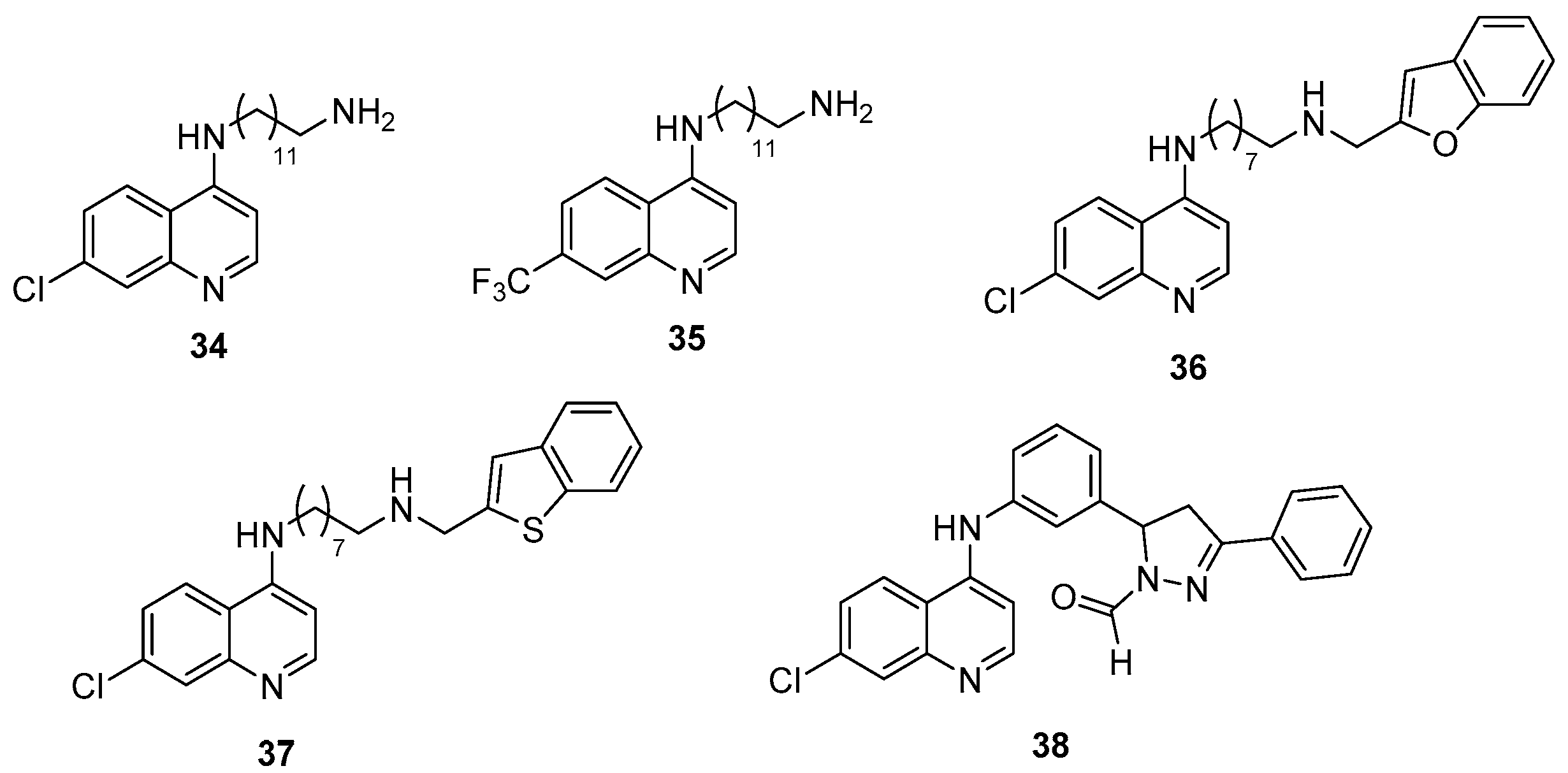
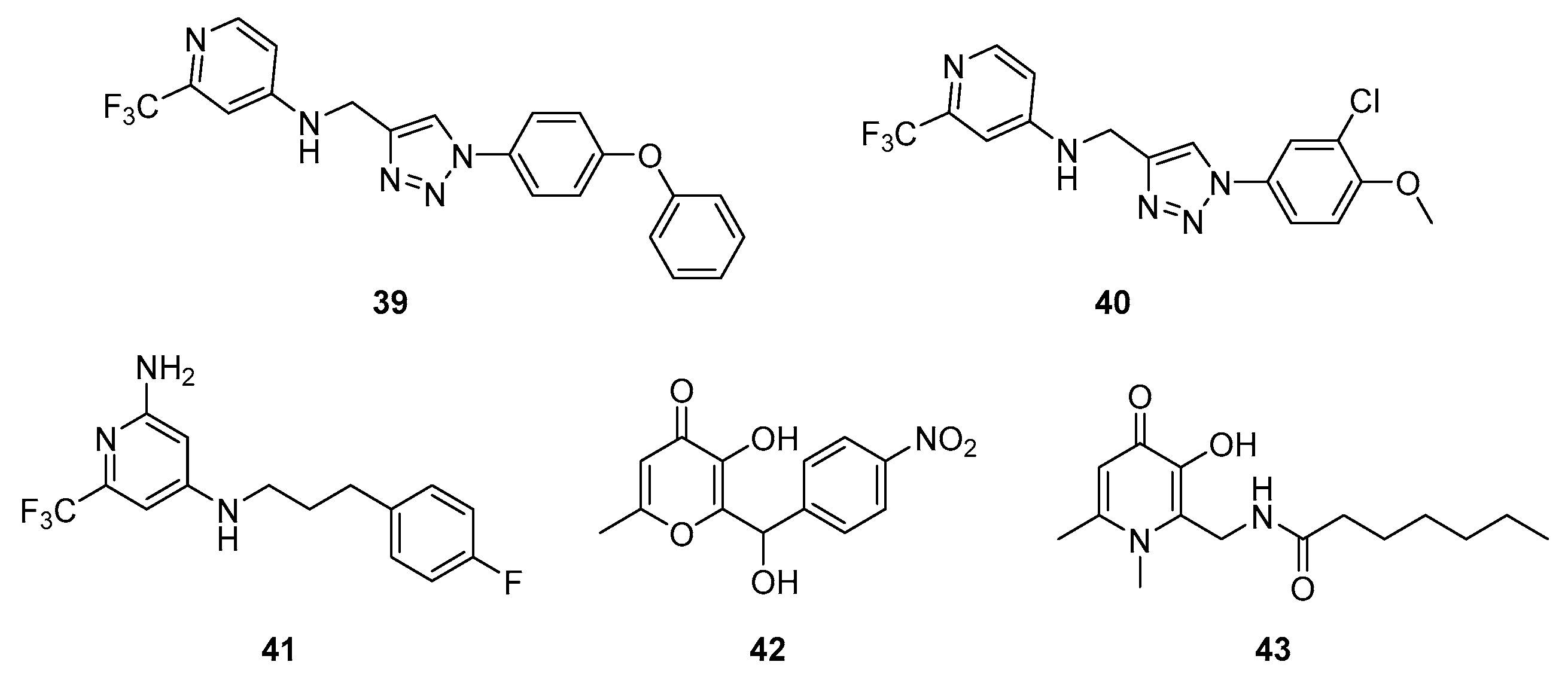

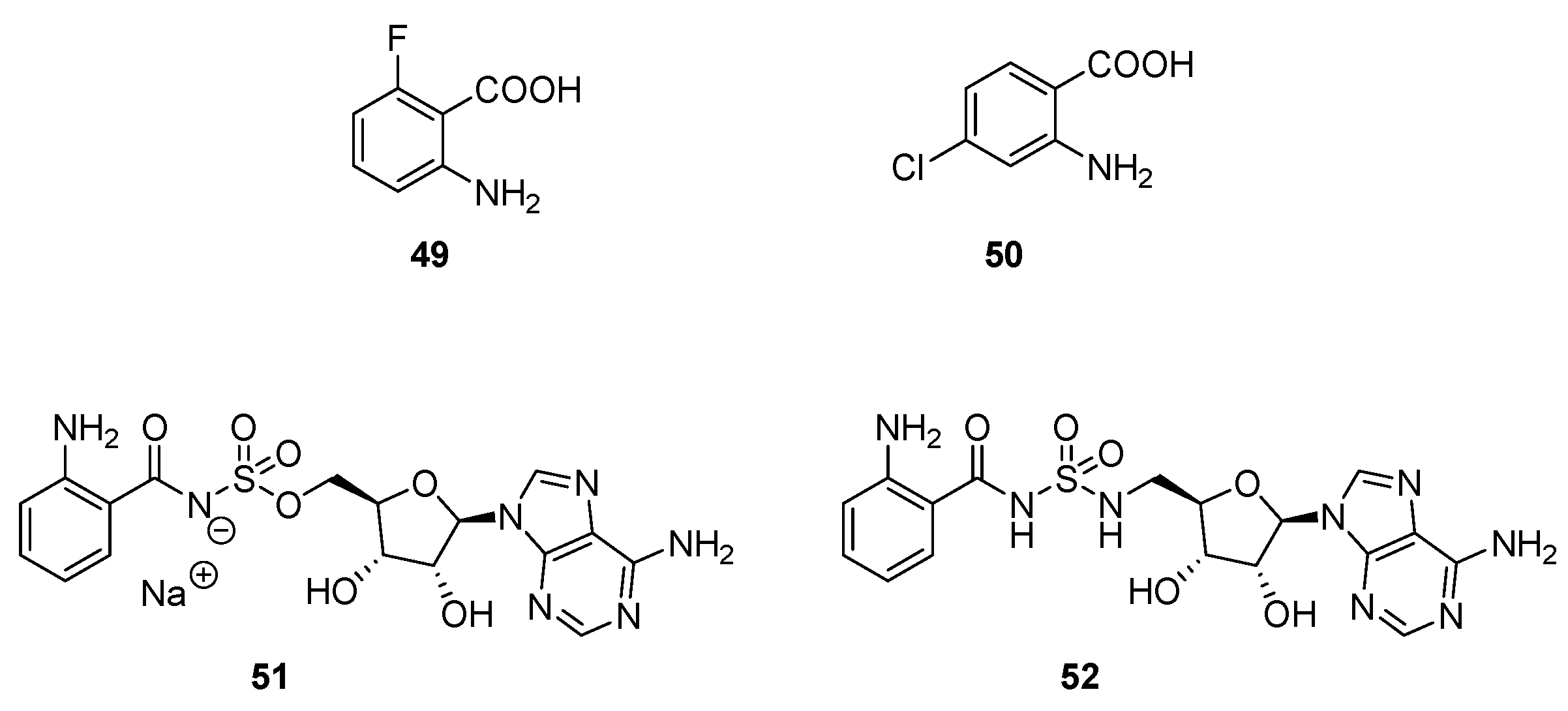
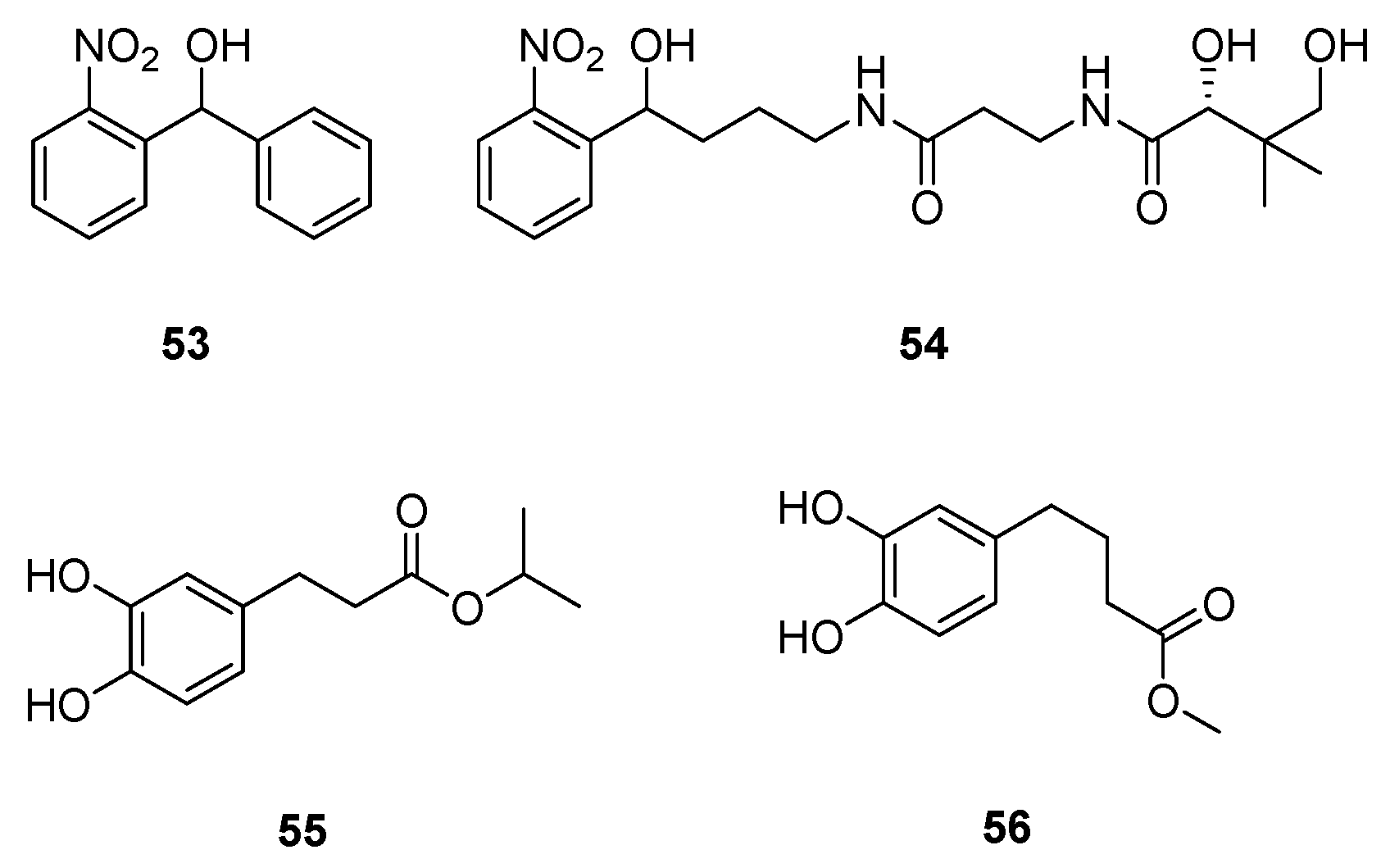
| Class | QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | ||||
|---|---|---|---|---|---|---|---|---|
| Target | % Inhibition (IC50) | Anti-Biofilm Activity | Anti-Elastase Activity | Anti-Rhamnolipid Activity | Anti-Pyocyanin Activity | |||
| % Inhibition | % Inhibition | % Inhibition | % Inhibition | |||||
| Ia | 1 | LasR | 80 µM (a) | NA at 250 µM (f) | 77% at 250 µM (m) | 87% at 250 µM (m) | 64% at 250 µM (m) | [81] |
| RhlR | 90 µM (a) | |||||||
| 2 | LasR | ≈70% at 100 µM (b) | ND | NA | ND | 50% at 25–100µM (k) | [83] | |
| RhlR | NA at 50 µM (c) | |||||||
| 3 | LasR | 35% at 100 µM (b) | ≈100% at 50 µM (g) | 60% at 50 μM (j) | ND | ≈100% at 100 µM (k) | ||
| RhlR | 60% at 50 µM (c) | |||||||
| Ib | 4 | LasR | 60% at 10 µM and 90% at 100 μM (b) | Weak inhibition at 50 µM (g) | 50% at 10 µM (j) | ND | NA at 100 µM (k) | [82] |
| RhlR | 80% at 50 μM (c) | |||||||
