Abstract
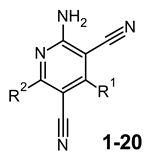
The amino-3,5-dicyanopyridine derivatives belong to an intriguing series of adenosine receptor (AR) ligands that has been developed by both academic researchers and industry. Indeed, the studies carried out to date underline the versatility of the dicyanopyridine scaffold to obtain AR ligands with not only a wide range of affinities but also with diverse degrees of efficacies at the different ARs. These observations prompted us to investigate on the structure–activity relationships (SARs) of this series leading to important previously reported results. The present SAR study has helped to confirm the 1H-imidazol-2-yl group at R2 position as an important feature for producing potent AR agonists. Moreover, the nature of the R1 substituent highly affects not only affinity/activity at the hA1 and hA2B ARs but also selectivity versus the other subtypes. Potent hA1 and hA2B AR ligands were developed, and among them, the 2-amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-[4-(prop-2-en-1-yloxy)phenyl]pyridine-3,5-dicarbonitrile (3) is active in the low nanomolar range at these subtypes and shows a good trend of selectivity versus both the hA2A and hA3 ARs. This combined hA1/hA2B partial agonist activity leads to a synergistic effect on glucose homeostasis and could potentially be beneficial in treating diabetes and related complications.
1. Introduction
Adenosine is a ubiquitous autacoid that has been reported to play important roles in various tissues by acting through four different adenosine receptors (ARs) classified as A1, A2A, A2B, and A3 receptors [1]. These ARs belong to the G protein–coupled receptors superfamily, and each of them has different functions although with some overlap. Depending on their coupling to G-protein, they can either increase or decrease intracellular cAMP levels by affecting adenylate cyclase (AC) activity. However, other effector systems have been proposed [2]. Adenosine has the highest affinity for the A1 and A2A ARs, the lowest for A2BAR, and intermediate affinity for the A3 subtype. Selective agonists and antagonists have been developed for each of the ARs [3,4], and their discovery opened up new avenues for potential drugs treatment of a variety of conditions such as neurodegenerative disorders, chronic inflammatory diseases, asthma, and many other pathophysiological states that are known to be associated with changes in adenosine levels [3].
Dysregulated intracellular nucleotide concentrations can be produced by altered and defective cellular metabolism leading to important metabolic pathologies such as diabetes mellitus [3,5,6]. This disease is increasing worldwide due to aging, incorrect lifestyle, and eating habits, and its complications are of great clinical and socioeconomic impact. All four ARs are involved in the regulation of islet hormone secretion and have the potential of being drug targets for the treatment of diabetes [3]. Currently, much attention has been devoted to A2BAR agonists as potential new drugs for diabetes treatment, even if there is discrepancy in the studies reported in literature, which may be ascribed to the different experimental conditions employed [3,7]. However, it is reported that agonists of this AR subtype may be useful for the therapy of this pathology [3].
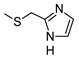
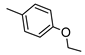
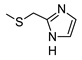
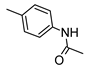
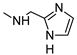
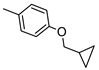
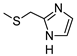
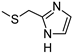
The role of the A1AR in diabetes seems to be clearer [3,8]. In fact, improvement of insulin sensitivity and glucose homeostasis by A1 activation has been well recognized for many years, thus the promotion of A1AR agonists as drug candidates [5,6]. Since full A1AR agonists are generally characterized by cardiovascular side effects, partial A1AR agonists could represent the best choice to avoid cardiovascular complications [9]. For this reason, a synergistic effect on glucose homeostasis targeting both A1 and A2B ARs could be of interest for treating diabetes and its complications. It is also worth noting that A1AR is involved in lipid metabolism, so the development of A1AR agonists as lipolytic agents is reported to be useful in cardiovascular disease–associated pathologies [10]. While the pathophysiological roles of the A1AR are well known, the A2B subtype is the least characterized because of the paucity of potent and selective agonists. BAY-606583 (2-{[6-Amino-3,5-dicyano-4-(4-(cyclopropylmethoxy)phenyl)pyridin-2-yl]thio}acetamide) is one of the most potent and selective A2BAR agonists known to date [11]. However, we recently produced a new series of amino-3,5-dicyanopyridines as BAY-606583 analogues [12]; most of the compounds are potent human (h) A2B AR partial agonists. Among them, the 2-{[(1H-imidazol-2-yl)methyl]thio}-6-amino-4-[4-(cyclopropylmethoxy)phenyl]pyridine-3,5-dicarbonitrile (P453, Figure 1) turns out to be three-fold more active than BAY-606583, although it is less selective. Compound P453 was used for experiments in in vitro models of short-term synaptic plasticity and ischemic-like condition [13]. In particular, compound P453 was shown to be a functional A2BAR agonist whose activation decreases PPF (paired pulse facilitation) by increasing glutamate release at presynaptic terminals and delays A1AR-mediated fEPSP inhibition during a two-minute oxygen and glucose deprivation (OGD) insult in the rat CA1 hippocampus.

Figure 1.
Lead structures and currently reported amino-3,5-dicyanopyridine-based adenosine receptor ligands.
From a structure–activity point of view, the new findings confirm the 1H-imidazol-2-yl group in the R2 pendant as an important feature for producing potent AR agonists. Certain 2-amino-4-aryl-6-(1H-imidazol-2-yl-methylsulfanyl)-pyridine-3,5-dicarbonitrile compounds, belonging to the LUF series (Figure 1), were already reported to display nanomolar affinity for all the ARs, including the A2B subtype for which they showed, in general, considerable affinity and efficacy [14]. The growing interest in this class is also dictated by the fact that non-nucleoside AR agonists seem to be more versatile for pharmacological studies, showing fewer species differences than the adenosine-like ones [4]. Moreover, the studies carried out to date are sufficient to underline the versatility of the amino-3,5-dicyanopyridine scaffold for producing AR ligands with not only a wide range of affinities but, interestingly, with different degrees of efficacy at the different ARs [11,12,14,15,16,17,18,19].
On this basis, the 1H-imidazol-2-yl group was used to design other amino-3,5-dicyanopyridine derivatives in order to deeply investigate the influence of the R1 substituent at the 4-position on AR affinity and selectivity. Moreover, other modifications were performed at R2 to enrich what is known about how the nature of the substituent at this position modulates AR affinity and selectivity.
2. Results
2.1. Chemistry
Compounds 1–4, 6–20, and their corresponding intermediates were synthesized according to the procedure reported in Scheme 1, Scheme 2 and Scheme 3. Following the same procedures, the 2-(((1H-Imidazol-2-yl)methyl)thio)-6-amino-4-(4-ethoxyphenyl)pyridine-3,5-dicarbonitrile 5, previously reported in reference [12], was prepared. For the synthesis of derivatives 1–4, 6–13, and 20, the strategic intermediates are represented by the 6-sulfanyl-substituted compounds 28–40 [12,14,18,20,21,22,23], which were prepared as depicted in Scheme 1. The suitable aldehyde was reacted with malononitrile and thiophenol, in boiling water, in the presence of a catalytic amount of basic alumina to yield the 4-aryl- and 4-benzyl-6-phenylsulfanyl-derivatives 21–27 [18,24,25,26], with moderate to good yield. When these latter were treated with anhydrous sodium sulfide in anhydrous DMF at 80 °C, followed by acidification with HCl, the free thiol 28–38 [12,18,20,21], 40 [14] were obtained in high yields. The preparation of the 4-methyl-6-sulfanyl compound 39 [22,23] was performed by reacting the (1-ethoxyethylidene)propanedinitrile 41 [22] with 2-cyanothioacetamide in boiling ethanol containing EtONa. The target dicyanopyridines 1–4, 6–13, and 20 were obtained by reaction of the 6-sulfanyl intermediates 28–40 with 2-(bromomethyl)-1H-imidazole hydrobromide (1–4, 6–13) or methyl chloroacetate (20) in the presence of sodium hydrogen carbonate at room temperature (rt).

Scheme 1.
Reagents and conditions. (a) malononitrile, thiophenol, basic Al2O3, H2O, 100 °C; (b) Na2S, anhydrous DMF, 80 °C; 1N HCl, room temperature (rt); (c) 2-cyanothioacetamide, EtONa, absolute EtOH, reflux; d) RCH2Cl, NaHCO3, anhydrous DMF, rt.

Scheme 2.
Reagents and conditions. (a) 1-(1H-imidazol-2-yl)methanamine for 14, ethanolamine for 15, 2-(4-methoxyphenyl)ethanamine for 16, or imidazole (for 17), triethylamine, THF/EtOH 2:1, reflux; (b) H2, 10% Pd/C, absolute EtOH, 30 Psi.

Scheme 3.
Reagents and conditions. (a) 2-aminoethanol, DMF, 100 °C.
To replace the thiomethyl linker at the 6-position with an aminomethyl bridge, compounds 14–16 were synthesized (Scheme 2) starting from the 6-chloro derivative 42, obtained with a reported procedure [27], and the suitable commercially available amine. The reaction was carried out in the presence of triethylamine and in a refluxing mixture of THF/EtOH. The same procedure was applied for the synthesis of the 6-(1H-imidazol-1-yl)-derivative 17, which was obtained in high yield. Elimination of the 6-substituent was realized in compound 18 which was synthesized by catalytic reduction (10% Pd/C) of 42 with hydrogen at 30 Psi in absolute EtOH.
