Machine Learning-Based Diabetic Neuropathy and Previous Foot Ulceration Patients Detection Using Electromyography and Ground Reaction Forces during Gait
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset Description
2.2. Signal Processing
2.3. Feature Extraction
2.3.1. EMG Feature Extraction
2.3.2. GRF Feature Extraction
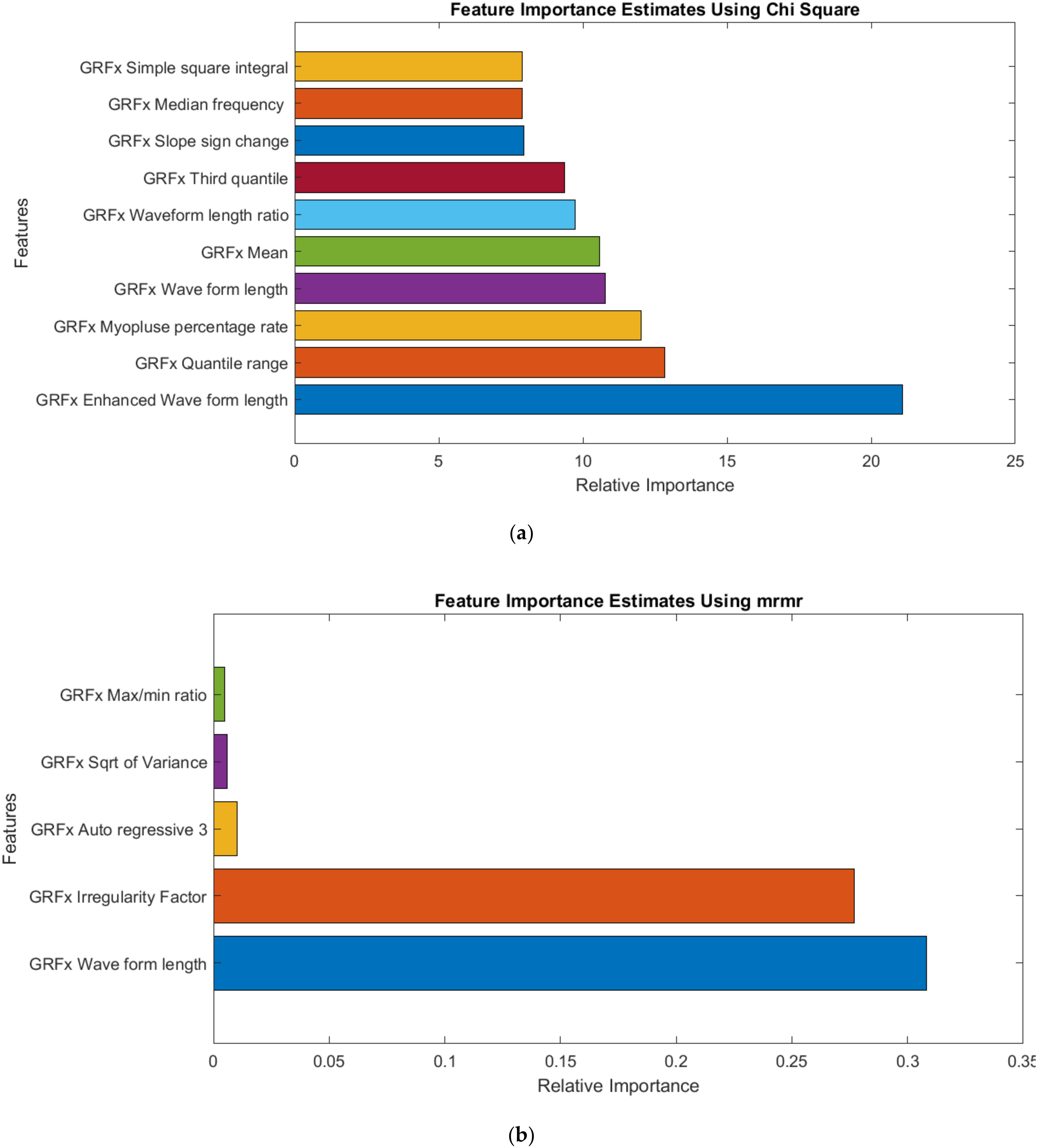
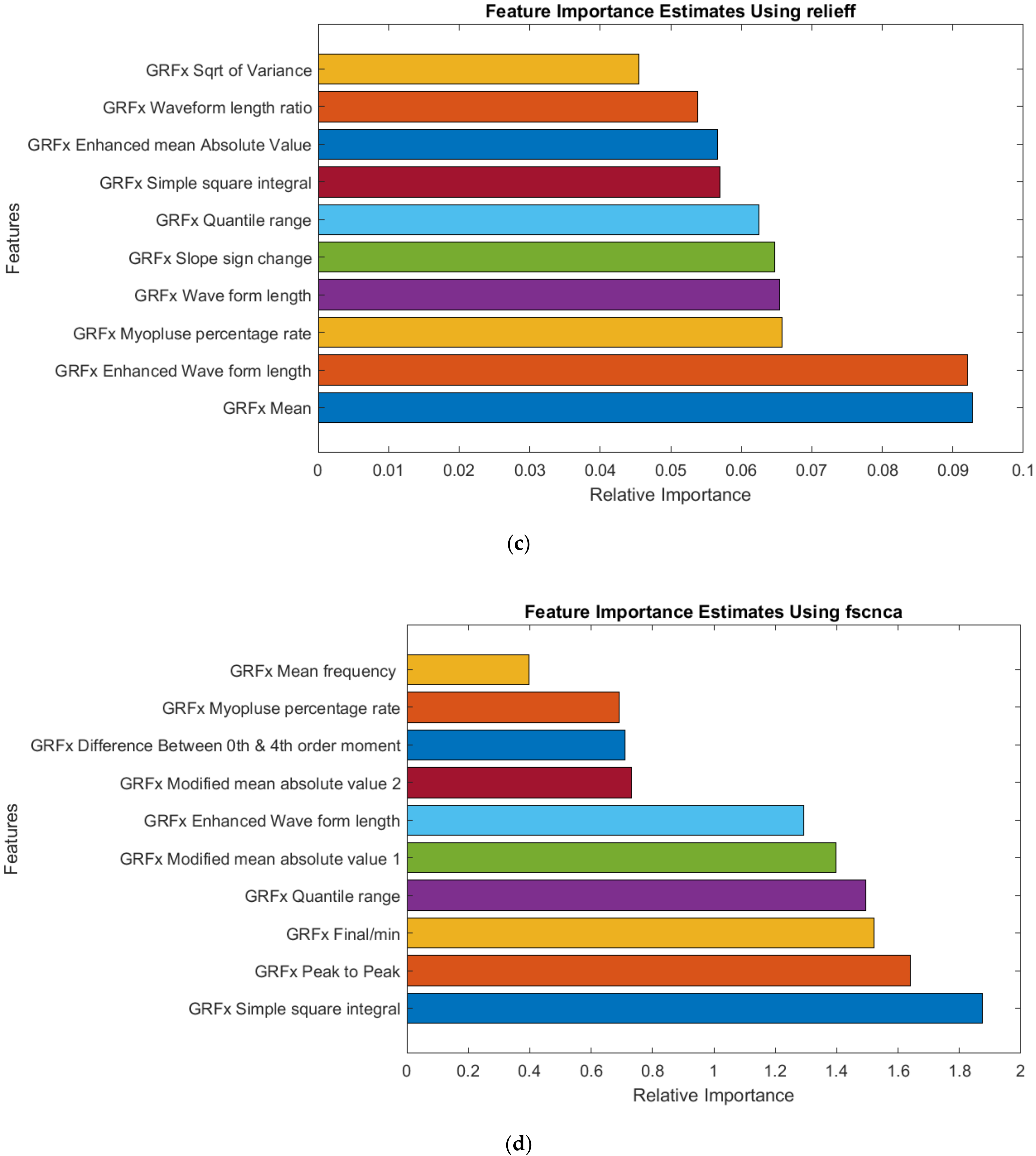
2.4. Feature Selection
2.5. Dimensionality Reduction
2.6. Machine Learning Models and Hyperparameter Tuning