| 5 | ND | ND | ND | 63% at 200 µM (m) | ND | 93% at 200 µM (m) | [85] | |
| 6 | ND | ND | ND | ND | ND | 67% at 200 µM (m) | ||
| 7 | ND | ND | ND | 34% at 200 µM (m) | ND | 73% at 200 µM (m) | ||
| IIa | 8 | LasR | 154 μM (d) | ND | ND | ND | 34% à 100 µM (l) | [86] |
| IIb | 9 | LasR | 12.5 µM (b) | Strong inhibition at 50 µM (h) | 66% at 200 μM (m) | ND | ND | [80,87,88] |
| 10 | LasR | ≈25 µM (b) | Strong inhibition at 50 µM (h) | 77% at 200 µM (m) | ND | ≈57% à 50 µM (k) | ||
| 11 | LasR | ND | 36.2% at 15 µM (i) | 13.7% at 15 µM (m) | 28.1% at 15 µM (m) | 34.5% at 15 µM (m) | [89] | |
| IIIa | 12 | LasR | 0.79 µM (e) | ND | ND | ND | ND | [90] |
| 13 | LasR | 0.14 µM (e) | ND | ND | ND | ND | ||
| IIIb | 14 | LasR | 0.40 µM (e) | ND | ND | ND | ND | |
| IV | 15 | ND | ND | 40% at 162.5 µM (i) | 16% at 162.5 µM (m) | 57% at 162.5 µM (m) | 22% at 162.5 µM (m) | [91] |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | |||
|---|---|---|---|---|---|---|
| Target | % Inhibition (IC50) | Anti-Biofilm Activity | Anti-Elastase Activity | Anti-Pyocyanin Activity | ||
