Bioactive Diterpenes, Norditerpenes, and Sesquiterpenes from a Formosan Soft Coral Cespitularia sp.
Abstract
:1. Introduction
2. Results
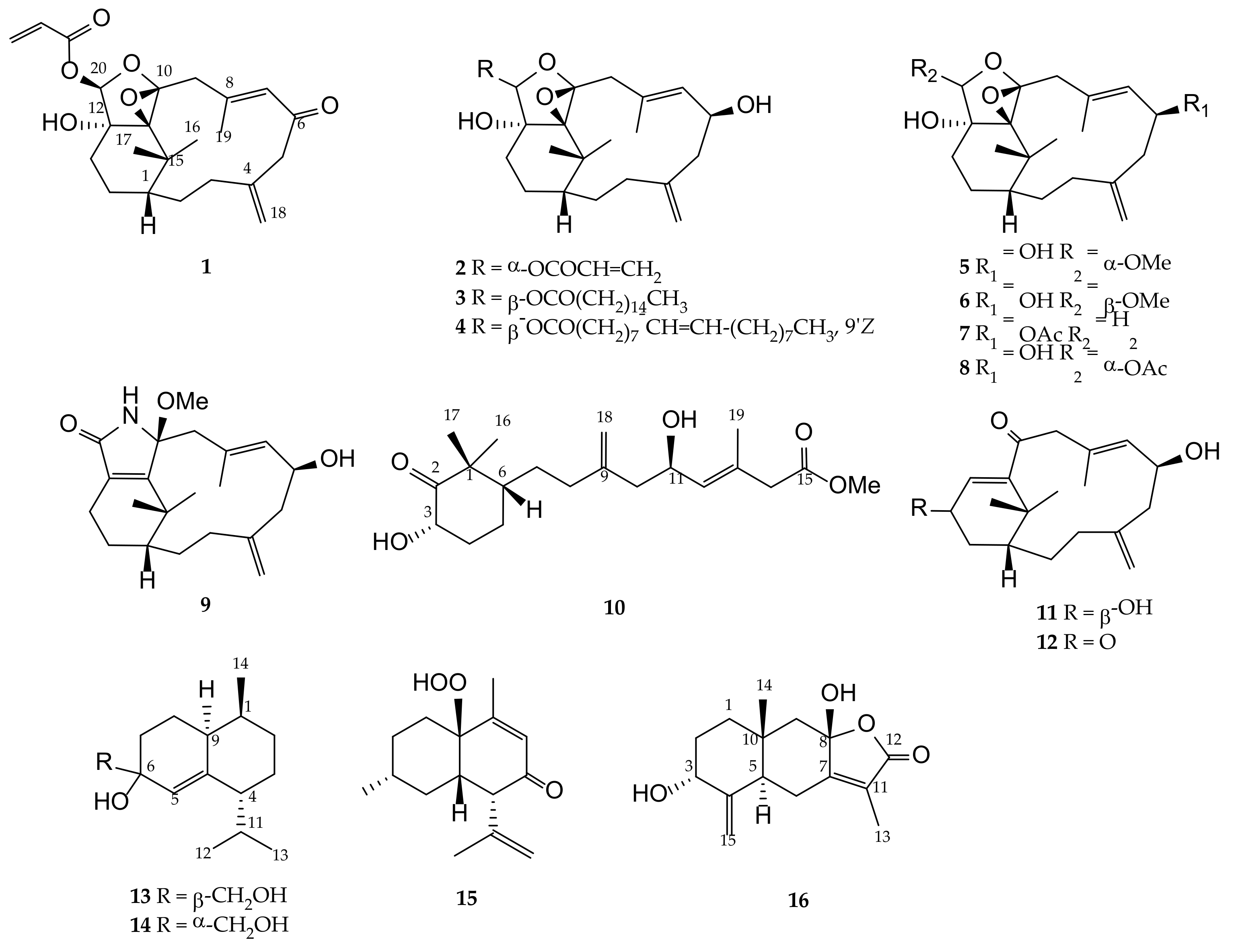
2.1. Structure Elucidation of the Verticillane-Type Diterpenes 1–9
2.2. Structure Elucidation of a Novel Norditerpene 10 and the Verticillane-Type Norditerpenes 11 and 12
2.3. Structure Elucidation of the Cadinane-Type Sesquiterpenes 13–15 and the Eudesmane-Type Sesquiterpenoid 16
2.4. Anti-Inflammatory Activities of the EtOAc Extract and the Isolated Compounds 1–28
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Animal Material
4.3. Extraction and Isolation
4.4. In Vitro Anti-Inflammatory Assay
4.4.1. Measurement of Cytokine Production by Dendritic Cells (DCs)
4.4.2. Measurement of Nitric Oxide (NO) Production by DCs
4.4.3. Measurement of Pro-Inflammatory Inducible NO Synthase (iNOS) and Cyclooxygenase-2 (COX-2) Gene Expression by DCs
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parameswaran, N.; Patial, S. Tumor Necrosis Factor-α Signaling in Macrophages. Crit. Rev. Eukaryot. Gene Expr. 2010, 20, 87–103. [Google Scholar] [CrossRef]
- Tang, T.; Scambler, T.E.; Smallie, T.; Cunliffe, H.E.; Ross, E.A.; Rosner, D.R.; O’Neil, J.D.; Clark, A.R. Macrophage responses to lipopolysaccharide are modulated by a feedback loop involving prostaglandin E2, dual specificity phosphatase 1 and tristetraprolin. Sci. Rep. 2017, 7, 4350. [Google Scholar] [CrossRef]
- Ling-Chien, H.; Wang, W.-H.; Chen, S.-H.; Chang, Y.-W.; Hung, L.-C.; Chen, C.-Y.; Chen, Y.-H. Lipopolysaccharide-Induced Nitric Oxide, Prostaglandin E2, and Cytokine Production of Mouse and Human Macrophages Are Suppressed by Pheophytin-b. Int. J. Mol. Sci. 2017, 18, 2637. [Google Scholar] [CrossRef] [Green Version]
- Wei, W.-C.; Su, Y.-H.; Chen, S.-S.; Sheu, J.-H.; Yang, N.-S. GM-CSF plays a key role in zymosan-stimulated human dendritic cells for activation of Th1 and Th17 cells. Cytokine 2011, 55, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Si, C.; Zhang, R.; Wu, T.; Lu, G.; Hu, Y.; Zhang, H.; Xu, F.; Wei, P.; Chen, K.; Tang, H.; et al. Dendritic cell-derived nitric oxide inhibits the differentiation of effector dendritic cells. Oncotarget 2016, 7, 74834–74845. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.K.; Yu, Y.L.; Chen, K.C.; Chang, W.T.; Lee, M.S.; Yang, M.J.; Cheng, H.C.; Liu, C.H.; Chen, D.C.; Chu, C.L. Kaempferol from Semen cuscutae Attenuates the Immune Function of Dendritic Cells. Immunobiology 2011, 216, 1103–1109. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.-H.; You, W.-J.; Lin, C.-C.; El-Shazly, M.; Liao, Z.-J.; Su, J.-H. Anti-Inflammatory Cembranoids from the Soft Coral Lobophytum crassum. Mar. Drugs 2017, 15, 327. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, A.F.; Chen, Y.-W.; Huang, C.-Y.; Tseng, Y.-J.; Lin, C.-C.; Dai, C.-F.; Wu, Y.-C.; Sheu, J.-H. Isolation and Structure Elucidation of Cembranoids from a Dongsha Atoll Soft Coral Sarcophyton stellatum. Mar. Drugs 2018, 16, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, C.-Y.; Sung, P.-J.; Uvarani, C.; Su, J.-H.; Lu, M.-C.; Hwang, T.-L.; Dai, C.-F.; Wu, S.-L.; Sheu, J.-H. Glaucumolides A and B, Biscembranoids with New Structural Type from a Cultured Soft Coral Sarcophyton glaucum. Sci. Rep. 2015, 5, 15624. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, H.B.; Hung, W.L.; Qian, H.; Li, Y. TNF-α Inhibitors with Anti-Oxidative Stress Activity from Natural Products. Curr. Top. Med. Chem. 2012, 12, 1408–1421. [Google Scholar] [CrossRef]