Finally, the 6-[(2-hydroxyethyl)amino]-derivative 19 was synthesized (Scheme 3) starting from the intermediate 27 [24] by exploiting the good property of the 6-phenylthio function as the leaving group in nucleophilic substitution [15]. The reaction was performed at 100 °C in DMF using an excess of 2-aminoethanol as nucleophile.
2.2. Pharmacological Assays
The amino-3,5-dicyanopyridines 1–20 were tested for their affinity at hA1, hA2A, and hA3 ARs, stably transfected in Chinese Hamster Ovary (CHO) cells, and were also studied as hA2B agonists by evaluating their stimulatory effect on cAMP production in CHO cells, stably expressing the hA2B AR. Compounds 3, 8, and 11, the most interesting in terms of combined affinity/activity at hA1 and hA2B ARs, were evaluated for their A1 pharmacological profile in the cAMP assay, where each compound was tested to assess its capability to modulate Forskolin-stimulated cAMP levels in the absence or presence of the A1AR agonist 2-chloro-N6-cyclopentyladenosine (CCPA) (1 nM). All pharmacological data are reported in Table 1 and Table 2.

Table 1.
Binding affinities (Ki) at hA1, hA2A, and hA3 ARs and potencies (EC50) at hA2B ARs.

Table 2.
Modulation of Forskolin-stimulated cAMP levels of selected amino-3,5-dicyanopyridine derivatives on cyclic cAMP assay in hA1 CHO cells.a.
2.3. Molecular Docking Studies
To simulate the binding mode of the herein reported dicyanopyridines at hA1 and hA2B ARs, molecular docking studies were performed on the cryo-EM structure of the agonist-bound hA1AR (pdb code: 6D9H; 3.6-Å resolution [28]) and on a homology model of the hA2BAR developed by using the crystal structure of agonist-bound hA2AAR as a template (pdb code: 2YDO; 3.0-Å resolution [29]). The obtained hA2BAR homology model was checked using the Protein Geometry Monitor application within MOE (Molecular Operating Environment 2019.0101) [30] by inspecting the structural quality of the protein model (backbone bond lengths, angles, and dihedrals, Ramachandran φ-ψ dihedral plots, and quality of side chain rotamer and non-bonded contact). The two AR structures were then used as target for the docking analysis of the synthesized derivatives. The docking studies were performed using the MOE docking tool (induced fit docking and optimization protocol) and Gold software [31]. For each compound, the top-score docking pose at each AR model, according to at least two out of three scoring functions, was selected for final ligand-target interaction analysis.
Docking conformations at hA1 and hA2A ARs of selected dicyanopyridine compounds are reported in Figure 2 and Figure 3, respectively (see the Discussion Section).

Figure 2.
Docking conformations at hA1AR. The simulated binding mode of compound 8 is shown (A) with detail of key receptor residues involved in ligand-target interaction or (B) with representation of the binding cavity as molecular surface.

Figure 3.
Docking conformations at hA2BAR. The simulated binding mode of compound 6 is shown, with detail of key receptor residues involved in ligand–target interaction.
3. Discussion
3.1. Structure–Activity Relationships
The biological and pharmacological data for the newly synthesized amino-3,5-dicyanopyridine derivatives 1–20 are reported in Table 1 and Table 2 together with those of the reference compounds LUF5833 (2-amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-phenylpyridine-3,5-dicarbonitrile) [14] and P453 [12]. All the derivatives that interact with the hA2BAR (2, 3, 5–8, 10–12) display EC50 values from 12 to 260 nM behaving as partial agonists, with the only exception being compound 6, previously reported as trifluoroacetate salt [32], which shows a full agonist profile. Some compounds (17–20), selected among those that had no activity at the hA2BAR were also evaluated to investigate their antagonistic effect on cAMP production stimulated by 5′-(N-ethylcarboxamido)adenosine (NECA) 100 nM in hA2BCHO cells. These functional studies reveal their inability to inhibit NECA-stimulated cAMP levels (I% ≤ 3) (data not shown). Most of the compounds have generally modest to null hA2A and hA3 AR affinity with the only exception of the two pairs of compounds 6 and 10 and 5 and 6 that bind, respectively, the hA2A and hA3 ARs with nanomolar affinity. Best results have been obtained in the hA1AR binding experiments. In particular, all the 6-(imidazole-2-ylmethyl)sulfanyl derivatives reported herein (1–13), with the only exception being compounds 1, 2, 9, and 12, have from high to good hA1AR affinity. In particular, some compounds showed Ki values in the low nanomolar range and in general below 10 nM. Functional studies at the hA1 subtype (data are reported in Table 2) indicated that the tested compounds 3, 8, and 11 behave as partial agonists and that their relative potencies correlate well with the Ki values obtained in the hA1 binding experiments. All the newly synthesized compounds showed different and not easily predictable behavior depending on both R1 and R2 substituents. However, the data shown in Table 1 indicate that the binding at the hA2BAR subtype depends strictly on the substitution at the R1 position [12].
Modifications of the para-substituent at the R1 pendant of the lead compound P453 were performed to evaluate the influence on hA2BAR activity and selectivity. 6-(Imidazole-2-ylmethyl)sulfanyl derivatives 1–5, bearing different cycloalkyl- or cycloalkenyl-methyloxy substituents at R1 position, were synthesized. Only compounds 3 and 5 maintain the hA2BAR activity in the nanomolar range, while derivative 2 and derivatives 1 and 4 are, respectively, scarcely active or inactive, probably due to the different steric hindrance of the para-substituent with respect to P453. In particular, derivative 5 is endowed with hA2BAR activity comparable to that of the lead P453, despite a low or null selectivity versus the other AR subtypes. Introduction of a para-acetamido substituent on the 4-phenyl moiety led to compound 6, which is the only amino-3,5-dicyanopyridine in the whole series having a full agonist profile at the hA2BAR. The para-acetamido substituent, in addition to possessing an oxygen atom as H-bond acceptor, also contains the NH donor group that could have some influence in determining the pharmacological profile of this derivative as observed for the dicyanopyridine of the LUF series (Figure 1) [33,34].
Activity at the hA2BAR is also affected by the electronic effects of the para-substituent on the 4-phenyl ring. In fact, the corresponding EC50 values fall in the low nanomolar range for compounds 8 (para-fluoro) and 11 (para-thiomethyl) and move to higher values by passing to derivatives 10 (para-methyl) and 7 (para-chloro), up to the total inactivity of 9, bearing a para-trifuoromethyl group. Homologation of the 4-phenyl moiety of LUF5833 led to the 4-benzyl-susbstituted derivative 12, which maintains modest hA2B activity. In contrast, its replacement with a methyl group was highly detrimental leading to compound 13, which is completely avoid of any activity at the hA2BAR subtype. In this first set of compounds (1–13), bearing a 6-(imidazole-2-ylmethyl)sulfanyl moiety on the dicyanopyridine scaffold, the hA1AR affinity depends on the para-phenyl substitution as observed for hA2B receptor activity trend. In fact, many compounds show mixed hA1/hA2B combined affinity/activity. Looking at Table 1, we can observe different activity profiles—for example, compound 6, whose biological data fall in the same range of values (EC50 at hA2BAR and Ki at the other subtypes) can be considered in effect as a pan ligand. Compound 3 is interesting in terms of its combined hA1/hA2B AR partial agonist activity, exhibiting a good trend of selectivity versus both the hA2A and hA3 subtypes. The data of derivatives 8 and 11, which lose a little in selectivity but behave as hA1/hA2B partial agonists, are also noteworthy. Moreover, compound 8, binding the hA1AR in the sub-nanomolar range, is the most potent hA1AR ligand among the herein reported dicyanopyridine series. When the 6-sulfanyl linker of LUF5833 was replaced by a 6-amino bridge, compound 14 was obtained. A dramatic decrease of hA2BAR activity was observed with respect to the LUF derivative but the effect was detrimental also for the other ARs. Replacement of the imidazolyl moiety at R2 position with different substituents in terms of both steric hindrance and capability to engage hydrogen-bonding led to derivatives 15–20, which show null affinity/activity for each of the AR subtypes. The only exception are compounds 15 and 20 endowed with hA1AR binding affinity in the micromolar range. Compounds 17–20, besides being inactive as hA2B agonists, do not even work as antagonists.