2.7. ML Model Development
- Single Channel: Initially, the ML model was trained for individual muscle or GRF component data. So, we trained ML models for three individual muscle features (GL, VL, and TA) and three GRF component features (GRFx, GRFy, and GRFz) separately and observed the classification performance.
- Two Channel: Second, the ML model was trained with features combined from two muscles (GL and VL; GL and TA; and TA and VL) or two GRF components (GRFx and GRFy; GRFx and GRFz; and GRFy and GRFz).
- Three Channel: Last, we observed the performance of the ML models with all three muscles (GL, VL, and TA) or three GRF (GRFx, GRFy, and GRFz) components combined.
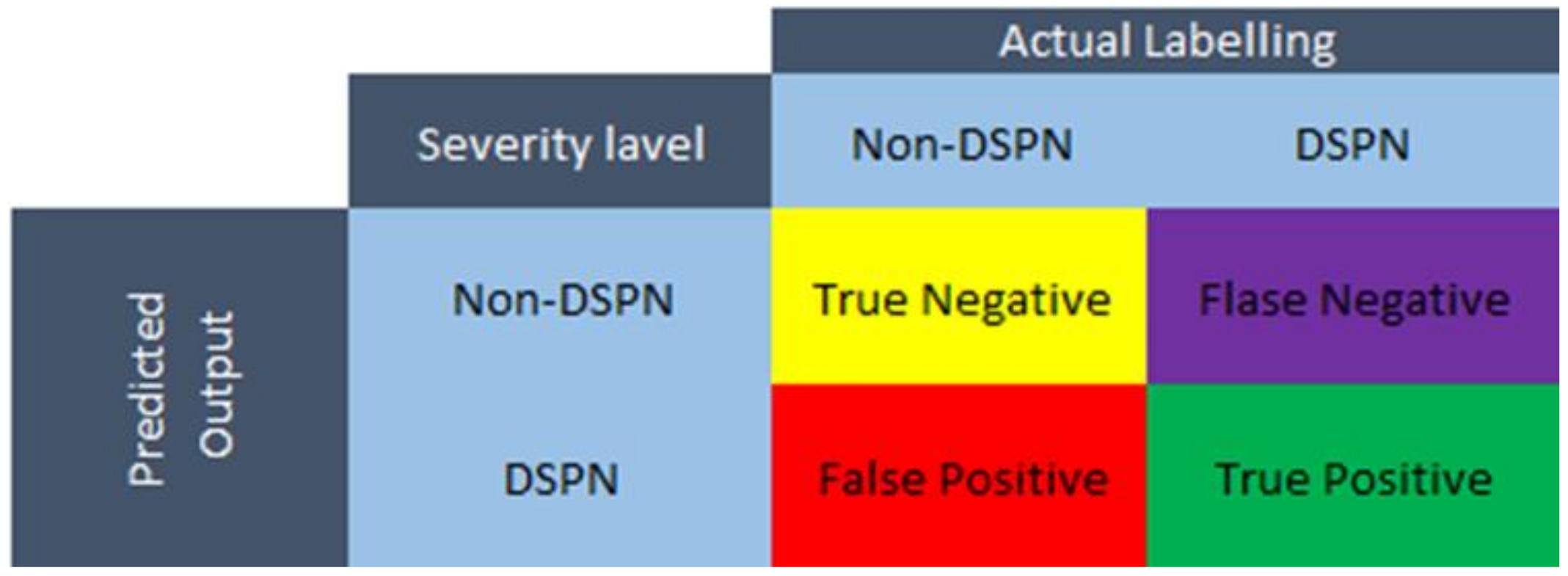
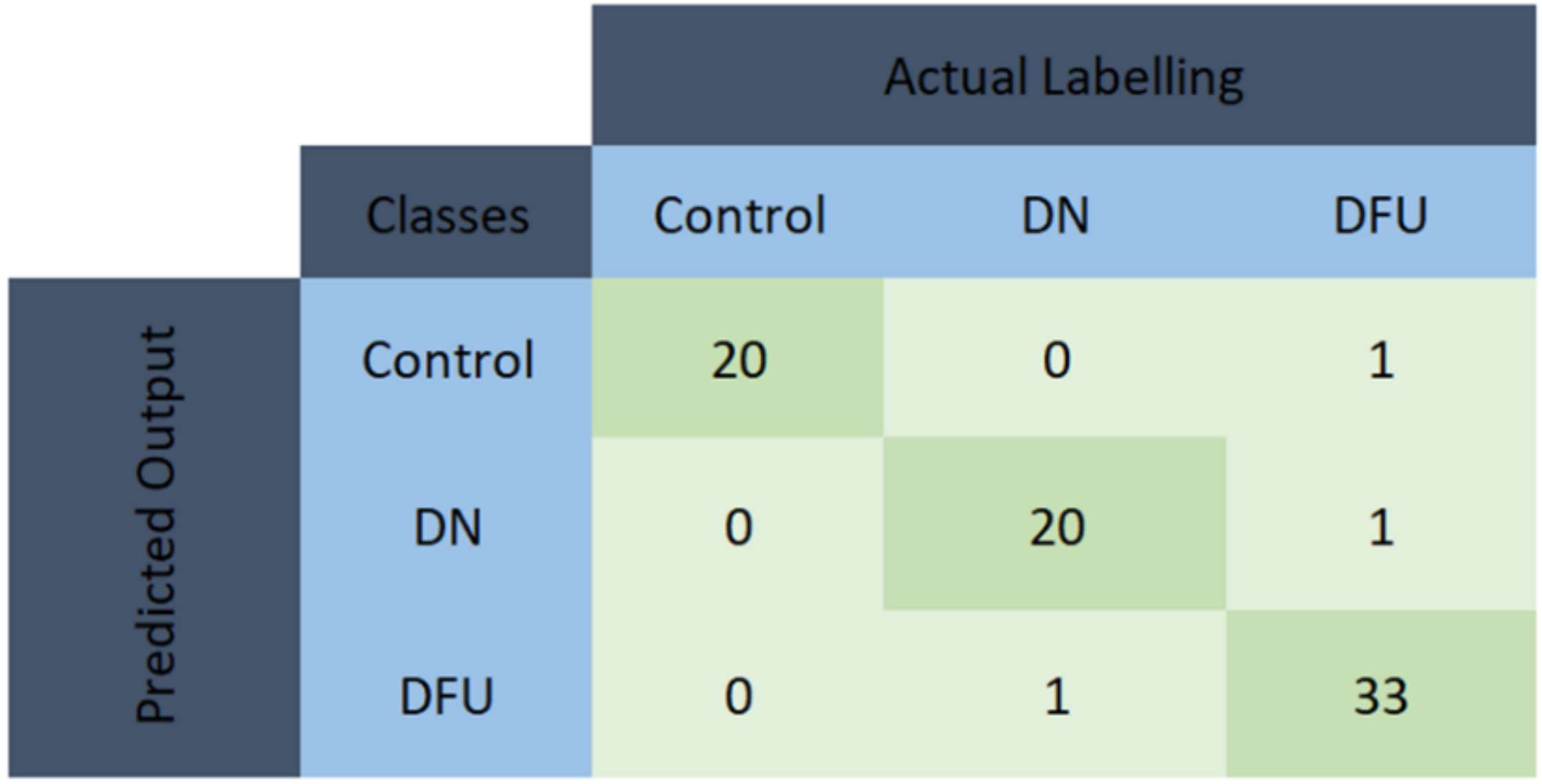
2.8. Performance Evaluation of ML Models
- True positive (TP): True DN patients
- True negative (TN): True Non-DN
- False-positive (FP): Non-DN patients, classified as DN patients.
- False-negative (FN): DN patients, classified as non-DN patients.