- Azab, A.; Nassar, A.; Azab, A.N. Anti-Inflammatory Activity of Natural Products. Molecules 2016, 21, 1321. [Google Scholar] [CrossRef]
- Kiemer, A.K.; Hartung, T.; Huber, C.; Vollmar, A.M. Phyllanthus amarus has Anti-Inflammatory Potential by Inhibition of iNOS, COX-2, and Cytokines via the NF-kB Pathway. J. Hepatol. 2003, 38, 289–297. [Google Scholar] [CrossRef]
- Kim, D.H.; Li, H.; Han, Y.E.; Jeong, J.H.; Lee, H.J.; Ryu, J.-H. Modulation of Inducible Nitric Oxide Synthase Expression in LPS-Stimulated BV-2 Microglia by Prenylated Chalcones from Cullen corylifolium (L.) Medik. through Inhibition of I-κBα Degradation. Molecules 2018, 23, 109. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.C.; Yang, L.L.; Lee, T.J.F. Oroxylin A Inhibition of Lipopolysaccharide-Induced iNOS and COX-2 Gene Expression via Suppression of Nuclear Factor-kB Activation. Biochem. Pharmacol. 2000, 59, 1445–1457. [Google Scholar] [CrossRef]
- Elshamy, A.I.; Nassar, M.I.; Mohamed, T.A.; Hegazy, M.-E.F. Chemical and biological profile of Cespitularia species: A mini review. J. Adv. Res. 2016, 7, 209–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.-C.; Lo, K.-L.; Kuo, Y.-H.; Kuo, Y.-C.; Chen, C.-H.; Khalil, A.T. Cespihypotins Q−V, Verticillene Diterpenoids from Cespitularia hypotentaculata. J. Nat. Prod. 2008, 71, 1993–1997. [Google Scholar] [CrossRef]
- Duh, C.-Y.; El-Gamal, A.A.H.; Wang, S.-K.; Dai, C.-F. Novel Terpenoids from the Formosan Soft Coral Cespitularia hypotentaculata. J. Nat. Prod. 2002, 65, 1429–1433. [Google Scholar] [CrossRef]
- Duh, C.-Y.; Li, C.-H.; Wang, S.-K.; Dai, C.-F. Diterpenoids, Norditerpenoids, and Secosteroids from the Formosan Soft Coral Cespitularia hypotentaculata. J. Nat. Prod. 2006, 69, 1188–1192. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Wang, S.-S.; Chen, C.-H.; Kuo, Y.-H.; Shen, Y.-C. Cespitulones A and B, Cytotoxic Diterpenoids of a New Structure Class from the Soft Coral Cespitularia taeniata. Mar. Drugs 2014, 12, 3477–3486. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.-C.; Ho, C.-J.; Kuo, Y.-H.; Lin, Y.-S. Cespitulactones A and B, new diterpenoids from Cespitularia taeniata. Bioorganic Med. Chem. Lett. 2006, 16, 2369–2372. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-C.; Lin, Y.-S.; Kuo, Y.-H.; Cheng, Y.-B. Cespitulactams A, B, and C, three new nitrogen-containing diterpenes from Cespitularia taeniata May. Tetrahedron Lett. 2005, 46, 7893–7897. [Google Scholar] [CrossRef]
- Shen, Y.-C.; Cheng, Y.-B.; Kobayashi, J.; Kubota, T.; Takahashi, Y.; Mikami, Y.; Ito, J.; Lin, Y.-S. Nitrogen-Containing Verticillene Diterpenoids from the Taiwanese Soft Coral Cespitularia taeniata. J. Nat. Prod. 2007, 70, 1961–1965. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.K.; Maarisit, W.; Roy, M.C.; Taira, J.; Ueda, K. Five New Diterpenoids from an Okinawan Soft Coral, Cespitularia sp. Mar. Drugs 2012, 10, 2741–2748. [Google Scholar] [CrossRef]
- Cheng, Y.-B.; Chen, C.-Y.; Kuo, Y.-H.; Shen, Y.-C. New Nitrogen-Containing Sesquiterpenoids from the Taiwanese Soft Coral Cespitularia taeniata May. Chem. Biodivers. 2009, 6, 1266–1272. [Google Scholar] [CrossRef]
- Roy, P.K.; Roy, M.C.; Taira, J.; Ueda, K. Structure and bioactivity of a trisnorditerpenoid and a diterpenoid from an Okinawan soft coral, Cespitularia sp. Tetrahedron Lett. 2014, 55, 1421–1423. [Google Scholar] [CrossRef]