3.2. Molecular Docking Investigation and In Silico ADMET Prediction.
The simulated binding mode at both the hA1 and hA2B AR structures, generally associated to the best score, presents the herein reported compounds oriented similarly to analogous derivatives previously reported as hAR ligands (Figure 2A) [12,35,36]. In detail, the pyridine scaffold is located in the AR cavity in correspondence to the purine moiety of Ado observed at the hA1AR cryo-EM structure and at the hA2AAR template, and interacts with the side chains of hA1AR residues Phe171 extracellular loop (EL) 2 and Ile2747.39 (Phe173 and Ile2767.39 in hA2BAR). For clarity, in this section, derivative 8 has been considered the reference compound to define the position of the substituents on the pyridine nucleus. Thus, starting from the N1 position, the amino and the sulfanyl functions occupy positions 2 and 6, respectively. The 3-cyano and 2-amino groups make H-bonds with the amide function of Asn2546.55. An additional H-bond is observed between the 2-amino group and the hA1AR residue Glu172 (EL2, Glu174 in hA2BAR), while the 5-cyano group points toward transmembrane (TM) 2 (i.e., Ala662.61 in hA1AR, Ala642.61 in hA2BAR) and TM7 (i.e., His2787.43 in hA1AR, His2807.43 in hA2BAR) residues. The 2-sulfanyl substituent is located at the entrance of the binding cavity, providing interaction with residues of TM1, TM2, and TM7. Binding data show that in general an aryl moiety at R1 is the type of substituent providing highest affinity at ARs. Its replacement with a smaller methyl group (compound 13) or a larger (and differently oriented) benzyl substituent (12) leads to a significant decrease (if not loss) of affinity. In this sense, the 4-aryl moiety well fits the depth of the binding cavity at both receptors, by occupying the available space in a more complete and efficient way with respect to the methyl (13) or benzyl group (12) (Figure 2B). Looking at the para-position of the 4-aryl moiety, biological data show that the size of the substituent at this level is critical for compound affinity, with the highest obtained with a small group. At the hA1AR receptor, the space available around the para-position is limited, allowing the insertion of small residues and not well tolerating ramified substituents. Accordingly, compounds bearing a bulky, non-linear para-substituent (compounds 4 and 6) are endowed with lower hA1AR affinity with respect to other compounds of the series active in the low nanomolar range.
At the hA2BAR, the impact of the size of the para-substituent on compound affinity appears analogous to that observed at the hA1AR, with the exception of compound 6 bearing a para-acetamido group. This compound is among the most potent derivatives of the series at the hA2BAR. Figure 3 shows the binding mode of compound 6 at the hA2BAR, with indication of the key residues involved in ligand interaction. The para-acetamido function engages a double H-bond interaction with Thr893.36 and His2516.52. This double polar interaction with the receptor is also given by the 5′-amide groups of the hA2AAR agonists NECA [29], UK-432097 (6-[2,2-di(phenyl)ethylamino]-9-[(2R,3R,4S,5S)-5-(ethylcarbamoyl)-3,4-dihydroxyoxolan-2-yl]-N-[2-[(1-pyridin-2-ylpiperidin-4-yl)carbamoylamino]ethyl]purine-2-carboxamide) [37] and CGS-21680 (4-[2-[[6-amino-9-(N-ethyl-β-D-ribofuranuronamidosyl)-9H-purin-2-yl]amino]ethyl]benzenepropanoic acid) [38], as observed from X-ray structures of the hA2AAR in complex with these two compounds.
The insertion of an amide group at the 5′-position of nucleosides is a typical modification of the Ado scaffold to improve its affinity and potency at the hA2A and hA2B ARs. This observation helps to explain the high activity of compound 6 at the hA2BAR.
The potential use of derivatives belonging to this series as therapeutic agents depends not only on their pharmacodynamics but also pharmacokinetics, the latter including Adsorption, Distribution, Metabolism, Excretion, and Toxicity (ADME/T). A preliminary study of the theoretical ADMET profiles of compound 3 was performed using the methodology developed by pKCSM, a novel approach to the prediction of pharmacokinetic and toxicity properties [39]. A complete summary of all the parameters evaluated for the selected compound is reported in the Supplementary Materials. It is worth noting that no violation of the Lipinski’s rule of five (RO5) (molecular weight (MW) < 500; log P < 5; number of H.-bond donors (HBD) < 5, and acceptors (HBA) < 10) was evidenced, making derivative 3 a promising drug-like candidate [40]. Nevertheless, while some bioavailability and toxicity parameters fall in the safety range, others indicate some points of concerns for compound 3. In interpreting the contrasting results, it is important to consider that in silico evaluation of the ADMET parameters for a group of approved antidiabetic drugs [41] evidenced not only violation of some of the RO5 (25%) but also the toxicity parameters as the major failures (2–33%).
4. Conclusions
This study, which has led to some amino-3,5-dicyanopyridines differently substituted at R1 and R2 positions, provides greater insight into the SARs of this series as AR ligands. In particular, it has emerged that the affinity/activity at the hA1 and hA2B ARs depends strictly on the R1 substitution which also affects the selectivity versus the other subtypes. Moreover, the 1H-imidazol-2-yl group at R2 position was confirmed to be an essential feature for potent AR agonists. In fact, looking at the biological and pharmacological data of this series, it is usual to find a combined AR affinity/activity, even if most of the compounds have generally modest to null hA2A and hA3 AR affinity. Many of the dicyanopyridines herein reported interact with the hA2B subtype with IC50 values from 12 to 260 nM behaving as partial agonists. The best results were obtained in the hA1AR binding assays where most of the compounds show from high to good affinity at this receptor, the 2-amino-4-(4-fluorophenyl)-6-[(1H-imidazol-2-ylmethyl)sulfanyl]pyridine-3,5-dicarbonitrile (8) emerging as the most active compound with a partial agonist profile. The compounds showing a combined hA1/hA2B AR partial agonist activity, as derivatives 3, 8, and 11, deserve the greatest attention. In particular, the 2-amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-[4-(prop-2-en-1-yloxy)phenyl]pyridine-3,5-dicarbonitrile (3) is active in the low nanomolar range at both these subtypes and shows a good trend of selectivity versus both the hA2A and hA3 ARs. This combined hA1/hA2B partial agonist activity, leading to a synergistic effect on glucose homeostasis, could be of interest in the development of potentially useful agents for treating diabetes and its complications. The in silico calculated ADMET profiles indicated that some bioavailability and toxicity parameters fall in the safety range, while others suggest some points of concerns for compound 3. Thus, further studies will be devoted in the future to thoroughly investigate on the toxicity of compound 3. We are however confident in considering 3 as a promising lead for the development of new antidiabetics with improved pharmacokinetics and safer therapeutic potential.
5. Materials and Methods
5.1. Chemistry
5.1.1. General Methods
Analytical silica gel plates (Merck F254, Sigma-Aldrich, Milan, Italy), preparative silica gel plates (Merck F254, 2 mm), and silica gel 60 (Merck, 70-230 mesh) were used for analytical and preparative TLC, and for column chromatography, respectively. All melting points were determined on a Gallenkamp (U.K) melting point apparatus and are uncorrected. Elemental analyses were performed with a Flash E1112 Thermo Finnigan elemental analyzer for C, H, abd N (Thermo Fisher Scientific, Milan, Italy), and the results were within ±0.4% of the theoretical values. All final compounds revealed purity not less than 95%. The IR spectra were recorded with a Perkin-Elmer Spectrum RX I spectrometer (Perkin-Elmer, Milan, Italy) in Nujol mulls and are expressed in cm-1. NMR spectra were recorded on a Bruker Avance 400 spectrometer (Bruker, Milan, Italy) (400 MHz for 1H NMR and 100 MHz for 13C NMR). The chemical shifts are reported in δ (ppm) and are relative to the central peak of the residual non-deuterated solvent, which was DMSO-d6. The following abbreviations are used: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad, Ar = aromatic protons. Compounds 28–32 were synthesized as reported in reference [12]. When available, melting point and/or 1H NMR values were in accordance to literature data.
5.1.2. Synthesis of the target compounds 1–4, 6–13, and 20
Sodium hydrogen carbonate (2 mmol) and the suitable halomethyl-derivative (1 mmol) were added to a solution of the suitable mercapto-compound (28-40 [12,14,18,20,21,22,23], 1 mmol) in anhydrous DMF (1 mL). The reaction mixture was stirred at room temperature (rt) until disappearance of the starting material (TLC monitoring). Then, water was added (25 mL) to precipitate a solid that was collected by filtration and washed with water. The crude product was triturated with Et2O (5 mL), collected by filtration and purified.