3. Results
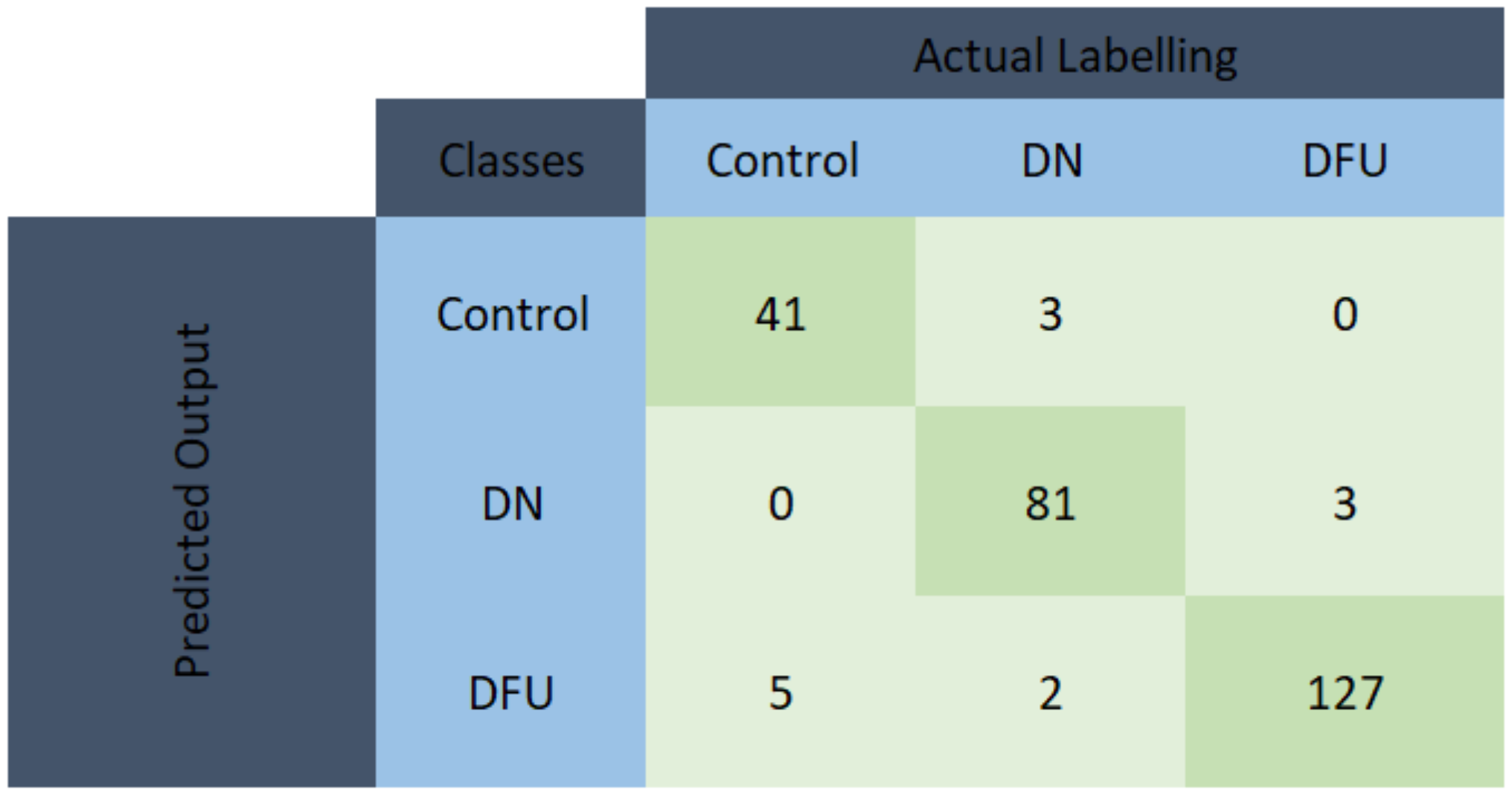
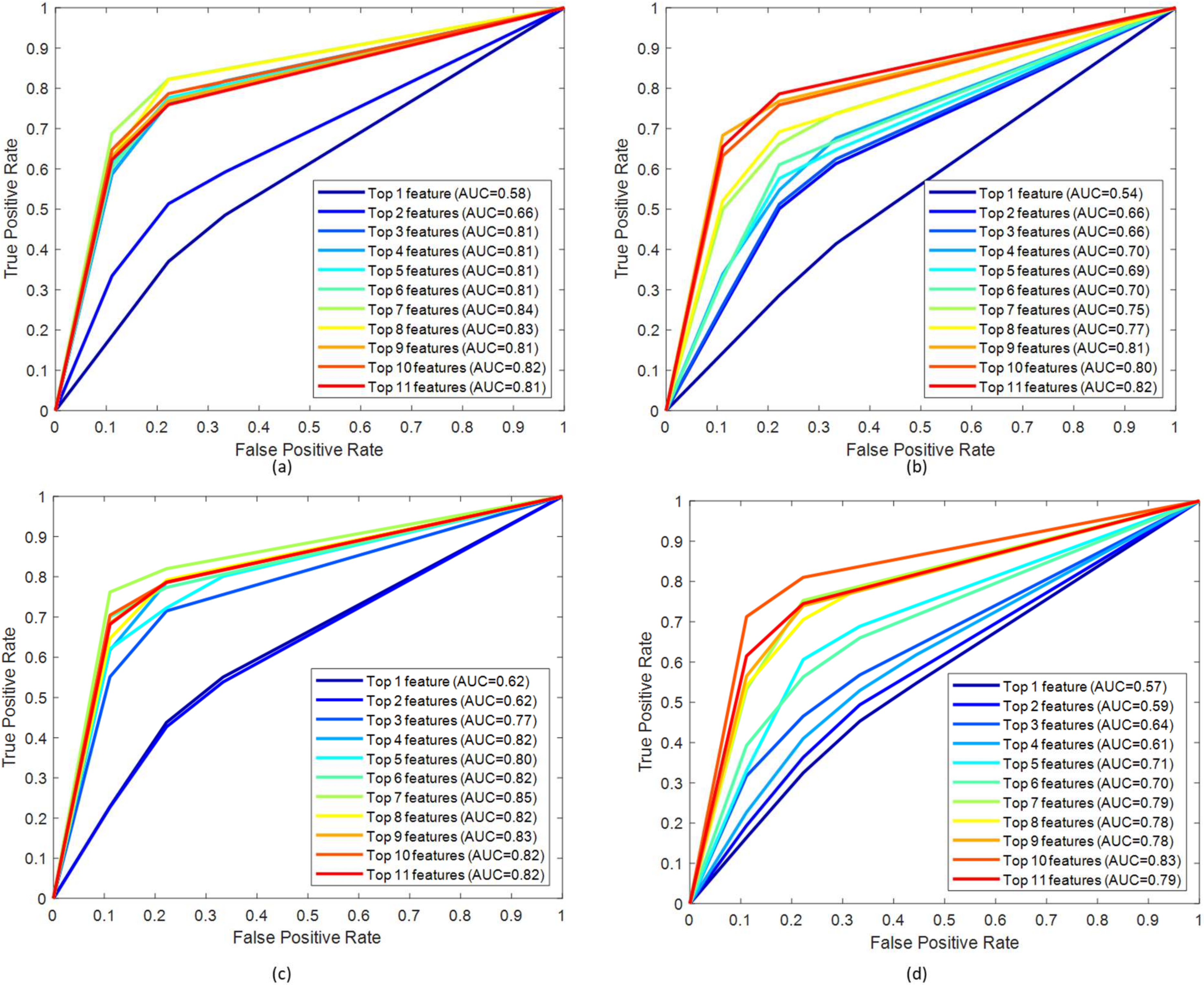
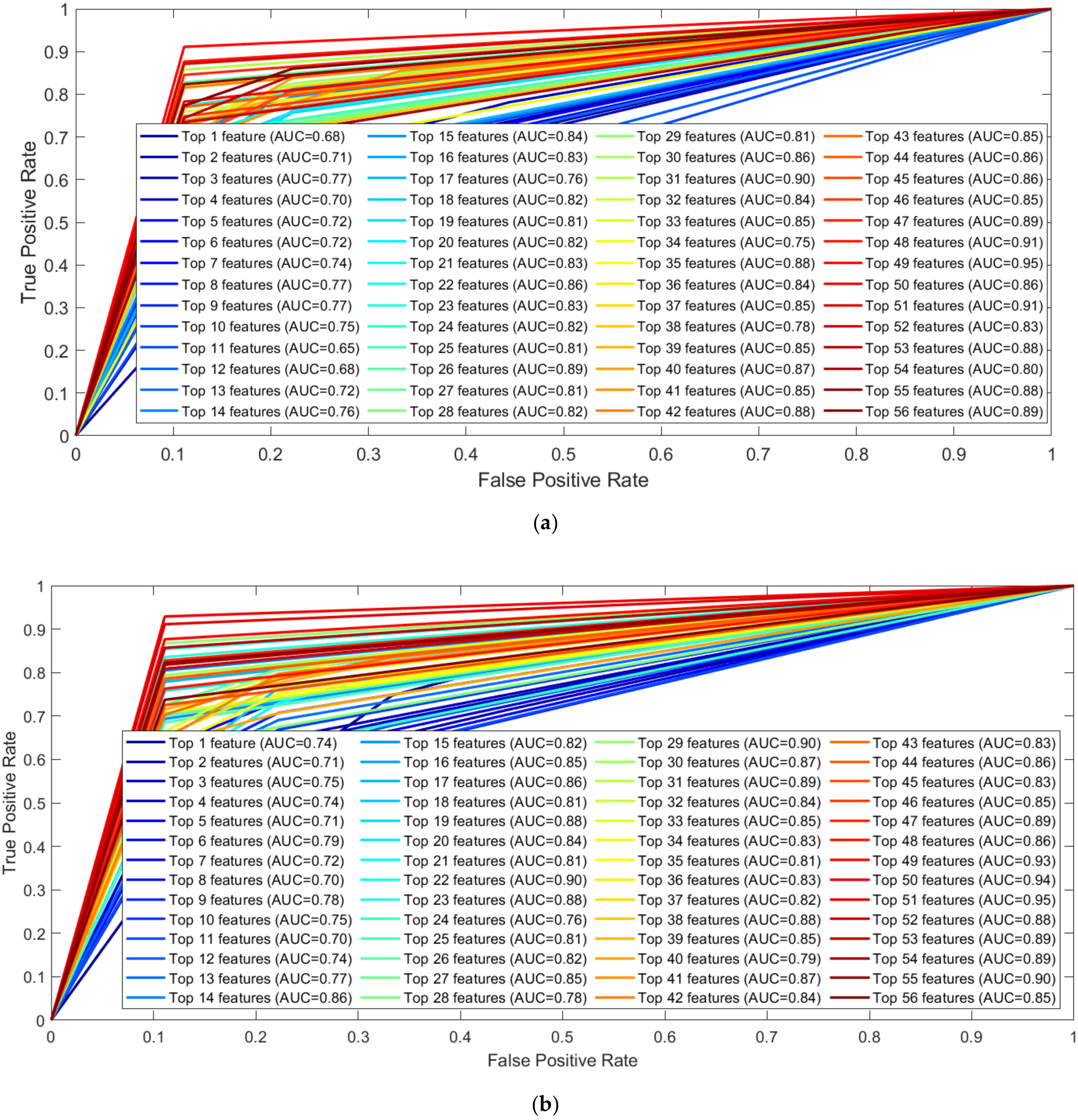
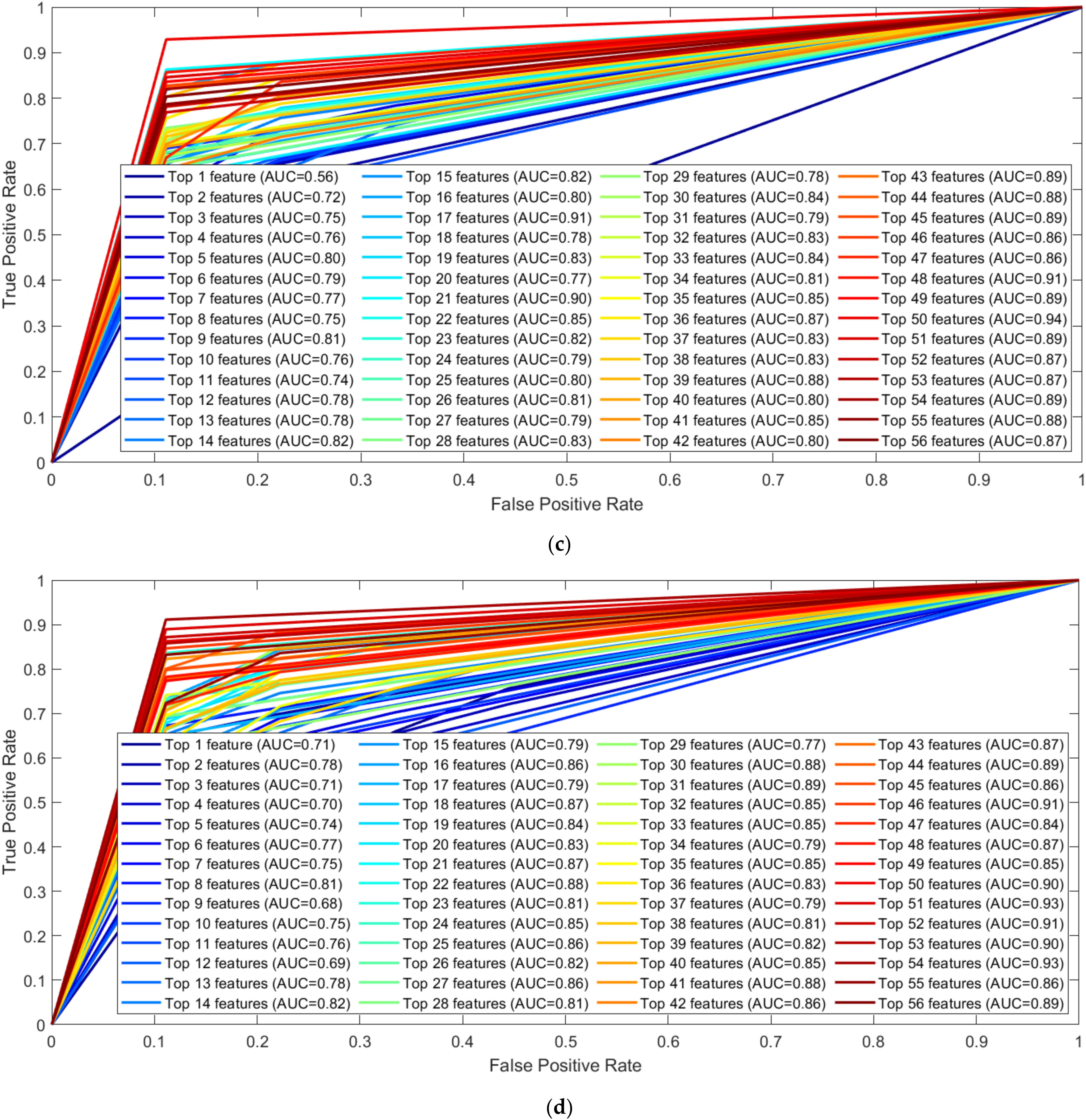