| % Inhibition | % Inhibition | % Inhibition | ||||
| 16 (C56) | LasR | >50% at 11 µM (a) | 60% at 0.5 µM (g) | Active at 0.5 µM (j) | ND | [92,93] |
| 17 (C30) | ND | ND | ND | ND | ND | [93] |
| 18 | LasR | Active above 40 µM (b) | 74% at 256 µg/mL (h) | ND | 10% at 64 µg/mL (k) | [94] |
| 19 | LasR | 10 µM (c) | ND | 60% at 100 µM (l) | 90% at 100 µM (l) | [95] |
| 20 | LasR | 30 nM (c) | ND | 20% at 10 µM (l) | 40% at 10 µM (l) | |
| 21 | LasR | 50 µM (d) | ND | ND | ND | [96] |
| 22 | LasR | 70% at 4 mg/mL (e) | 76% at 4 mg/mL (i) | 75% (LasA) and 65% (LasB) at 4 mg/mL (l) | 68% at 4 mg/mL (l) | [97] |
| 23 | LasR | 64% at 62.5 µM (f) | ND | ND | ND | [98] |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | |||
|---|---|---|---|---|---|---|
| Target | % Inhibition/Activation (IC50/EC50) | Anti-Biofilm Activity | Anti-Rhamnolipid Activity | Anti-Pyocyanin Activity | ||
| % Inhibition | % Inhibition | IC50 | ||||
| 24 | RhlR (agonist) | 43% at 1 mM (a) | ≈40% at 100 µM (e) | ND | 8 µM (g) | [100] |
| 80% at 20 µM (b) | ||||||