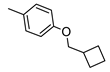
2-Amino-4-[4-(cyclobutylmethoxy)phenyl]-6-[(1H-imidazol-2-ylmethyl)sulfanyl]pyridine-3,5-dicarbonitrile (1)
The crude product was purified by preparative TLC using EtOAc/cyclohexane/MeOH 6:4:1 as eluting system. Yield 20%; mp 214-216 °C. 1H NMR (DMSO-d6) 11.87 (s, 1H, NH); 8.10 (s, 2H, NH2); 7.46 (d, 2H, Ar, J = 7.8 Hz); 7.09 (d, 2H, Ar, J = 7.8 Hz); 7.08 (s, 1H, Ar); 6.96 (s, 1H, Ar); 4.49 (s, 2H, CH2); 4.03 (d, 2H, OCH2, J = 6.0 Hz); 2.70-2.60 (m, 1H, CH); 2.08-2.07 (m, 2H, CH2); 1.91-1.88 (m, 4H, 2CH2). IR 3385; 3320; 3217; 2210. Anal. Calc. for C22H20N6OS
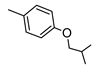
2-Amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-[4-(2-methylpropoxy)phenyl]pyridine-3,5-dicarbonitrile (2)
The crude product was purified by preparative TLC using EtOAc/cyclohexane /MeOH 5:5:1 as eluting system. Yield 31%; mp 230-232 °C. 1H NMR (DMSO-d6) 11.84 (s, 1H, NH); 8.14 (s, 2H, NH2), 7.47 (d, 2H Ar, J = 8.7 Hz); 7.10 (d, 2H Ar, J = 8.6 Hz); 7.08 (s, 1H, Ar); 6.85 (s, 1H, Ar); 4.50 (s, 2H, CH2); 3.84 (d, 2H, OCH2, J = 6.3 Hz); 2.09-2.02 (m, 1H, CH); 1.00 (d, 6H, 2 CH3, J = 6.7 Hz); IR 3387; 3315; 3215; 2208. Anal. Calc. for C21H20N6OS
2-Amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-[4-(prop-2-en-1-yloxy)phenyl]pyridine-3,5-dicarbonitrile (3)
The crude product was purified by preparative TLC using DCM/MeOH 9:1 as eluting system. Yield 21%; mp 216-218 °C. 1H NMR (DMSO-d6) 11.86 (s, 1H, NH); 8.09 (s, 2H, NH2); 7.48 (d, 2H Ar, J = 8.5 Hz); 7.12 (d, 2H, Ar, J = 8.6 Hz); 7.07 (s, 1H, Ar); 6.85 (s, 1H, Ar); 6.11-6.04 (m, 1H, CH); 5.40 (d, 1H, CH); 5.30 (d, 1H, CH); 4.62 (d, 2H, OCH2, J = 5.2 Hz); 4.48 (s, 2H, CH2). Anal. Calc. for C20H16N6OS
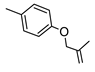
2-Amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-{4-[(2-methylprop-2-en-1-yl)oxy]phenyl} pyridine-3,5-dicarbonitrile (4)
The crude product was purified by column chromatography using EtOAc/cyclohexane /MeOH 6:3:1 as eluting system. Yield 36%; mp 224-226 °C. 1H NMR (DMSO-d6) 11.85 (s, 1H, NH); 8.08 (s, 2H, NH2); 7.48 (d, 2H, Ar, J = 8.4 Hz); 7.12 (d, 2H Ar, J = 8.4 Hz); 7.07 (s, 1H, Ar); 6.85 (s, 1H, Ar); 5.09 (s, 1H, CH); 4.99 (d, 1H, CH); 4.56 (s, 2H, CH2); 4.49 (s, 2H, OCH2); 1.79 (s, 3H, CH3); 13C NMR (DMSO-d6) 167.19; 161.03; 160.95; 159.11; 143.71; 141.67; 131.75; 131.28; 126.93; 116.58; 115.93; 115.80; 113.79; 94.22; 87.00; 72.14; 27.75; 20.32; IR 3404; 3377; 3336; 2210. Anal. Calc. for C21H18N6OS
N-(4-{2-Amino-3,5-dicyano-6-[(1H-imidazol-2-ylmethyl)sulfanyl]pyridin-4-yl}phenyl) acetamide (6)
The crude product was purified by column chromatography using EtOAc/cyclohexane /MeOH 8.2:1.5:0.9 as eluting system. Yield 61%; mp 275-278 °C. 1H NMR (DMSO-d6) 11.84 (s, 1H, NH); 10.22 (s, 1H, NH); 8.11 (s, 2H, NH2); 7.73 (d, 2H, Ar, J = 8.3 Hz); 7.46 (d, 2H, ar, J = 8.4 Hz); 7.05 (s, 1H, Ar); 6.86 (s, 1H, Ar); 4.49 (s, 2H, CH2); 2.09 (s, 3H, CH3); IR 3387; 3211; 2214; 1683. Anal. Calc. for C19H15N7OS
2-Amino-4-(4-chlorophenyl)-6-[(1H-imidazol-2-ylmethyl)sulfanyl]pyridine-3,5-dicarbonitrile (7)
Yield 42%; mp 229-231 °C dec (EtOAc). 1H NMR (DMSO-d6) 11.87 (br s, 1H, NH); 8.2 (br s, 2H, NH2); 7.65 (d, 2H, Ar, J = 8.0 Hz); 7.59 (d, 2H, Ar, J = 8.0 Hz); 7.08 (s, 1H, Ar); 6.85 (s, 1H, Ar); 4.50 (s, 2H, CH2); IR 3336; 3244; 2220; 1683. Anal. Calc. for C17H11ClN6S
2-Amino-4-(4-fluorophenyl)-6-[(1H-imidazol-2-ylmethyl)sulfanyl]pyridine-3,5-dicarbonitrile (8)
Yield 80%; mp 223-225 °C dec (EtOAc). 1H NMR (DMSO-d6) 11.9 (br s, 1H, NH); 8.2 (br s, 2H, NH2); 7.60 (m, 2H, Ar); 7.45 (t, 2H, Ar, J = 8.8 Hz); 7.0 (br s, 2H, Ar); 4.50 (s, 2H, CH2); IR 3314; 3145; 2229. Anal. Calc. for C17H11FN6S
2-Amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-[4-(trifluoromethyl)phenyl]pyridine-3,5-dicarbonitrile (9)
Yield 80%; mp 233-235 °C dec (EtOH). 1H NMR (DMSO-d6) 11.85 (br s, 1H, NH); 8.22 (br s, 2H, NH2); 7.97 (d, 2H, Ar, J = 7.8 Hz); 7.79 (d, 2H, Ar, J = 7.8 Hz); 7.08 (s, 1H, Ar); 6.86 (s, 1H, Ar);4.55 (s, 2H, CH2); IR 3527; 3185; 2437. Anal. Calc. for C18H11F3N6S
2-[(1H-Imidazol-2-il)metiltio]-6-ammino-4(4-metilfenil)piridina-3,5-dicarbonitrile (10)
Yield 67%; mp 259-261 °C dec (EtOH). 1H NMR (DMSO-d6) 11.85 (br s, 1H, NH); 8.14 (br s, 2H, NH2); 7.40 (d, 2H, Ar, J = 8.0 Hz); 7.38 (d, 2H, Ar, J = 8.0 Hz); 7.07 (br s, 2H, Ar); 4.50 (s, 2H, CH2); 2.39 (s, 3H, CH3). IR 3518; 3129; 2213. Anal. Calc. for C18H14N6S
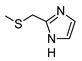
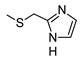
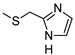
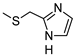
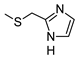
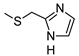
2-Amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-[4-(methylsulfanyl)phenyl]pyridine-3,5-dicarbonitrile (11)
Yield 95%; mp 257-259 °C dec (MeOH). 1H NMR (DMSO-d6) 11.87 (br s, 1H, NH); 8.12 (br s, 2H, NH2); 7.47 (d, 2H, Ar, J = 8.0 Hz); 7.42 (d, 2H, Ar, J = 8.0 Hz); 7.08 (br s, 1H, Ar); 6.85 (br s, 1H, Ar); 4.50 (s, 2H, CH2); 2.51 (s, 3H, CH3). 13C NMR (DMSO-d6) 166.65; 160.26; 158.33; 143.00; 142.31; 130.19; 129.43; 125.69; 115.81; 93.54; 86.35; 27.23; 14.47. IR 3420; 3390; 2223. Anal. Calc. for C18H14N6S2
2-Amino-4-benzyl-6-[(1H-imidazol-2-ylmethyl)sulfanyl]pyridine-3,5-dicarbonitrile (12)
Yield 70%; mp 246-248 °C dec (EtOH). 1H NMR (DMSO-d6) 11.82 (br s, 1H, NH); 8.14 (br s, 2H, NH2); 7.34 (d, 2H, Ar, J = 8.0 Hz); 7.24 (m, 3H, Ar); 6.95 (br s, 2H, Ar); 4.47 (s, 2H, CH2); 4.08 (s, 2H, CH2). 13C NMR (DMSO-d6) 166.73; 160.33; 159.10; 142.89; 136.61; 129.38; 128.78; 127.64; 115.61; 93.99; 86.95; 38.84; 27.15; 14.47. IR 3431; 3357; 2377. Anal. Calc. for C18H14N6S
2-Amino-6-[(1H-imidazol-2-ylmethyl)sulfanyl]-4-methylpyridine-3,5-dicarbonitrile (13)
Yield 66%; mp 267-269 °C dec (EtOH). 1H NMR (DMSO-d6) 11.87 (br s, 1H, NH); 8.12 (br s, 2H, NH2); 7.02 (br s, 2H, Ar); 4.51 (s, 2H, CH2), 2.43 (s, 3H, CH3). IR 3455; 3363; 2473. Anal. Calc. for C12H10N6S
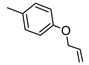
Methyl{[6-amino-3,5-dicyano-4-(4-methoxyphenyl)pyridin-2-yl]sulfanyl}acetate (20)
Yield 80%; mp 218-220 °C (EtOH). 1H NMR (DMSO-d6) 7.96 (br s, 2H, NH2); 7.51 (d, 2H, Ar, J = 8.8 Hz); 7.12 (d, 2H, Ar; J = 8.8 Hz)); 4.20 (s, 2H, CH2), 3.84 (s, 3H, CH3), 3.70 (s, 3H, CH3). IR 3426; 3397; 3325; 3224; 2209. Anal. Calc. for C17H14N4O3S
5.1.3. Synthesis of the Target Compounds 14–17
The suitable commercially available amino-derivative (1-(1H-imidazol-2-yl)methanamine, for compound 14; ethanolamine, for 15; 2-(4-methoxyphenyl)ethanamine, for 16; or imidazole (for compound 17 (1.18 mmol), and triethylamine (1.18 mmol) were sequentially added to a solution of the 2-chloro-derivative 42 [27] (0.59 mmol) in a mixture THF/EtOH 2:1 (7.5 mL). The reaction mixture was heated at reflux until the disappearance of the starting material (TLC monitoring) and then cooled at rt. After dilution with water (20 mL), a solid precipitated was collected and washed with water and Et2O.