3.1. Performance Evaluation
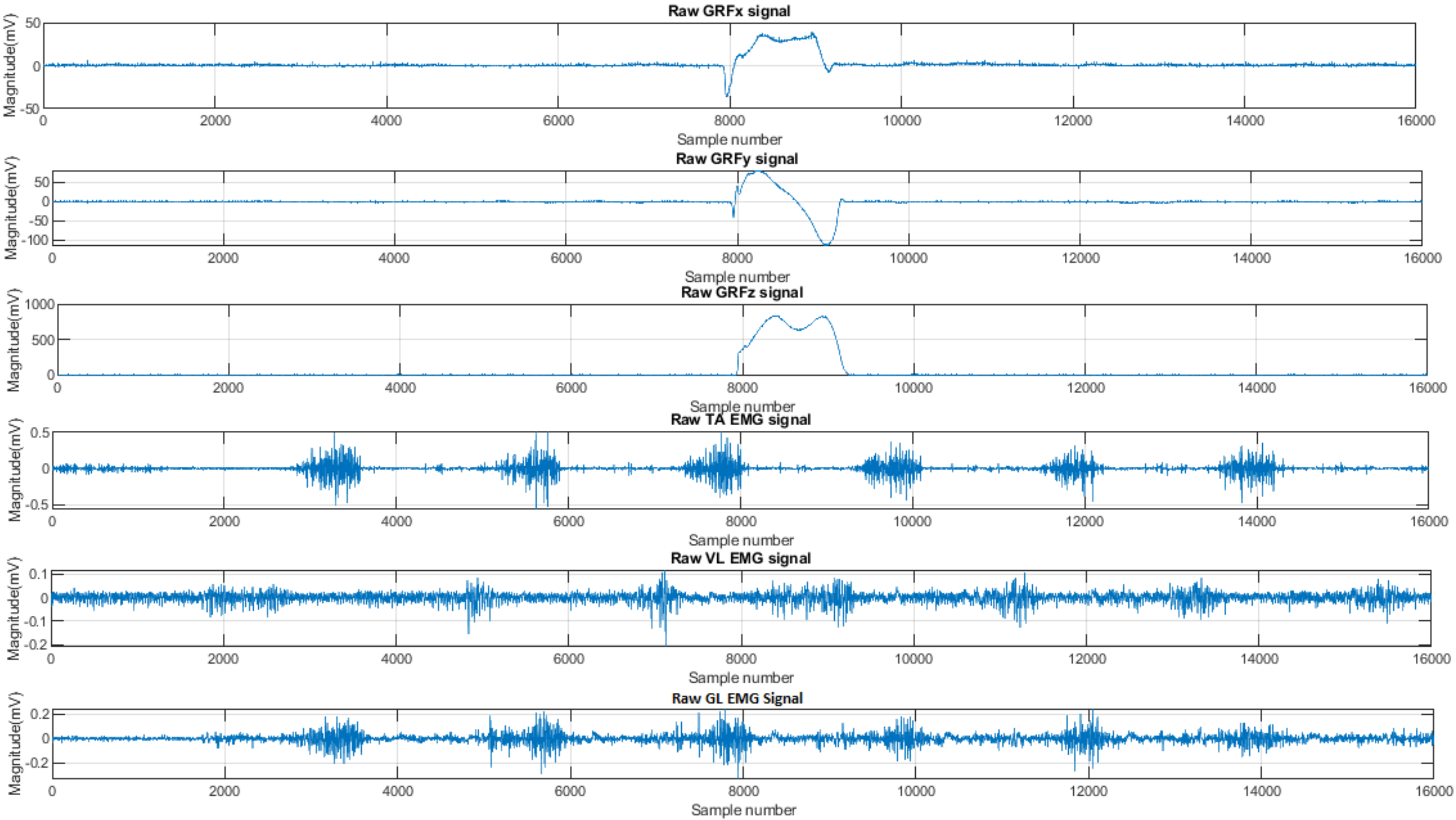
3.2. EMG Signal Analysis
3.3. GRF Signal Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- International Diabetes Federation. IDF Diabetes Atlas 10th ed. 2021. Available online: www.diabetesatlas.org (accessed on 7 March 2022).