- Cheng, S.-Y.; Lin, E.-H.; Wen, Z.-H.; Chiang, M.Y.-N.; Duh, C.-Y. Two New Verticillane-Type Diterpenoids from the Formosan Soft Coral Cespitularia hypotentaculata. Chem. Pharm. Bull. 2010, 58, 848–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, J.-Y.; Fazary, A.; Lin, Y.-C.; Hwang, T.-L.; Shen, Y.-C. New Verticillane Diterpenoids from Cespitularia taeniata. Chem. Biodivers. 2012, 9, 654–661. [Google Scholar] [CrossRef]
- Shen, Y.-C.; Wu, Y.-R.; Lin, J.-J.; Lo, K.-L.; Kuo, Y.-C.; Khalil, A.T. Eight new diterpenoids from soft coral Cespitularia hypotentaculata. Tetrahedron 2007, 63, 10914–10920. [Google Scholar] [CrossRef]
- Shen, Y.-C.; Lin, J.-J.; Wu, Y.-R.; Chang, J.-Y.; Duh, C.-Y.; Lo, K.L. New norditerpenoids from Cespitularia hypotentaculata. Tetrahedron Lett. 2006, 47, 6651–6655. [Google Scholar] [CrossRef]
- Duan, J.-A.; Wang, L.; Qian, S.; Su, S.; Tang, Y. A new cytotoxic prenylated dihydrobenzofuran derivative and other chemical constituents from the rhizomes of Atractylodes lancea DC. Arch. Pharmacal Res. 2008, 31, 965–969. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, F.; Dutta, L.N.; Knauf, W.; Robinson, H.; King, R.M. Neue sesquiterpenlactone aus Aster umbellatus. Phytochemistry 1980, 19, 433–436. [Google Scholar] [CrossRef]
- Hoang, L.S.; Tran, M.H.; Lee, J.-S.; Ngo, Q.M.T.; Woo, M.H.; Min, B.S. Inflammatory Inhibitory Activity of Sesquiterpenoids from Atractylodes macrocephala Rhizomes. Chem. Pharm. Bull. 2016, 64, 507–511. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yang, X.-W. New eudesmane-type sesquiterpenoids from the processed rhizomes of Atractylodes macrocephala. J. Asian Nat. Prod. Res. 2014, 16, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Izumi, S.; Ito, D.I.; Iwaeda, T.; Utsumi, R.; Hirata, T. Secretion of Allelochemicals from the Cultured Suspension Cells of Marchantia polymorpha. Chem. Lett. 1996, 3, 205–206. [Google Scholar] [CrossRef]
- Halgren, T.A.; Nachbar, R.B. Merck molecular force field. IV. conformational energies and geometries for MMFF94. J. Comput. Chem. 1996, 17, 587–615. [Google Scholar] [CrossRef]
- Lainer, J.; Dawid, C.; Dunkel, A.; Gläser, P.; Wittl, S.; Hofmann, T. Characterization of Bitter-Tasting Oxylipins in Poppy Seeds (Papaver somniferum L.). J. Agric. Food Chem. 2020, 68, 10361–10373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-T.; Munot, Y.S. Direct Atom-Efficient Esterification between Carboxylic Acids and Alcohols Catalyzed by Amphoteric, Water-Tolerant TiO(acac)2. J. Org. Chem. 2005, 70, 8625–8627. [Google Scholar] [CrossRef]
- Leuenberger, M.G.; Engeloch-Jarret, C.; Woggon, W.D. The Reaction Mechanism of the Enzyme-Catalyzed Cleavage of β-Carotene to Retinal. Angew. Chem. Int. Ed. 2001, 40, 2613–2617. [Google Scholar] [CrossRef]
- Jang, D.-I.; Lee, A.-H.; Shin, H.-Y.; Song, H.-R.; Park, J.-H.; Kang, T.-B.; Lee, S.-R.; Yang, S.-H. The Role of Tumor Necrosis Factor Alpha (TNF-α) in Autoimmune Disease and Current TNF-α Inhibitors in Therapeutics. Int. J. Mol. Sci. 2021, 22, 2719. [Google Scholar] [CrossRef] [PubMed]
- Kurach, Ł.; Kulczycka-Mamona, S.; Kowalczyk, J.; Skalicka-Woźniak, K.; Boguszewska-Czubara, A.; El Sayed, N.; Osmani, M.; Iwaniak, K.; Budzyńska, B. Mechanisms of the Procognitive Effects of Xanthotoxin and Umbelliferone on LPS-Induced Amnesia in Mice. Int. J. Mol. Sci. 2021, 22, 1779. [Google Scholar] [CrossRef]
- Król, M.; Kepinska, M. Human Nitric Oxide Synthase—Its Functions, Polymorphisms, and Inhibitors in the Context of Inflammation, Diabetes and Cardiovascular Diseases. Int. J. Mol. Sci. 2020, 22, 56. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-Y.; Kao, S.-H.; Hung, L.-C.; Chien, H.-J.; Wang, W.-H.; Chang, Y.-W.; Chen, Y.-H. Lipopolysaccharide-Induced Nitric Oxide and Prostaglandin E2 Production Is Inhibited by Tellimagrandin II in Mouse and Human Macrophages. Life 2021, 11, 411. [Google Scholar] [CrossRef] [PubMed]