2-Amino-6-[(1H-imidazol-2-ylmethyl)amino]-4-phenylpyridine-3,5-dicarbonitrile (14)
Yield 57%; mp 208-210 °C (EtOH). 1H NMR (DMSO-d6) 11.64 (br s, 1H, NH); 8.39 (br s, 2H, NH2); 7.88 (t, 1H, NH, J = 5.5 Hz); 7.61-7.45 (m, 5H, Ar); 7.04 (br s, 1H, Ar); 6.83 (br s, 1H, Ar);4.56 (d, 2H, CH2, J = 5.6 Hz). IR 3360; 3322; 3186; 2207. Anal. Calc. for C17H13N7
2-Amino-6-[(2-hydroxyethyl)amino]-4-phenylpyridine-3,5-dicarbonitrile (15)
Yield 64%; mp 230-231 °C (EtOH). 1H NMR (DMSO-d6) 7.55-7.53 (m, 3H, Ar); 7.47-7.45 (m, 2H, ar); 7.37 (br s, 2H, NH2); 7.20 (t, 1H, NH, J = 5.1 Hz); 4.74 (t, 1H, OH, J = 5.2 Hz); 3.54 (m, 2H, CH2); 3.47 (m, 2H, CH2). IR 3472; 3345; 3223; 2204. Anal. Calc. for C15H13N5O
2-Amino-6-{[2-(4-methoxyphenyl)ethyl]amino}-4-phenylpyridine-3,5-dicarbonitrile (16)
Yield 80%; mp ˃ 300 °C (EtOH/DMF). 1H NMR (DMSO-d6) 7.54-7.53 (m, 3H, Ar); 7.51-7.45 (m, 3H, 2 Ar + NH); 7.39 (br s, 2H, NH2); 7.20 (d, 2H, Ar, J = 8.4 Hz); 6.87 (d, 2H, Ar, J = 8.4 Hz); 3.73 (s, 3H, CH3); 3.57-3.51 (m, 2H, CH2); 2.80 (t, 2H, CH2, J = 8.0 Hz). IR 3451; 3342; 3244; 2201. Anal. Calc. for C22H19N5O
2-Amino-6-(1H-imidazol-1-yl)-4-phenylpyridine-3,5-dicarbonitrile (17)
Yield 77%; mp 253-255 °C (EtOH). 1H NMR (DMSO-d6) 8.40 (br s, 2H, NH2), 8.36 (s, 1H, Ar), 7.81 (s, 1H, Ar), 7.63 (m, 5H, Ar), 7.18 (s, 1H, Ar). IR 3399; 2220; 1681;1466. Anal. Calc. for C16H10N6
5.1.4. Synthesis of 2-amino-4-phenylpyridine-3,5-dicarbonitrile (18)
Ten per cent Pd/C (0.1 g) was added to a warm solution of the 2-chloro-derivative 42 [27] (0.98 mmol, 0.25 g) in absolute EtOH (50 mL). The mixture was hydrogenated in a Parr apparatus, at 30 Psi for 6 h. The catalyst was filtered off and the solvent was distilled to a small volume under reduced pressure to yield a yellow solid, which was collected and washed with Et2O. Yield 33%; mp 244-246 °C (2- Propanol). 1H NMR (DMSO-d6) 8.66 (s, 1H, ar), 8.10 (br s, 2H, NH2), 7.52-7.60 (m, 5H, ar). IR 3328; 3140; 2224; 1660. Anal. Calc. for C13H8N4
5.1.5. 2-Amino-6-[(2-hydroxyethyl)amino]-4-(4-methoxyphenyl)pyridine-3,5-dicarbonitrile (19)
A suspension of the 6-phenylsulfanyl derivative 27 [24] (1.81 mmol) and an excess of ethanolamine (7.25 mmol) in anhydrous DMF (7.8 mL) was heated at 100 °C for 5h. After cooling at rt, water (30 mL) was added. A solid precipitated, which was collected by filtration and washed with water and Et2O. Yield 90%; mp 248-250 °C (EtOH). 1H NMR (DMSO-d6) 7.43 (d, 2H, Ar, J = 8.8 Hz); 7.32 (br s, 2H, NH2); 7.14 (t, 1H, NH), 7.09 (d, 2H, Ar, J = 8.8 Hz)); 4.74 (t, 1H, OH), 3.84 (s, 3H, CH3), 3.56-3.53 (m, 2H, CH2), 3.50-3.47 (m, 2H, CH2). IR 3331; 3226; 2204. Anal. Calc. for C16H15N5O2
5.1.6. Synthesis of 4-substituted-2-amino-6-(phenylsulfanyl)pyridine-3,5-dicarbonitriles 21–27 [18,24–26]
A suspension of the suitable aldehyde (8.9 mmol), malononitrile (20.8 mmol), thiophenol (10.4 mmol), and basic alumina (0.019 mol) in water (20 mL) was stirred at 100 °C until the disappearance of the starting material (TLC monitoring). After cooling at rt, the aqueous layer was separated by decantation from the yellow sticky mass. The residue was treated with a mixture EtOH/Et2O 1:2 (3 mL) yielding a solid which was collected by filtration and extracted with boiling EtOAc (100 mLx2). After distillation of the combined and dried (Na2SO4) organic layers, the crude product was triturated with a little of Et2O/EtOH 1:1 and collected by filtration. The compounds were used without further purification unless otherwise stated.
2-Amino-4-(4-chlorophenyl)-6-(phenylsulfanyl)pyridine-3,5-dicarbonitrile (21) [24]
Yield 75%; mp 236-237 °C (lit mp 236-237 °C)
2-Amino-4-(4-fluorophenyl)-6-(phenylsulfanyl)pyridine-3,5-dicarbonitrile (22) [25]
Yield 78%; mp 220-222 °C (lit mp 221-223 °C)
2-Amino-6-(phenylsulfanyl)-4-[4-(trifluoromethyl)phenyl]pyridine-3,5-dicarbonitrile (23) [18]
Yield 64%; mp 199-201 °C (EtOH). 1HNMR (DMSO-d6) 7.98 (d, 2H, J = 7.8 Hz); 7.92 (br s, 2H, NH2); 7.82 (d, 2H, Ar, J = 7.8 Hz); 7.63-7.61 (m, 2H, Ar); 7.52-7.51 (m, 3H, Ar). IR 3320; 3211; 2243. Anal. Calc. for C20H11F3N4S
2-Amino-4-(4-methylphenyl)-6-(phenylsulfanyl)pyridine-3,5-dicarbonitrile (24) [24]
Yield 74%; mp 220-221 °C (lit mp 215-216 °C).
2-Amino-4-[4-(methylsulfanyl)phenyl]-6-(phenylsulfanyl)pyridine-3,5-dicarbonitrile (25) [26]
Yield 75%; mp 244-246 °C (lit mp 254-256 °C).
2-Amino-4-benzyl-6-(phenylsulfanyl)pyridine-3,5-dicarbonitrile (26)
Yield 70%; mp 247-249 °C (MeOH). 1HNMR (DMSO-d6) 7.81 (br s, 2H, NH2); 7.60-7.57 (m, 2H, Ar); 7.47-7.50 (m, 3H, Ar); 7.39-7.35 (m, 2H, Ar); 7.29-7.25 (m, 3H, Ar); 4.12 (s, 2H, CH2). IR 3329; 3225; 2198. Anal. Calc. for C20H14N4S
2-Amino-4-(4-methoxyphenyl)-6-(phenylsulfanyl)pyridine-3,5-dicarbonitrile (27) [24]
Yield 66%; mp 252-254 °C (lit mp 254-255 °C).
5.1.7. Synthesis of 2-amino-4-substituted-6-sulfanylpyridine-3,5-dicarbonitriles 33–40 [14,18,20,21]
An excess of anhydrous sodium sulfide (33 mmol) was added to a stirred solution of the suitable sulfanyl derivative 21-27 [18,24,25,26] (10 mmol) in anhydrous DMF (1 mL), maintained at rt and under nitrogen atmosphere. The reaction mixture was heated at 80 °C until the disappearance of the starting material (TLC monitoring). Then, 6N HCl (25 mL) was drop by drop added to obtain a precipitate, which was collected by filtration and washed with water (20 mL) and Et2O (5 mL). Compounds were used without further purification unless otherwise stated.