| 25 | RhlR (agonist) | 4.5 µM (c) | ND | ND | ND | [101] |
| 26 | RhlR (agonist) | 7.2 µM (c) | ||||
| 27 | RhlR (antagonist) | 20% at 10 µM (d) | 15% at 10 µM (f) | ≈40% at 10 µM (h) | NA (h) | [105] |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | |||
|---|---|---|---|---|---|---|
| IC50 | Anti-Biofilm Activity | Anti-Elastase Activity | Anti-Rhamnolipid Activity | Anti-Pyocyanin Activity | ||
| % Inhibition (IC50) | % Inhibition (IC50) | % Inhibition (IC50) | % Inhibition (IC50) | |||
| 28 | 51 nM (a) | ND | ND | ND | ND | [106] |
| 29 | 54 nM (a) | ND | NA at 5 µM (m) | NA at 5 µM (m) | 74% at 3 mM (m) | |
| 30 | 51 nM (a) | ND | ND | ND | 1.9 µM (m) | [107] |
| 1.322 µM (b) | ||||||
| 31 | 5 µM (c) | ND | ND | ND | 50 µM (n) | [109] |
| 32 | 313 nM (d) | ND | ND | ND | 23% à 939 nM (o) | [110] |
| 342 nM (e) | ||||||
| 33 | 289 nM (d) | ND | ND | ND | 36% at 867 nM (o) | |
| 265 nM (e) | ||||||
| 34 | ND | 40% at 140 µM (h) | ND | ND | 140 µM (m) | [112] |
| 35 | ND | 50% at 125 µM (h) | ND | ND | 2.5 µM (m) | |