- Tesfaye, S.; Boulton, A.J.M.; Dyck, P.J.; Freeman, R.; Horowitz, M.; Kempler, P.; Lauria, G.; Malik, R.A.; Spallone, V.; Vinik, A.; et al. Diabetic neuropathies: Update on definitions, diagnostic criteria, estimation of severity, and treatments. Diabetes Care 2010, 33, 2285–2293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pop-Busui, R.; Boulton, A.J.M.; Feldman, E.L.; Bril, V.; Freeman, R.; Malik, R.A.; Sosenko, J.M.; Ziegler, D. Diabetic neuropathy: A position statement by the American diabetes association. Diabetes Care 2017, 40, 136–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haque, F.; Reaz, M.B.I.; Ali, S.H.; Arsad, N.; Enamul, M.; Chowdhury, H. Performance analysis of noninvasive electrophysiological methods for the assessment of diabetic sensorimotor polyneuropathy in clinical research: A systematic review and meta - analysis with trial sequential analysis. Sci. Rep. 2020, 1–16. [Google Scholar] [CrossRef]
- Akashi, P.M.H.; Sacco, I.C.N.; Watari, R.; Hennig, E. The effect of diabetic neuropathy and previous foot ulceration in EMG and ground reaction forces during gait. Clin. Biomech. 2008, 23, 584–592. [Google Scholar] [CrossRef] [PubMed]
- Sacco, I.C.N.; Amadio, A.C. A study of biomechanical parameters in gait analysis and sensitive cronaxie of diabetic neuropathic patients. Clin. Biomech. 2000, 15, 196–202. [Google Scholar] [CrossRef]
- Onodera, A.N.; Gomes, A.A.; Pripas, D.; Mezzarane, R.A.; Sacco, I.C.N. Lower limb electromygraphy and kinematics of neuropathic diabetic patients during real-life activities: Stair negotiation. Muscle and Nerve 2011, 44, 269–277. [Google Scholar] [CrossRef]
- Sacco, I.C.; Akashi, P.M.; Hennig, E.M. A comparison of lower limb EMG and ground reaction forces between barefoot and shod gait in participants with diabetic neuropathic and healthy controls. BMC Musculoskelet. Disord. 2010, 11. [Google Scholar] [CrossRef]
- Haque, F.; Reaz, M.B.I.; Chowdhury, M.E.H.; Hashim, F.H.; Arsad, N.; Ali, S.H.M. Diabetic Sensorimotor Polyneuropathy Severity Classification Using Adaptive Neuro Fuzzy Inference System. IEEE Access 2021, 9, 7618–7631. [Google Scholar] [CrossRef]
- Haque, F.; Reaz, M.B.I.; Chowdhury, M.E.H.; Srivastava, G.; Ali, S.H.M.; Bakar, A.A.A.; Bhuiyan, M.A.S. Performance analysis of conventional machine learning algorithms for diabetic sensorimotor polyneuropathy severity classification. Diagnostics 2021, 11, 801. [Google Scholar] [CrossRef]
- Gomes, A.A.; Onodera, A.N.; Otuzi, M.E.I.; Pripas, D.; Mezzarane, R.A.; Sacco, I.C.N. Electromyography and kinematic changes of gait cycle at different cadences in diabetic neuropathic individuals. Muscle and Nerve 2011, 44, 258–268. [Google Scholar] [CrossRef]
- Watari, R.; Sartor, C.D.; Picon, A.P.; Butugan, M.K.; Amorim, C.F.; Ortega, N.R.S.; Sacco, I.C.N. Effect of diabetic neuropathy severity classified by a fuzzy model in muscle dynamics during gait. J. Neuroeng. Rehabil. 2014, 11, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bacarin, T.A.; Sacco, I.C.N.; Hennig, E.M. Plantar pressure distribution patterns during gait in diabetic neuropathy patients with a history of foot ulcers. Clinics 2009, 64, 113–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawacha, Z.; Spolaor, F.; Guarneri, G.; Contessa, P.; Carraro, E.; Venturin, A.; Avogaro, A.; Cobelli, C. Abnormal muscle activation during gait in diabetes patients with and without neuropathy. Gait Posture 2012, 35, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Sawacha, Z.; Gabriella, G.; Cristoferi, G.; Guiotto, A.; Avogaro, A.; Cobelli, C. Diabetic gait and posture abnormalities: A biomechanical investigation through three dimensional gait analysis. Clin. Biomech. 2009, 24, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Spolaor, F.; Sawacha, Z.; Guarneri, G.; Del Din, S.; Avogaro, A.; Cobelli, C. Altered EMG patterns in diabetic neuropathic and not neuropathic patients during step ascending and descending. J. Electromyogr. Kinesiol. 2016, 31, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Fernando, M.; Crowther, R.; Lazzarini, P.; Sangla, K.; Cunningham, M.; Buttner, P.; Golledge, J. Biomechanical characteristics of peripheral diabetic neuropathy: A systematic review and meta-analysis of findings from the gait cycle, muscle activity and dynamic barefoot plantar pressure. Clin. Biomech. 2013, 28, 831–845. [Google Scholar] [CrossRef]