2-Amino-4-(4-chlorophenyl)-6-sulfanylpyridine-3,5-dicarbonitrile (33) [20,21]
Yield 80%; mp 202-204 °C (lit mp 110°C). 1HNMR (DMSO-d6) 13.12 (br s, 1H, SH); 8.01 (sl, 2H, NH2); 7.65 (d, 2H, Ar, J = 8.0 Hz); 7.58 (d, 2H, Ar, J = 8.0 Hz). IR 3320; 3192; 2220. Anal. Calc. for C13H7ClN4S
2-Amino-4-(4-fluorophenyl)-6-sulfanylpyridine-3,5-dicarbonitrile (34) [18]
Yield 86%; mp 234-236 °C. 1HNMR (DMSO-d6) 13.03 (br s, 1H, SH); 8.01(br s, 2H, NH2); 7.58-7.52 (m, 2H, Ar); 7.38 (m, 2H, Ar). IR 3319; 3182; 2224.
2-Amino-6-sulfanyl-4-[4-(trifluoromethyl)phenyl]pyridine-3,5-dicarbonitrile (35) [18]
Yield 90%; mp 212-214 °C (MeOH). 1HNMR (DMSO-d6) 13.15 (br s, 1H, SH); 8.07 (d, 2H, Ar, J = 8.0 Hz); 7.8 (d, 2H, Ar, J = 8.0 Hz). IR 3310; 3193; 2225.
2-Amino-4-(4-methylphenyl)-6-sulfanylpyridine-3,5-dicarbonitrile (36) [18]
Yield 83%; mp 228-230 °C. 1HNMR (DMSO-d6) 13.03 (br s, 1H, SH); 8.0 (br s, 2H, NH2); 7.41-7.37 (m, 4H, Ar). IR 3322; 3190; 2215.
2-Amino-4-[4-(methylsulfanyl)phenyl]-6-sulfanylpyridine-3,5-dicarbonitrile (37)
Yield 82%; mp 188-190 °C (EtOH). 1HNMR (DMSO-d6) 13.1 (br s, 1H, SH); 8.03 (br s, 2H, NH2), 7.46 (d, 2H, Ar, J = 8.0 Hz); 7.41 (d, 2H, Ar J = 8.0 Hz); 2.51 (s, 3H, CH3). IR 3311; 3195; 2222. Anal. Calc. for C14H10N4S2
2-Amino-4-benzyl-6-sulfanylpyridine-3,5-dicarbonitrile (38)
Yield 80%; mp 231-233 °C (EtOH). 1HNMR (DMSO-d6) 12.96 (br s, 1H, SH); 7.82 (br s, 2H); 7.37-7.35 (m, 2H, Ar); 7.30-7.28 (m, 3H, Ar); 4.01 (s, 2H, CH2). IR 3319; 3280; 2229. Anal. Calc. for C14H10N4S
2-Amino-4-(4-methoxyphenyl)-6-sulfanylpyridine-3,5-dicarbonitrile (40) [14]
Yield 80%; mp 261-262 °C (EtOH). 1H NMR (DMSO-d6) 7.96 (br s, 2H, NH2); 7.52 (d, 2H, Ar, J = 8.8 Hz); 7.13 (d, 2H, Ar; J = 8.8 Hz)); 3.85 (s, 3H, CH3). IR 3289; 3182; 2213.
5.1.8. Synthesis of 2-amino-4-methyl-6-sulfanylpyridine-3,5-dicarbonitrile (39) [22,23]
Compound 41 [22] (23 mmol) and 2-cyanothioacetamide (22.3 mmol) were consequentially added to an ethanolic solution of EtONa (570 mg of Na in 50 mL absolute EtOH). The reaction mixture was heated at reflux for 4 h. After evaporation of the solvent at small volume, a yellow solid precipitated which was collected under suction, washed with water and recrystallized. Yield 90%; mp 226-228 °C (EtOH) (lit mp 225 °C [23]; lit mp 120-123 °C [22]). 1H NMR (DMSO-d6) 12.94 (br s, 1H, SH); 7.1 (br s, 2H, NH2); 2.25 (s, 3H, CH3). IR 3338; 3253; 2248.
5.1.9. Synthesis of 2-amino-6-chloro-4-phenylpyridine-3,5-dicarbonitrile (42) [27]
Malononitrile (15.2 mmol) was portion-wise added to a solution of triethyl orthobenzoate (8mmol) and pyridine (0.6 mL) at 100 °C. Heating was continued for 6 h, and after cooling at 0 °C, 12 M HCl (2.7 mL) was drop by drop added to the mixture. The suspension was heated at 100 °C for 2 h, and the resulting precipitate was then collected by filtration and suspended in NaHCO3 saturated solution (5 mL). After stirring at room temperature for 30 min, the solid was filtered by suction and washed with water. Yield 61%; mp ˃ 300 °C (MeOH) (lit mp ˃ 250 °C). 1H NMR (DMSO-d6) 8.41 (br s, 2H, NH2), 7.61-7.57 (m, 5H, Ar); IR 3356; 3179; 2225;1660;1564.
5.2. Pharmacological Assays
5.2.1. Cell Culture and Membrane Preparation.
CHO cells transfected with hA1, hA2A, hA2B, and hA3 ARs were grown adherently and maintained in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, Milan, Italy) with nutrient mixture F12, containing 10% fetal calf serum, penicillin (100 U/mL), streptomycin (100 μg/mL), l-glutamine (2 mM) and geneticin (G418; 0.2 mg/mL) at 37°C in 5% CO2/95% air until the use in cAMP assays [42]. For membrane preparation, the culture medium was removed, and the cells were washed with phosphate-buffered saline and scraped off T75 flasks in ice-cold hypotonic buffer (5 mM Tris HCl, 1 mM EDTA, pH 7.4). The cell suspension was homogenized with a Polytron, centrifuged for 30 min at 40000 g at 4°C, and the resulting membrane pellet was used for competition binding experiments [42].
5.2.2. Competition Binding Experiments
All synthesized compounds have been tested for their affinity to hA1, hA2A, and hA3 ARs. Competition experiments to hA1 ARs were carried out incubating 1 nM [3H]-8-cyclopentyl-1,3-dipropylxanthine ([3H]-DPCPX) with membrane suspension (50 μg of protein/100 μL) and different concentrations of the examined compounds at 25°C for 90 min in 50 mM Tris HCl, pH 7.4. Non-specific binding was defined as binding in the presence of 1 μM DPCPX and was always < 10% of the total binding [42]. Inhibition experiments to hA2A ARs were performed incubating the radioligand [3H]-ZM241385 (4-(2-[7-Amino-2-(2-furyl)[1,2,4]triazolo[2,3-a][1,3,5]triazin-5-ylamino]ethyl)phenol) (1 nM) with the membrane suspension (50 μg of protein/100 μL) and different concentrations of the examined compounds for 60 min at 4°C in 50 mM Tris HCl (pH 7.4), 10 mM MgCl2. Non-specific binding was determined in the presence of ZM241385 (1 μM) and was about 20% of the total binding [43]. Competition binding experiments to A3 ARs were carried out incubating the membrane suspension (50 μg of protein/100 μL) with 0.5 nM [125I]-N6-(4-aminobenzyl)-N-methylcarboxamidoadenosine ([125I]-ABMECA) in the presence of different concentration of the examined compounds for an incubation time of 120 min at 4°C in 50 mM Tris HCl (pH 7.4), 10 mM MgCl2, and 1 mM EDTA. Non-specific binding was defined as binding in the presence of 1μM ABMECA and was always < 10% of the total binding [44]. Bound and free radioactivity were separated by filtering the assay mixture through Whatman GF/B glass fiber filters using a Brandel cell harvester (Brandel Instruments, Unterföhring, Germany). The filter bound radioactivity was counted by Packard Tri Carb 2810 TR scintillation counter (Perkin Elmer, Boston, MA, USA).
5.2.3. Cyclic AMP Assays.
CHO cells transfected with hAR subtypes were washed with phosphate-buffered saline, detached with trypsin and centrifuged for 10 min at 200 g. Cells were seeded in 96-well white half-area microplate (Perkin Elmer, Boston, USA) in a stimulation buffer composed of Hank Balanced Salt Solution (Sigma-Aldrich, Milan, Italy), 5 mM HEPES, 0.5 mM Ro 20-1724, and 0.1% BSA. cAMP levels were then quantified by using the AlphaScreen cAMP Detection Kit (Perkin Elmer) following the manufacturer’s instructions [45]. At the end of the experiments, plates were read with the Perkin Elmer EnSight Multimode Plate Reader.
5.2.4. Data Analysis
The protein concentration was determined according to a Bio-Rad method with bovine albumin as a standard reference. Inhibitory binding constant (Ki) values were calculated from those of IC50 according to Cheng & Prusoff equation Ki = IC50/(1+[C*]/KD*), where [C*] is the concentration of the radioligand and KD* its dissociation constant [44]. Ki and IC50 values were calculated by non-linear regression analysis using the equation for a sigmoid concentration-response curve (Graph-PAD Prism, San Diego, CA, USA).