| 36 | 25 µM (f) | 50 µM (i) | 35% at 50 µM (m or n) | ND | 12 µM (m) | [113] |
| 37 | 30 µM (f) | 20% at 100 µM (i) | 30% at 50 µM (m or n) | ND | 25 µM (m) | |
| 38 | 2.3 µM (e) | 8 µM (j) | ND | ND | 65% at 8 µM (m) | [114] |
| 12.4 µM (d) | 34 µM (k) | 80% at 34 µM (o) | ||||
| 39 | ≥50 nM (a) | ≥500 nM (l) | ND | ND | ≥250 nM (m) | [115] |
| 40 | ≥50 nM (a) | ≥500 nM (l) | ND | ND | Between 251 and 1000 nM (m) | |
| 41 | 0.14 µM (a) | ND | ND | ND | 5.9 µM (m) | [116] |
| 42 | Active from 2.5 to 40 µM (g) | 20 µM (i) | NA (n) | NA (n) | 60% at 40 µM (n) | [117] |
| 43 | Active from 1.25 to 20 µM (g) | 6.57 μM (i) | Active from 1.25 to 20 µM (n) | Active from 1.25 to 20 µM (n) | Active from 1.25 to 20 µM (m) | [118] |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | ||
|---|---|---|---|---|---|
| Target | IC50 | Anti-Biofilm Activity | Anti-Pyocyanin Activity | ||
| IC50 | % Inhibition (IC50) | ||||
| 44 (M64) | PqsR | 0.32 µM (a) | 1 µM (c) | 300 nM (d) | [119,120,124] |
| 1.22 µM (b) | |||||
| 45 | PqsR | 0.25 µM (a) | ND | 80% at 1 µM (e) | [120] |
| 0.34 µM (b) | |||||