- Needleman, P.; Manning, P. Interactions between the inducible cyclooxygenase (COX-2) and nitric oxide synthase (iNOS) pathways: Implications for therapeutic intervention in osteoarthritis. Osteoarthr. Cartil. 1999, 7, 367–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Joshi-Barve, S.; Barve, S.; Chen, L.H. Eicosapentaenoic Acid Prevents LPS-Induced TNF-α Expression by Preventing NF-kB Activation. J. Am. Coll. Nutr. 2004, 23, 71–78. [Google Scholar] [CrossRef]
- Singh, V.K.; Mehrotra, S.; Narayan, P.; Pandey, C.M.; Agarwal, S.S. Modulation of Autoimmune Diseases by Nitric Oxide. Immunol. Res. 2000, 22, 1–19. [Google Scholar] [CrossRef]
- Sonar, S.A.; Lal, G. The iNOS Activity During an Immune Response Controls the CNS Pathology in Experimental Autoimmune Encephalomyelitis. Front. Immunol. 2019, 10, 710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatzantoni, K.; Mouzaki, A. Anti-TNF-α Antibody Therapies in Autoimmune Diseases. Curr. Top. Med. Chem. 2006, 6, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.D.; Busquets-Cortés, C.; Capó, X.; Tejada, S.; Tur, J.A.; Pons, A.; Sureda, A. Cyclooxygenase-2 Inhibitors as a Therapeutic Target in Inflammatory Diseases. Curr. Med. Chem. 2019, 26, 3225–3241. [Google Scholar] [CrossRef] [PubMed]
- Kröncke, K.D.; Fehsel, K.; Kolb-Bachofen, V. Inducible nitric oxide synthase in human diseases. Clin. Exp. Immunol. 1998, 113, 147–156. [Google Scholar] [CrossRef]
- Minghetti, L. Cyclooxygenase-2 (COX-2) in Inflammatory and Degenerative Brain Diseases. J. Neuropathol. Exp. Neurol. 2004, 63, 901–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hori, M.; Kita, M.; Torihashi, S.; Miyamoto, S.; Won, K.-J.; Sato, K.; Ozaki, H.; Karaki, H. Upregulation of iNOS by COX-2 in muscularis resident macrophage of rat intestine stimulated with LPS. Am. J. Physiol. Gastrointest. Liver Physiol. 2001, 280, G930–G938. [Google Scholar] [CrossRef] [PubMed]
- Johnston, B.L.; Conly, J.M. Tumour Necrosis Factor Inhibitors and Infection: What is there to Know for Infectious Diseases Physicians? Can. J. Infect. Dis. Med. Microbiol. 2006, 17, 209–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smallwood, M.J.; Nissim, A.; Knight, A.R.; Whiteman, M.; Haigh, R.; Winyard, P.G. Oxidative stress in autoimmune rheumatic diseases. Free. Radic. Biol. Med. 2018, 125, 3–14. [Google Scholar] [CrossRef]
- Liou, C.-J.; Lai, Y.-R.; Chen, Y.-L.; Chang, Y.-H.; Li, Z.-Y.; Huang, W.-C. Matrine Attenuates COX-2 and ICAM-1 Expressions in Human Lung Epithelial Cells and Prevents Acute Lung Injury in LPS-Induced Mice. Mediat. Inflamm. 2016, 2016, 3630485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.-F.; Su, X.-H.; Li, L.-G.; Wang, W.; Zhang, M.-L.; Huo, C.-H.; Shi, Q.-W. Verticillane Derivatives from Natural Sources. Chem. Biodivers. 2009, 6, 1661–1673. [Google Scholar] [CrossRef]












| No. | 1 1 | 2 2 | 3 3 | 4 3 | 5 3 | 6 3 |
|---|---|---|---|---|---|---|
| 1 | 44.1, CH 4 | 44.0, CH | 44.1, CH | 44.1, CH | 44.2, CH | 44.2, CH |
| 2 | 34.4, CH2 | 33.8, CH2 | 33.8, CH2 | 33.8, CH2 | 34.0, CH2 | 33.6, CH2 |
| 3 | 38.7, CH2 | 37.7, CH2 | 37.7, CH2 | 37.7, CH2 | 37.8, CH2 | 37.8, CH2 |
| 4 | 144.1, C | 145.7, C | 145.7, C | 145.7, C | 145.8, C | 145.7, C |
| 5 | 55.2, CH2 | 45.8, CH2 | 45.8, CH2 | 47.5, CH2 | 45.9, CH2 | 45.6, CH2 |