5.3. Molecular Modelling
Homology modelling and energy minimization studies were carried out using MOE (Molecular Operating Environment, C.C.G., Inc., 1255 University St., Suite 1600, Montreal, Quebec, Canada, H3B 3X3, version 2019.0101) suite [30]. A homology model of the hA2B AR was built using the X-ray structure of the hA2A AR in complex with Ado as template (pdb code: 2YDO; 3.0-Å resolution [29]). A multiple alignment of the hAR primary sequences was built within MOE as preliminary step. The boundaries identified from the used X-ray crystal structure of hA2A AR were then applied for the corresponding sequences of the TM helices of the hA2B AR. The missing loop domains were built by the loop search method implemented in MOE. Once the heavy atoms were modelled, all hydrogen atoms were added, and the protein coordinates were then minimized with MOE using the AMBER10 force field until the Root Mean Square (RMS) gradient of the potential energy was less than 0.05 kJ mol−1 Å−1. Reliability and quality of the model were checked using the Protein Geometry Monitor application within MOE, which provides a variety of stereochemical measurements for inspection of the structural quality in a given protein, like backbone bond lengths, angles and dihedrals, Ramachandran φ-ψ dihedral plots, and quality of side chain rotamer and non-bonded contact. The cryo-EM structure of the agonist-bound hA1 AR (pdb code: 6D9H; 3.6-Å resolution [28]) was added of hydrogen atoms and energetically minimized following the same protocol above described; the coordinates of the heavy atoms were kept fixed in this task.
All compound structures were docked into the binding site of the hA2B and hA1 ARs structures using as docking tools the Induced Fit docking protocol of MOE and the genetic algorithm docking tool of CCDC Gold [31]. The Induced Fit docking protocol of MOE is divided into the following several stages:
- Conformational Analysis of ligands. The algorithm generated conformations from a single 3D conformation by conducting a systematic search. In this way, all combinations of angles were created for each ligand.
- Placement. A collection of poses was generated from the pool of ligand conformations using Alpha Triangle placement method. Poses were generated by superposition of ligand atom triplets and triplet points in the receptor binding site. The receptor site points are alpha sphere centres which represent locations of tight packing. At each iteration, a random conformation was selected, a random triplet of ligand atoms and a random triplet of alpha sphere centres were used to determine the pose.
- Scoring. Poses generated by the placement methodology were scored using the Alpha HB scoring function, which combines a term measuring the geometric fit of the ligand to the binding site and a term measuring hydrogen bonding effects.
- Induced Fit. The generated docking conformations were subjected to energy minimization within the binding site and the protein sidechains are included in the refinement stage. In detail, the protein backbone is set as rigid while the side chains are not set to “free to move” but are set to “tethered”, where an atom tether is a distance restraint that restrains the distance not between two atoms but between an atom and a fixed point in space.
- Rescoring. Complexes generated by the Induced Fit methodology stage were scored using the Alpha HB scoring function. Gold tool was used with default efficiency settings through MOE interface, by selecting GoldScore as scoring function [31].
Supplementary Materials
The following are available online at https://www.mdpi.com/1424-8247/12/4/159/s1: Table S1. Combustion analysis data of the newly synthesized compounds 1–4, 6–20, 26, 37, 38, and derivative 33. Table S2. Calculated molecular properties and ADMET parameters for compound 3.
Author Contributions
D.C. conceived and supervised the research work, and wrote the manuscript; F.V. (Flavia Varano) designed and performed the synthetic and analytical experiments; V.C. supervised the synthetic and analytical experiments, and analyze the data; K.V. and F.V. (Fabrizio Vincenzi) designed the pharmacological experiments, and process the data; S.P. performed the pharmacological experiments; D.D.B and R.V. conceived and performed the docking studies, and analyzed the results.
Funding
The work was financially supported by the Italian Ministry for University and Research (MIUR, PRIN 2010-2011, 20103W4779_004 project) and an intramural grant from the University of Florence (ex 60%).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ABMECA | N6-(4-Aminobenzyl)-N-methylcarboxamidoadenosine |
| AC | Adenylate Cyclase |
| ADMET | Absorption, Distribution, Metabolism, Excretion, Toxicity |
| Ado | Adenosine |
| AR | Adenosine Receptor |
| BSA | Bovine Serum Albumin |
| CCPA | 2-Chloro-N6-cyclopentyladenosine |
| CHO | Chinese Hamster Ovary |
| CPA | N6-Cyclopentyladenosine |
| DPCPX | 8-Cyclopentyl-1,3-dipropylxanthine |
| EL | Extracellular loop |
| EtOAc | Ethyl Acetate |
| DMF | Dimethylformamide |
| DMSO | Dimethylsulfoxide |
| EDTA | Ethylendiaminotetracetic Acid |
| HEPES | 4-(2-Hydroxyethyl)-1-piperazine-1-ethane sulfonic Acid |
| MOE | Molecular operating environment; |
| NECA | 5′-(N-Ethylcarboxamido)adenosine |
| rt | room temperature |
| SARs | Structure-Activity Relationships |
| THF | Tetrahydrofuran |
| TM | Transmembrane |
References
- Borea, P.A.; Gessi, S.; Merighi, S.; Varani, K. Adenosine as a multisignalling guardian angel in human diseases: When, where and how does it exert its protective effects? Trends Pharmacol. Sci. 2016, 37, 419–434. [Google Scholar] [CrossRef]
- Borea, P.A.; Gessi, S.; Merighi, S.; Vincenzi, F.; Varani, K. Pharmacology of adenosine receptors: The state of the art. Physiol. Rev. 2018, 98, 1591–1625. [Google Scholar] [CrossRef]
- Chen, J.F.; Eltzschig, H.K.; Fredholm, B.B. Adenosine receptors as drug targets—what are the challenges? Nature Rev. Drug Discov. 2013, 12, 265–286. [Google Scholar] [CrossRef]
- Müller, C.E.; Jacobson, K.A. Recent developments in adenosine receptor ligands ad their potential as novel drugs. Biochim. Biophys. Acta 2011, 1808, 1290–1308. [Google Scholar] [CrossRef] [PubMed]
- Dhalla, A.K.; Chisholm, J.W.; Reaven, G.M.; Belardinelli, L. A1 adenosine receptor: Role in diabetes and obesity. Handb. Exp. Pharmacol. 2009, 193, 271–295. [Google Scholar]
- Koupenova, M.; Ravid, K. Adenosine, adenosine receptors and their role in glucose homeostasis and lipid metabolism. J. Cell. Physiol. 2013, 228, 1703–1712. [Google Scholar] [CrossRef] [PubMed]
- Eisenstein, A.; Patterson, S.; Ravid, K. The many faces of the A2B adenosine receptor in cardiovascular and metabolic diseases. J. Cell. Physiol. 2015, 230, 2891–2897. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, M.; Zieleniak, A.; Wozniak, L.A. New insight into A1 adenosine receptors in diabetes treatment. Curr. Pharm. Design 2010, 16, 4237–4242. [Google Scholar] [CrossRef] [PubMed]
- Gessi, S.; Merighi, S.; Fazzi, D.; Stefanelli, A.; Varani, K.; Borea, P.A. Adenosine receptor targeting in health and disease. Expert Opin. Investig. Drug. 2011, 20, 1591–1609. [Google Scholar] [CrossRef] [PubMed]
- Johansson, S.M.; Lindgren, E.; Yang, J.N.; Herling, A.W.; Fredholm, B.B. Adenosine A1 receptors regulate lipolysis and lipogenesis in mouse adipose tissue-interactions with insulin. Eur. J. Pharmacol. 2008, 597, 92–101. [Google Scholar] [CrossRef]
- Rosentreter, U.; Henning, R.; Bauser, M.; Krämer, T.; Vaupel, A.; Hübsch, W.; Dembowsky, K.; Salcher-Schraufstätter, O.; Stasch, J.P.; Krahn, T.; et al. Substituted 2-Thio-3,5-Dicyano-4-Aryl-6-Aminopyridines and the use Thereof as Adenosine Receptor Ligands. WO/2001/025210. 2001. Available online: https://worldwide.espacenet.com/ (accessed on 15 October 2019).
- Betti, M.; Catarzi, D.; Varano, F.; Falsini, M.; Varani, K.; Vincenzi, F.; Dal Ben, D.; Lambertucci, C.; Colotta, V. The aminopyridine-3,5-dicarbonitrile core for the design of new non-nucleoside-like agonists of the human adenosine A2B receptor. Eur. J. Med. Chem. 2018, 150, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Fusco, I.; Cherchi, F.; Catarzi, D.; Colotta, V.; Varano, F.; Pedata, F.; Pugliese, A.M.; Coppi, E. Functional characterization of a novel adenosine A2B receptor agonist on short-term plasticity and synaptic inhibition during oxygen and glucose deprivation in the rat CA1 hippocampus. Brain Res. Bull. 2019, 151, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Beukers, M.W.; Chang, L.C.W.; von Frijtag Drabbe Kunzel, J.K.; Mulder-Krieger, T.; Spanjersberg, R.F.; Brussee, J.; IJzerman, A.P. New, non-adenosine, high-potency agonists for the human adenosine A2B receptor with an improved selectivity profile compared to the reference agonist N-ethylcarboxamidoadenosine. J. Med. Chem. 2004, 47, 3707–3709. [Google Scholar] [CrossRef] [PubMed]
- Betti, M.; Catarzi, D.; Varano, F.; Falsini, M.; Varani, K.; Vincenzi, F.; Pasquini, S.; di Cesare Mannelli, L.; Ghelardini, C.; Lucarini, E.; et al. Modifications on the amino-3,5-dicyanopyridine core to obtain multifaceted adenosine receptor ligands with antineuropathic activity. J. Med. Chem. 2019, 62, 6894–6912. [Google Scholar] [CrossRef] [PubMed]
- Baraldi, P.G.; Tabrizi, M.A.; Fruttarolo, F.; Romagnoli, R.; Preti, D. Recent improvement in the development of A2B adenosine receptor agonists. Purinergic Signal. 2009, 5, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Krahn, T.; Krämer, T.; Rosentreter, U.; Downey, J.M.; Solenkova, N. Use of substituted 2-thio-3,5-dicyano-4-phenyl-6-aminopyridines for the treatment of reperfusion injury and reperfusion damage. WO 2006099958 A1. 2006. Available online: https://worldwide.espacenet.com/ (accessed on 15 October 2019).