| 46 | ND | ND | ND | 0.05–0.25 µΜ (d) | [121] |
| 47 | PqsR | 12.5 µM (f) 23.6 µM (g) | ND | 87.2 µM (d) | [122,123] |
| 48 | PqsR | 7.5 µM (f) 38.5 µM (g) | ND | 46% at 250 μM (d) | [122] |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | |
|---|---|---|---|---|
| Anti-HHQ Activity | Anti-PQS Activity | Anti-Pyocyanin Activity | ||
| % Inhibition | % Inhibition | % Inhibition | ||
| 49 | ≈100% at 1.5 mM (a) | ≈100% at 1.5 mM (a) | ≈100% at 1.5 mM (b) | [126] |
| 50 | ≈100% at 1.5 mM (a) | ≈100% at 1.5 mM (a) | ≈100% at 1.5 mM (b) | |
| 51 | 67% at 1.5 mM (a) | 77% at 1.5 mM (a) | NA at 1 mM (b) | [127] |
| 52 | 90% at 1.5 mM (a) | 92% at 1.5 mM (a) | NA at 1 mM (b) | |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. |
|---|---|---|---|
| IC50 | Anti-Biofilm Activity | ||
| % Inhibition | |||
| 53 | 3.2 µM (a) | 38% at 500 µM (b) | [128,129] |
| 54 | 7.9 µM (a) | ND | |
| 55 | 8.6 µM (a) | ND | [130] |
| 56 | 7.9 µM (a) | ND |
| QSIs | Anti-QS Properties | Anti-Virulence Properties | Ref. | ||
|---|---|---|---|---|---|