| 6 | 197.7, C | 69.2, CH | 69.2, CH | 69.2, CH | 69.3, CH | 69.1, CH |
| 7 | 129.4, CH | 133.5, CH | 133.5, CH | 133.5, CH | 133.3, CH | 133.2, CH |
| 8 | 148.5, C | 132.4, C | 132.0, C | 132.4, C | 132.8, C | 132.8, C |
| 9 | 41.0, CH2 | 40.7, CH2 | 40.7, CH2 | 40.7, CH2 | 41.0, CH2 | 41.0, CH2 |
| 10 | 94.6, C | 94.7, C | 94.7, C | 94.6, C | 94.3, C | 94.4, C |
| 11 | 72.8, C | 72.4, C | 72.4, C | 72.4, C | 72.8, C | 72.9, C |
| 12 | 80.0, C | 79.8, C | 79.7, C | 79.7, C | 78.3, C | 79.9, C |
| 13 | 26.8, CH2 | 26.2, CH2 | 26.2, CH2 | 26.2, CH2 | 31.6, CH2 | 25.2, CH2 |
| 14 | 23.8, CH2 | 25.4, CH2 | 25.4, CH2 | 25.4, CH2 | 26.2, CH2 | 25.5, CH2 |
| 15 | 38.1, C | 37.6, C | 37.6, C | 37.6, C | 37.6, C | 37.7, C |
| 16 | 24.9, CH3 | 25.1, CH3 | 25.2, CH3 | 25.2, CH3 | 25.1, CH3 | 25.3, CH3 |
| 17 | 26.8, CH3 | 26.4, CH3 | 26.4, CH3 | 26.4, CH3 | 26.0, CH3 | 26.4, CH3 |
| 18 | 116.5, CH2 | 115.8, CH2 | 115.8, CH2 | 115.8, CH2 | 115.7, CH2 | 115.7, CH2 |
| 19 | 19.2, CH3 | 17.2, CH3 | 17.4, CH3 | 17.2, CH3 | 17.4, CH3 | 17.4, CH3 |
| 20 | 101.8, CH | 101.3, CH | 100.8, CH | 100.8, CH | 104.6, CH | 109.1, CH |
| 21 | 164.9, C | 164.8, C | 56.6, CH3 | 57.5, CH3 | ||
| 22 | 127.6, CH | 127.3, CH | ||||
| 23 | 132.6, CH2 | 132.7, CH2 | ||||
| 1′ | 172.8, C | 172.8, C | ||||
| 2′ | 34.1, CH2 | 34.1, CH2 | ||||
| 3′ | 24.7, CH2 | 24.7, CH2 | ||||
| 4′-13′ | 29.7 × 2, 29.6 × 3, 29.4 × 3, 29.2, 29.0, each CH2 | |||||
| 4′-7′/12′-15′ | 29.8, 29.7, 29.5, 29.3 × 2, 29.2 × 2, 29.0, each CH2 | |||||
| 8′/11′ | 27.2/27.1, each CH2 | |||||
| 9′-10′ | 130.1/129.7, each CH | |||||
| 14′ | 31.9, CH2 | |||||
| 15′ | 22.7, CH2 | |||||
| 16′ | 14.1, CH3 | 31.9, CH2 | ||||
| 17′ | 22.7, CH2 | |||||
| 18′ | 14.1, CH3 |
| No. | 1 1 | 2 2 | 3 3 | 4 3 | 5 3 |
|---|---|---|---|---|---|
| 1 | 1.19, m | 1.48, m | 1.49, m | 1.49, m | 1.46, m |
| 2 | 1.66, m | 1.82, m | 1.81, m | 1.81, m | 1.84, m |
| 1.38, m | 1.11, td (14.0, 5.0) | 1.11, td (14.5, 5.0) | 1.11, td (14.0, 5.5) | 1.09, td (14.5, 4.5) | |
| 3 | 1.94, td (13.0, 4.0) 4 | 2.22, m | 2.22 m | 2.22, m | 2.20, td (14.0, 4.5) |
| 1.82, td (13.0, 4.0) | 2.08, td (14.0, 3.2) | 2.08, td (14.0, 4.0) | 2.08, m | 2.08, dd (14.0, 4.5) | |
| 5 | 2.94, d (11.0) | 2.65, dd (12.5, 3.2) | 2.66, dd (13.0, 3.0) | 2.66, dd (12.5, 2.5) | 2.65, dd (13.0, 3.0) |
| 2.85, d (11.0) | 2.24, m | 2.25, m | 2.25, m | 2.22, m | |
| 6 | 4.49, t (8.0) | 4.50, quint (3.0) | 4.50, m | 4.50, br t (8.5) | |
| 7 | 6.13, s | 5.47, d (8.0) | 5.47, d (8.5) | 5.47, d (8.0) | 5.46, d (8.0) |
| 9 | 3.00, d (16.0) | 3.10, d (14.4) | 3.09, d (14.5) | 3.09, d (14.0) | 3.03, d (14.5) |
| 2.21, d (16.0) | 2.55, d (14.4) | 2.54, d (14.5) | 2.54, d (14.0) | 2.52, d (14.5) | |
| 13 | 1.57, td (14.0, 3.5) | 1.71, td (14.0, 3.6) | 1.67, m | 1.66, m | 1.70, br d (14.0) |
| 1.46, m | 1.58, m | 1.50, m | 1.50, m | 1.58, m | |
| 14 | 2.16, m | 2.28, m | 2.28, m | 2.27, m | 2.31, tt (17.5, 3.5) |
| 1.11, ddd (14.0, 6.0, 3.5) | 1.37, m | 1.37, m | 1.37, m | 1.33, m | |
| 16 | 0.72, 3H, s | 0.94, 3H, s | 0.98, 3H, s | 0.98, 3H, s | 0.95, 3H, s |
| 17 | 1.35, 3H, s | 1.34, 3H, s | 1.33, 3H, s | 1.33, 3H, s | 1.31, 3H, s |
| 18 | 5.09, 4.77, both s | 4.93, 2H, s | 4.94, 2H, s | 4.94, 2H, s | 4.93, 2H, s |
| 19 | 2.11, 3H, s | 1.81, 3H, s | 1.81, 3H, s | 1.81, 3H, s | 1.83, 3H, s |
| 20 | 5.96, s | 5.70, s | 5.63, s | 5.63, s | 4.36, s |
| 21 | 3.47, 3H, s | ||||
| 22 | 5.77, dd (17.5, 10.5) | 6.14, dd (17.2, 10.4) | |||
| 23 | 6.15, br d (17.5) | 6.47, d (17.2) | |||