- Chang, L.C.W.; von Frijtag Drabbe Künzel, J.K.; Mulder-Krieger, T.; Spanjersberg, R.F.; Roerink, S.F.; van den Hout, G.; Beukers, M.W.; Brussee, J.; IJzerman, A.P. A series of ligand displaying a remarkable agonistic-antagonistic profile at the adenosine A1 receptor. J. Med. Chem. 2005, 48, 2045–2053. [Google Scholar] [CrossRef]
- Meibom, D.; Albrecht-Küpper, B.; Diedrichs, N.; Hübsch, W.; Kast, R.; Krämer, T.; Krenz, U.; Lerchen, H.-J.; Mittendorf, J.; Nell, P.G.; et al. Neladenoson bialanate hydrochloride: A prodrug of a partial adenosine A1 receptor agonist for the chronic treatment of heart disease. ChemMedChem. 2017, 12, 728–737. [Google Scholar] [CrossRef]
- El-Torgoman, A.M.; El-Kousy, S.M.; El-Shahat, K.Z. Nitriles in heterocyclic synthesis: The reaction of 2-(thiocarbamoyl)cinnamonitriles with active methylene reagents. Z. Naturforsch. B Chem. Sci. 1987, 42, 107–111. [Google Scholar] [CrossRef]
- Alinaghizadeh, F.; Zahedifar, M.; Seifi, M.; Sheibani, H. Cascade synthesis of thieno[2,3-b]pyridines by using intramolecular cyclization reactions of 3-cyano-2-(organylmethylthio)pyridines. J. Braz. Chem. Soc. 2016, 27, 663–669. [Google Scholar] [CrossRef]
- Ibrahim, D.A.; Ismail, N.S.M. Design, synthesis and biological study of novel pyrido[2,3-d]pyrimidine as anti-proliferative CDK2 inhibitors. Eur. J. Med. Chem. 2011, 46, 5825–5832. [Google Scholar] [CrossRef]
- Geies, A.A.; Kamal El-Dean, A.M.; Abdel Monem, M.I. Reinvestigation of the reaction of arylidenemalonitriles with cyanothioacetamide: New approach for the synthesis of pyridine derivatives. Z. Naturforsch. B. 1992, 47, 1438–1440. [Google Scholar] [CrossRef]
- Kambe, S.; Saito, K.; Sakurai, A.; Midorikawa, H. Synthetic studies using α,β-unsaturated nitriles: Facile synthesis of pyridine derivatives. Synthesis 1981, 7, 531–533. [Google Scholar] [CrossRef]
- Guo, K.; Thompson, M.J.; Reddy, T.R.K.; Mutter, R.; Chen, B. Mechanistic studies leading to a new procedure for rapid, microwave assisted generation of pyridine-3,5-dicarbonitrile libraries. Tetrahedron 2007, 63, 5300–5311. [Google Scholar] [CrossRef]
- Ranu, B.C.; Jana, R.; Sowmiah, S. An improved procedure for the three-component synthesis of highly substituted pyridines using ionic liquid. J. Org.Chem. 2007, 72, 3152–3154. [Google Scholar] [CrossRef]
- Murray, T.J.; Zimmerman, S.C.; Kolotuchin, S.V. Synthesis of heterocyclic compounds containing three contiguous hydrogen bonding sites in all possible arrangements. Tetrahedron 1995, 51, 635–648. [Google Scholar] [CrossRef]
- Draper-Joyce, C.J.; Khoshouei, M.; Thal, D.M.; Liang, Y.L.; Nguyen, A.T.N.; Furness, S.G.B.; Venugopal, H.; Baltos, J.A.; Plitzko, J.M.; Danev, R.; et al. Structure of the adenosine-bound human adenosine A1 receptor-Gi complex. Nature 2018, 558, 559–563. [Google Scholar] [CrossRef]
- Lebon, G.; Warne, T.; Edwards, P.C.; Bennett, K.; Langmead, C.J.; Leslie, A.G.; Tate, C.G. Agonist-bound adenosine A2A receptor structures reveal common features of GPCR activation. Nature 2011, 474, 521–525. [Google Scholar] [CrossRef]
- Molecular Operating Environment, C.C.G., Inc.: Montreal, QC, Canada, H3B 3X3.
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Rosentreter, U.; Krämer, T.; Shimada, M.; Hübsch, W.; Diedrichs, N.; Krahn, T.; Henninger, K.; Stasch, J.P. Substituted 2-thio-3,5-dicyano-4-phenyl-6-aminopyridines and their use as adenosine receptor-selective ligands. WO/2003/008384. 2003. Available online: https://worldwide.espacenet.com/ (accessed on 15 October 2019).
- Thimm, D.; Schiedel, A.C.; Sherbiny, F.F.; Hinz, S.; Hochheiser, K.; Bertarelli, D.C.G.; Maass, A.; Müller, C.E. Ligand-Specific Binding and Activation of the Human Adenosine A2B Receptor. Biochemistry 2013, 52, 726–740. [Google Scholar] [CrossRef]
- Dal Ben, D.; Buccioni, M.; Lambertucci, C.; Thomas, A.; Volpini, R. Simulation and comparative analysis of binding modes of nucleoside and non-nucleoside agonists at the A2B adenosine receptor. In Silico Pharmacol. 2013, 1, 24. [Google Scholar] [CrossRef]
- Dal Ben, D.; Buccioni, M.; Lambertucci, C.; Marucci, G.; Santinelli, C.; Spinaci, A.; Thomas, A.; Volpini, R. Simulation and Comparative Analysis of Different Binding Modes of Non-nucleoside Agonists at the A2A Adenosine Receptor. Mol. Inform. 2016, 35, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, D.; Gao, Z.G.; Moss, S.M.; Jacobson, K.A.; Carlsson, J. Molecular docking screening using agonist-bound GPCR structures: Probing the A2A adenosine receptor. J. Chem. Inf. Model. 2015, 55, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Wu, H.; Katritch, V.; Han, G.W.; Jacobson, K.A.; Gao, Z.G.; Cherezov, V.; Stevens, R.C. Structure of an agonist-bound human A2A adenosine receptor. Science 2011, 332, 322–327. [Google Scholar] [CrossRef] [PubMed]
- Lebon, G.; Bennett, K.; Jazayeri, A.; Tate, C.G. Thermostabilisation of an agonist-bound conformation of the human adenosine A2A receptor. J. Mol. Biol. 2011, 409, 298–310. [Google Scholar] [CrossRef]
- Pires, D.E.; Blundell, T.L.; Ascher, D.B. pkCSM: Predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015, 58, 4066–4072. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001, 46, 3–26. [Google Scholar] [CrossRef]
- Pereira, A.S.P.; den Haan, H.; Peña-García, J.; Moreno, M.M.; Pérez-Sánchez, H.; Apostolides, Z. Exploring african medicinal plants for potential anti-diabetic compounds with the DIA-DB inverse virtual screening web server. Molecules 2019, 24, 2002. [Google Scholar] [CrossRef]
- Vincenzi, F.; Targa, M.; Romagnoli, R.; Merighi, S.; Gessi, S.; Baraldi, P.G.; Borea, P.A.; Varani, K. TRR469, a potent A1 adenosine receptor allosteric modulator, exhibits anti-nociceptive properties in acute and neuropathic pain models in mice. Neuropharmacology 2014, 81, 6–14. [Google Scholar] [CrossRef]
- Varani, K.; Massara, A.; Vincenzi, F.; Tosi, A.; Padovan, M.; Trotta, F.; Borea, P.A. Normalization of A2A and A3 adenosine receptor up-regulation in rheumatoid arthritis patients by treatment with anti-tumor necrosis factor alpha but not methotrexate. Arthritis Rheum. 2009, 60, 2880–2891. [Google Scholar] [CrossRef]
- Varani, K.; Merighi, S.; Gessi, S.; Klotz, K.-N.; Leung, E.; Baraldi, P.G.; Cacciari, B.; Romagnoli, R.; Spalluto, G.; Borea, P.A. [3H]MRE 3008F20: A novel antagonist radioligand for the pharmacological and biochemical characterization of human A3 adenosine receptors. Mol. Pharmacol. 2000, 57, 968–975. [Google Scholar]
- Ravani, A.; Vincenzi, F.; Bortoluzzi, A.; Padovan, M.; Pasquini, S.; Gessi, S.; Merighi, S.; Borea, P.A.; Govoni, M.; Varani, K. Role and function of A2A and A3 adenosine receptors in patients with ankylosing spondylitis, psoriatic arthritis and rheumatoid arthritis. Int. J. Mol. Sci. 2017, 18, 697. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).