| Target | IC50 | Anti-Biofilm Activity | Anti-Pyocyanin Activity | ||
| IC50 | IC50 | ||||
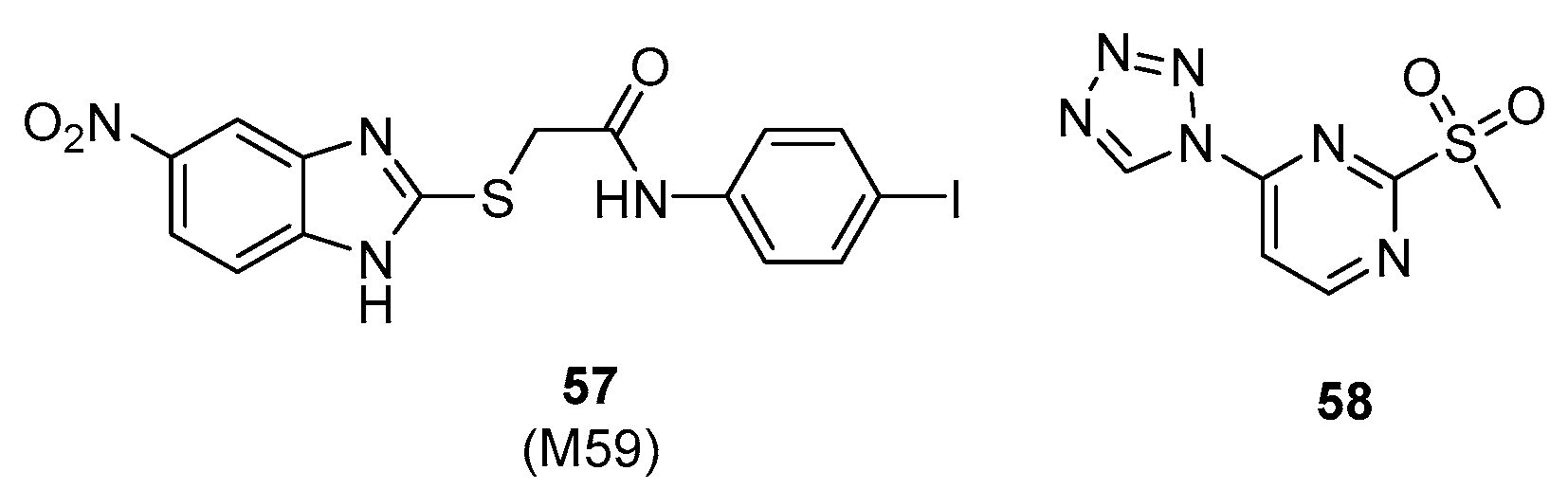
| 57 (M59) | PqsBC | 13.4 µM (a) | ND | ND | [132] |
| PqsR | ND | ||||
| 58 | PqsR | 15 µM (b) | 100 µM (d) | 86 µM (c) | [133,134] |
| PqsD | 21 µM (c) | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duplantier, M.; Lohou, E.; Sonnet, P. Quorum Sensing Inhibitors to Quench P. aeruginosa Pathogenicity. Pharmaceuticals 2021, 14, 1262. https://doi.org/10.3390/ph14121262
Duplantier M, Lohou E, Sonnet P. Quorum Sensing Inhibitors to Quench P. aeruginosa Pathogenicity. Pharmaceuticals. 2021; 14(12):1262. https://doi.org/10.3390/ph14121262
Chicago/Turabian StyleDuplantier, Marine, Elodie Lohou, and Pascal Sonnet. 2021. "Quorum Sensing Inhibitors to Quench P. aeruginosa Pathogenicity" Pharmaceuticals 14, no. 12: 1262. https://doi.org/10.3390/ph14121262
APA StyleDuplantier, M., Lohou, E., & Sonnet, P. (2021). Quorum Sensing Inhibitors to Quench P. aeruginosa Pathogenicity. Pharmaceuticals, 14(12), 1262. https://doi.org/10.3390/ph14121262