| 5.16, dd (10.5, 1.0) | 5.94, d (10.4) | ||||
| 2′ | 2.36, t (7.5) | 2.36, t (7.5) | |||
| 3′ | 1.62, 2H, m | 1.63, 2H, m | |||
| 4′-15′ | 1.20–1.31, 20H, m | ||||
| 4′-7′/12′-17′ | 1.25–1.34, 20H, m | ||||
| 8′/11′ | 2.01, H, m | ||||
| 9′/10′ | 5.34, dd (10.5, 6.5) | ||||
| 16′ | 0.88, 3H, t (7.0) | ||||
| 18′ | 0.88, 3H, t (7.0) | ||||
| 12-OH | 2.25, br s | 2.64, br s | 2.52, br s | 2.53, br s | 3.29, br s |
| No. | 6 1 | 7 1 | 8 1 | 9 1 | 11 1 | 12 1 |
|---|---|---|---|---|---|---|
| 1 | 1.45, m | 1.46, m | 1.49, m | 1.54, m | 1.81, m | 2.15, m |
| 2 | 1.79, m | 1.79, m | 1.84, m | 1.55, m | 1.92, m | 2.46, m |
| 1.11, td (14.0, 5.0) 2 | 1.11, m | 1.11, td (14.0, 5.0) | 1.36, m | 1.52, m | 1.73, m | |
| 3 | 2.20, m | 2.23, m | 2.22, m | 2.27, m | 2.59, dd | 1.94, m |
| (15.0, 11.0) | ||||||
| 2.09, m | 2.01, m | 2.08, m | 2.13, m | 1.86, m | 1.56, m | |
| 5 | 2.64, dd | 2.60, dd | 2.65, dd | 2.44, dd | 2.48, dd | 2.43, m |
| (13.0, 3.0) | (13.0, 3.5) | (13.0, 3.0) | (13.5, 2.5) | (13.0, 7.0) | ||
| 2.23, m | 2.30, d (13.0) | 2.24, m | 2.33, m | 2.28, dd (13.0, 2.5) | 2.34, dd (13.5, 3.0) | |
| 6 | 4.51, m | 5.49, td (9.0, 3.5) | 4.50, br t (8.0) | 4.38, m | 4.49, m | 4.44, m |
| 7 | 5.46, d (8.0) | 5.41, d (9.0) | 5.48, d (9.0) | 5.55, d (8.0) | 5.15, d (6.5) | 5.28, d (7.0) |
| 9 | 3.03, d (14.5) | 3.04, d (14.5) | 3.05, d (14.5) | 3.00, d (14.5) | 3.43, d (15.0) | 3.43, d (15.5) |
| 2.54, d (14.5) | 2.55, d (14.5) | 2.54, d (14.5) | 2.66, d (14.5) | 3.08, d (15.0) | 3.24, d (15.5) | |
| 12 | 6.10, d (3.5) | 6.07, s | ||||
| 13 | 1.67, m | 1.76, m | 1.81, m | 2.34, m | 4.50, m | |
| 1.56, m | 1.63, m | 1.62, m | 2.18, m | |||
| 14 | 2.21, m | 2.25, m | 2.30, tt | 2.19, m | 2.13, 2H, m | 3.02, dd |
| (14.5, 4.0) | (18.5, 7.0) | |||||
| 1.35, m | 1.33, m | 1.40, m | 1.63, m | 2.45, m | ||
| 16 | 0.98, 3H, s | 1.00, 3H, s | 0.98, 3H, s | 1.47, 3H, s | 1.09, 3H, s | 1.23, 3H, s |
| 17 | 1.31, 3H, s | 1.33, 3H, s | 1.33, 3H, s | 1.24, 3H, s | 1.40, 3H, s | 1.51, 3H, s |
| 18 | 4.92, 2H, s | 4.96, 4.92, both s | 4.94, 2H, s | 4.84, 2H, s | 4.87, 4.83, both s | 4.84, 4.78, both s |
| 19 | 1.84, 3H, s | 1.88, 3H, s | 1.83, 3H, s | 1.58, 3H, s | 1.76, 3H, s | 1.78, 3H, s |
| 20 | 4.47, s | 3.59, 3.43, both d (9.0) | 5.76, s | |||
| 21 | 3.46, 3H, s | 3.13, 3H, s | ||||
| 22 | 2.02, 3H, s | 2.14, 3H, s | ||||
| 12-OH | 2.63, br s | 1.96, br d (2.0) | 2.68, br s | |||
| N-H | 5.46, br s |
| No. | 7 1 | 8 1 | 9 1 | 11 1 | 12 1 |
|---|---|---|---|---|---|
| 1 | 44.3, CH 2 | 44.1, CH | 44.8, CH | 45.8, CH | 45.4, CH |
| 2 | 34.2, CH2 | 33.7, CH2 | 32.4, CH2 | 29.5, CH2 | 30.7, CH2 |
| 3 | 37.8, CH2 | 37.7, CH2 | 34.4, CH2 | 31.0, CH2 | 30.6, CH2 |
| 4 | 145.3, C | 145.5, C | 146.1, C | 146.0, C | 145.4, C |
| 5 | 43.4, CH2 | 45.8, CH2 | 43.9, CH2 | 44.8, CH2 | 44.2, CH2 |
| 6 | 72.2, CH | 69.2, CH | 68.3, CH | 69.0, CH | 69.5, CH |
| 7 | 128.7, CH | 133.7, CH | 135.7, CH | 133.6, CH | 135.3, CH |
| 8 | 134.5, C | 132.5, C | 131.3, C | 130.0, C | 129.1, C |
| 9 | 41.3, CH2 | 40.6, CH2 | 48.5, CH2 | 51.9, CH2 | 52.4, CH2 |
| 10 | 95.9, C | 95.1, C | 93.9, C | 202.6, C | 202.2, C |
| 11 | 74.0, C | 73.0, C | 160.1, C | 150.3, C | 166.0, C |
| 12 | 78.7, C | 78.5, C | 133.8, CH | 134.6, CH | 128.6, CH |
| 13 | 31.1, CH2 | 30.6, CH2 | 17.5, CH2 | 65.9, CH | 199.0, C |
| 14 | 26.2, CH2 | 26.0, CH2 | 24.8, CH2 | 34.9, CH2 | 40.0, CH2 |
| 15 | 37.5, C | 37.4, C | 37.7, C | 35.8, C | 37.0, C |
| 16 | 25.2, CH3 | 25.0, CH3 | 24.8, CH3 | 23.8, CH3 | 23.6, CH3 |
| 17 | 26.3, CH3 | 26.0, CH3 | 34.1, CH3 | 33.4, CH3 | 32.3, CH3 |
| 18 | 115.8, CH2 | 115.9, CH2 | 114.8, CH2 | 112.6, CH2 | 113.4, CH2 |
| 19 | 17.5, CH3 | 17.4, CH3 | 17.6, CH3 | 18.8, CH3 | 18.9, CH3 |
| 20 | 75.5, CH2 | 96.1, CH | 171.6, C | ||
| 21 | 170.2, C | 169.7, C | 50.4, CH3 | ||
| 22 | 21.3, CH3 | 21.2, CH3 |
| No. | 10 | |
|---|---|---|
| 1H 1 | 13C 2 | |
| 1 | 48.4, C | |
| 2 | 214.6, C | |
| 3 | 4.45, m | 71.4, CH 4 |
| 4 | 2.31, m | 31.6, CH2 |
| 1.58, m | ||
| 5 | 2.13, m | 21.0, CH2 |
| 1.68, m | ||
| 6 | 1.79, m | 46.9, CH |
| 7 | 1.64, m | 25.7, CH2 |
| 1.13, m | ||
| 8 | 2.11, m | 33.8, CH2 |
| 1.92, m | ||
| 9 | 147.1, C | |
| 10 | 2.20, 2H, m | 43.9, CH2 |
| 11 | 4.47, m | 66.1, CH |
| 12 | 5.32, br d (8.5) 3 | 131.7, CH |
| 13 | 122.2, C | |
| 14 | 3.02, 2H, br s | 44.6, CH2 |
| 15 | 171.4, C | |
| 16 | 1.11, 3H, s | 21.8, CH3 |
| 17 | 1.32, 3H, s | 27.0, CH3 |
| 18 | 4.87, 2H, s | 113.2, CH2 |
| 19 | 1.77, 3H, s | 16.9, CH3 |
| 20 | 3.69, 3H, s | 51.8, CH3 |
| No. | 13 1 | 14 2 | 15 3 | 16 2 |
|---|---|---|---|---|
| 1 | 34.7, CH 4 | 33.6, CH | 162.7, C | 35.2, CH2 |
| 2 | 29.1, CH2 | 28.8, CH2 | 128.6, CH | 29.0, CH2 |
| 3 | 22.7, CH2 | 22.5, CH2 | 197.7, C | 72.7, CH |
| 4 | 51.1, CH | 50.9, CH | 57.1, CH | 149.8, C |
| 5 | 125.8, CH | 125.4, CH | 31.7, CH2 | 45.6, CH |
| 6 | 71.2, CH | 70.8, CH | 25.1, CH | 24.0, CH2 |
| 7 | 31.1, CH2 | 31.4, CH2 | 29.1, CH2 | 159.4, C |
| 8 | 23.2, CH2 | 22.3, CH2 | 29.8, CH2 | 103.1, C |
| 9 | 36.6, CH | 37.5, CH | 84.0, C | 51.0, CH2 |
| 10 | 147.0, C | 146.1, C | 36.8, CH | 36.6, C |
| 11 | 26.6, CH | 26.8, CH | 140.5, C | 123.8, C |
| 12 | 21.2, CH3 | 21.3, CH3 | 18.3, CH3 | 174.7, C |
| 13 | 21.7, CH3 | 21.6, CH3 | 117.1, CH2 | 8.34, CH3 |
| 14 | 14.4, CH3 | 14.4, CH3 | 18.0, CH3 | 15.9, CH3 |
| 15 | 68.9, CH2 | 69.9, CH2 | 22.1, CH3 | 110.1, CH2 |
| No. | 13 1 | 14 2 | 15 3 | 16 2 |
|---|---|---|---|---|
| 1 | 1.96, m | 1.97, m | 1.71 td (13.8, 4.2) | |
| 1.38, m | ||||
| 2 | 1.82, m | 1.82, m | 5.95, d (1.0) | 1.87 dt (14.4, 3.6) |
| 1.31, br t (10.8) 4 | 1.35, dd (10.8, 4.8) | 1.78 dtd (14.4, 4.2, 2.4) | ||
| 3 | 1.70, m | 1.70, m | 4.37, br s | |
| 1.66, m | 1.65, m | |||
| 4 | 1.63, m | 1.64, m | 3.26, d (13.5) | |
| 5 | 5.45, s | 5.50, s | 1.46, m | 2.45, br s |
| 6 | 1.61, m | 2.60, d (10.2) | ||
| 2.44, d (10.2) | ||||
| 7 | 1.83, m | 1.67, m | 1.54, m | |
| 1.46, d (9.2) | 1.46, dd (3.6, 1.8) | 1.22, m | ||
| 8 | 1.80, m | 1.64, m | 1.91, br d (14.5) | |
| 1.46, d (9.2) | 1.50, m | 1.71, td (14.5, 5.0) | ||
| 9 | 2.31, br d (4.4) | 2.24, td (13.2, 4.8) | 2.26, d (13.8) | |
| 1.65, d (13.8) | ||||
| 10 | 2.90, br d (13.5) | |||
| 11 | 1.84, m | 1.81, m | ||
| 12 | 0.92, 3H, d (6.8) | 0.92, 3H, d (6.6) | 1.66, 3H, s | |
| 13 | 0.78, 3H, d (6.8) | 0.70, 3H, d (6.6) | 5.08, 4.87, both s | 1.84, 3H, s |
| 14 | 0.84, 3H, d (6.8) | 0.90, 3H, d (6.6) | 2.05, 3H, s | 1.03, 3H, s |
| 15 | 3.49, 2H, qd (10.8, 4.8) | 3.49, d (10.8) | 0.90, d (6.5) | 5.11, s |
| 3.43, d (10.8) | 4.77, s | |||
| 9-OOH | 7.42, br s |
| No. | TNF-α Expression | NO Production | ||
|---|---|---|---|---|
| Inh % 1 | Inh % | |||
| S9-EA2 | 84.1 ± 4.2 | **** | 76.1 ± 1.4 | **** |
| 1 | 95.0 ± 0.2 | **** | 39.8 ± 0.6 | **** |
| 2 | 95.7 ± 0.4 | **** | 63.3 ± 1.6 | **** |
| 3 | 95.8 ± 0.1 | **** | 44.0 ± 0.9 | **** |
| 4 | 32.6 ± 11.6 | * | 51.1 ± 0.1 | **** |
| 5 | 39.5 ± 7.8 | ** | 56.8 ± 0.2 | **** |
| 6 | 29.8 ± 6.7 | 43.3 ± 2.0 | **** | |
| 7 | 48.4 ± 10.9 | *** | 48.6 ± 1.5 | **** |
| 8 | 29.7 ± 17.9 | 55.6 ± 1.4 | **** | |
| 9 | 46.6 ± 2.6 | *** | 50.7 ± 0.6 | **** |
| 10 | 44.1 ± 7.1 | ** | 46.7 ± 0.6 | **** |
| 11 | 34.3 ± 1.7 | * | 50.8 ± 0.6 | **** |
| 12 | 44.4 ± 2.7 | ** | 61.1 ± 0.5 | **** |
| 13 | 48.3 ± 12.6 | *** | 44.5 ± 0.1 | **** |
| 14 | 33.3 ± 4.8 | * | 55.6 ± 1.3 | **** |
| 15 | 19.5 ± 2.9 | 49.6 ± 0.4 | **** | |
| 16 | 42.2 ± 4.1 | ** | 41.9 ± 1.5 | **** |
| 17 | 36.1 ± 0.6 | **** | 58.6 ± 0.9 | **** |
| 18 | 52.3 ± 6.1 | **** | 58.5 ± 1.1 | **** |
| 19 | 46.3 ± 4.4 | **** | 63.7 ± 0.8 | **** |
| 20 | 42.7 ± 5.1 | **** | 56.3 ± 0.6 | **** |
| 21 | 41.9 ± 3.2 | **** | 61.7 ± 1.0 | **** |
| 22 | 4.9 ± 7.7 | 38.2 ± 1.1 | **** | |
| 23 | 5.3 ± 8.8 | 59.3 ± 9.4 | **** | |
| 24 | −0.6 ± 2.0 | 30.4 ± 2.9 | **** | |
| 25 | −4.7 ± 0.9 | 29.6 ± 8.9 | **** | |
| 26 | 3.8 ± 6.2 | 9.5 ± 4.3 | **** | |
| 27 | 5.8 ± 3.5 | 15.3 ± 2.5 | ||
| 28 | −3.5 ± 7.4 | 6.3 ± 6.0 | ||
| DEX 3 | 85.6 ± 3.4 | **** | 73.4 ± 1.3 | **** |
| No. | iNOS mRNA | COX-2 mRNA | ||
|---|---|---|---|---|
| Exp % 1 | Exp % | |||
| S9-EA2 | 15.9 ± 0.4 | ** | 28.6 ± 4.1 | ** |
| 1 | 3.6 ± 1.8 | *** | 4.2 ± 0.1 | **** |
| 2 | 0.3 ± 0.1 | *** | 2.9 ± 0.6 | **** |
| 3 | 64.6 ± 1.8 | 25.6 ± 12.7 | ** | |
| 4 | 67.4 ± 11.6 | 21.8 ± 1.1 | ** | |
| 5 | 71.0 ± 10.2 | 43.1 ± 25.8 | ||
| 6 | 281.2 ± 15.4 | **** | 65.4 ± 3.1 | |
| 7 | 115.9 ± 14.3 | 46.7 ± 4.4 | ||
| 8 | 29.0 ± 1.7 | ** | 15.5 ± 2.4 | *** |
| 9 | 35.2 ± 10.1 | * | 47.9 ± 1.8 | |
| 10 | 70.0 ± 1.8 | 36.9 ± 14.7 | * | |
| 11 | 36.0 ± 5.8 | * | 19.6 ± 4.1 | ** |
| 12 | 20.5 ± 3.4 | ** | 11.2 ± 1.7 | *** |
| 13 | 7.4 ± 2.9 | *** | 4.4 ± 3.5 | *** |
| 14 | 1.5 ± 0.8 | *** | 43.4 ± 26.2 | |
| 15 | 4.6 ± 2.9 | *** | 110.5 ± 29.7 | |
| 16 | 18.4 ± 4.4 | ** | 196.9 ± 55.1 | **** |
| 17 | 22.0 ± 3.8 | ** | 11.9 ± 3.7 | *** |
| 18 | 19.7 ± 11.3 | ** | 4.5 ± 0.5 | *** |
| 19 | 17.7 ± 1.3 | ** | 27.6 ± 1.9 | ** |
| 20 | 0.2 ± 0.1 | *** | 14.8 ± 6.1 | *** |
| 21 | 19.8 ± 11.6 | ** | 43.8 ± 7.9 | |
| 22 | 48.2 ± 7.9 | 2.1 ± 0.4 | **** | |
| 23 | 32.7 ± 14.9 | * | 35.4 ± 18.0 | * |
| 24 | 66.0 ± 38.8 | 21.7 ± 11.1 | ** | |
| 25 | 13.7 ± 1.9 | *** | 141.5 ± 30.1 | |
| 26 | 15.0 ± 4.1 | ** | 49.3 ± 25.0 | |
| 27 | 25.4 ± 12.6 | ** | 117.6 ± 43.8 | |
| 28 | 1.2 ± 0.5 | *** | 81.8 ± 42.4 | |
| DEX 3 | 44.3 ± 4.3 | * | 4.5 ± 0.5 | *** |
| Gene | Reverse Primer | Forward Primer |
|---|---|---|
| iNOS | 5′-CCAATGTTTCCCTGACTTTCCCA-3′ | 5′-CAGAGGGGTAGGCTTGTCTC-3′ |
| COX-2 | 5′-CAGGAGGATGGAGTTGTTGTAG-3′ | 5′-ACCAGCAGTTCCAGTATCAGA-3′ |
| GAPDH | 5′-TGTCATCATACTTGGCAGGTTTCT-3′ | 5′-CGTGTTCCTACCCCCAATGT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-C.; Lin, C.-C.; Chu, Y.-C.; Fu, C.-W.; Sheu, J.-H. Bioactive Diterpenes, Norditerpenes, and Sesquiterpenes from a Formosan Soft Coral Cespitularia sp. Pharmaceuticals 2021, 14, 1252. https://doi.org/10.3390/ph14121252
Lin Y-C, Lin C-C, Chu Y-C, Fu C-W, Sheu J-H. Bioactive Diterpenes, Norditerpenes, and Sesquiterpenes from a Formosan Soft Coral Cespitularia sp. Pharmaceuticals. 2021; 14(12):1252. https://doi.org/10.3390/ph14121252
Chicago/Turabian StyleLin, You-Cheng, Chi-Chien Lin, Yi-Chia Chu, Chung-Wei Fu, and Jyh-Horng Sheu. 2021. "Bioactive Diterpenes, Norditerpenes, and Sesquiterpenes from a Formosan Soft Coral Cespitularia sp." Pharmaceuticals 14, no. 12: 1252. https://doi.org/10.3390/ph14121252
APA StyleLin, Y.-C., Lin, C.-C., Chu, Y.-C., Fu, C.-W., & Sheu, J.-H. (2021). Bioactive Diterpenes, Norditerpenes, and Sesquiterpenes from a Formosan Soft Coral Cespitularia sp. Pharmaceuticals, 14(12), 1252. https://doi.org/10.3390/ph14121252