- Petropoulos, I.N.; Alam, U.; Fadavi, H.; Marshall, A.; Asghar, O.; Dabbah, M.A.; Chen, X.; Graham, J.; Ponirakis, G.; Boulton, A.J.M.; et al. Rapid automated diagnosis of diabetic peripheral neuropathy with in vivo corneal confocal microscopy. Investig. Ophthalmol. Vis. Sci. 2014, 55, 2062–2070. [Google Scholar] [CrossRef]
- Gomes, A.A.; Ackermann, M.; Ferreira, J.P.; Orselli, M.I.V.; Sacco, I.C.N. Muscle force distribution of the lower limbs during walking in diabetic individuals with and without polyneuropathy. J. Neuroeng. Rehabil. 2017, 14, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Sacco, I.C.N.; Amadio, A.C. Influence of the diabetic neuropathy on the behavior of electromyographic and sensorial responses in treadmill gait. Clin. Biomech. 2003, 18, 426–434. [Google Scholar] [CrossRef]
- Kwon, O.Y.; Minor, S.D.; Maluf, K.S.; Mueller, M.J. Comparison of muscle activity during walking in subjects with and without diabetic neuropathy. Gait Posture 2003, 18, 105–113. [Google Scholar] [CrossRef]
- Abboud, R.J.; Rowley, D.I.; Newton, R.W. Lower limb muscle dysfunction may contribute to foot ulceration in diabetic patients. Clin. Biomech. 2000, 15, 37–45. [Google Scholar] [CrossRef]
- Sawacha, Z.; Guarneri, G.; Avogaro, A.; Cobelli, C. A new classification of diabetic gait pattern based on cluster analysis of biomechanical data. J. Diabetes Sci. Technol. 2010, 4, 1127–1138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernando, M.E.; Crowther, R.G.; Lazzarini, P.A.; Sangla, K.S.; Buttner, P.; Golledge, J. Gait parameters of people with diabetes-related neuropathic plantar foot ulcers. Clin. Biomech. 2016, 37, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Mustapa, A.; Justine, M.; Mustafah, N.M.; Manaf, H. The Effect of Diabetic Peripheral Neuropathy on Ground Reaction Forces during Straight Walking in Stroke Survivors. Rehabil. Res. Pract. 2017, 2017, 1–9. [Google Scholar] [CrossRef]
- Savelberg, H.H.C.M.; Ilgin, D.; Angin, S.; Willems, P.J.B.; Schaper, N.C.; Meijer, K. Prolonged activity of knee extensors and dorsal flexors is associated with adaptations in gait in diabetes and diabetic polyneuropathy. Clin. Biomech. 2010, 25, 468–475. [Google Scholar] [CrossRef]
- Allen, L.; Powell-Cope, G.; Mbah, A.; Bulat, T.; Njoh, E. A Retrospective Review of Adverse Events Related to Diabetic Foot Ulcers e. Ostomy Wound Manag. 2017, 63, 30–33. [Google Scholar]
- Fernando, M.E.; Crowther, R.G.; Cunningham, M.; Lazzarini, P.A.; Sangla, K.S.; Golledge, J. Lower limb biomechanical characteristics of patients with neuropathic diabetic foot ulcers: The diabetes foot ulcer study protocol. BMC Endocr. Disord. 2015, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Weledji, E.P.; Fokam, P. Treatment of the diabetic foot - to amputate or not? BMC Surg. 2014, 14, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Reaz, M.B.I.; Hussain, M.S.; Mohd-Yasin, F. Techniques of EMG signal analysis: Detection, processing, classification and applications. Biol. Proced. Online 2006, 8, 11–35. [Google Scholar] [CrossRef] [Green Version]
- Velliangiri, S.; Alagumuthukrishnan, S.; Thankumar Joseph, S.I. A Review of Dimensionality Reduction Techniques for Efficient Computation. Procedia Comput. Sci. 2019, 165, 104–111. [Google Scholar] [CrossRef]
- Islam, M.J.; Ahmad, S.; Haque, F.; Reaz, M.B.I.; Bhuiyan, M.A.S.; Islam, M.R. A Novel Signal Normalization Approach to Improve the Force Invariant Myoelectric Pattern Recognition of Transradial Amputees. IEEE Access 2021. [CrossRef]
- Islam, M.J.; Ahmad, S.; Haque, F.; Reaz, M.B.I.; Bhuiyan, M.A.S.; Islam, M.R. Force-invariant improved feature extraction method for upper-limb prostheses of transradial amputees. Diagnostics 2021, 11, 843. [Google Scholar] [CrossRef] [PubMed]
- Buongiorno, D.; Cascarano, G.D.; De Feudis, I.; Brunetti, A.; Carnimeo, L.; Dimauro, G.; Bevilacqua, V. Deep learning for processing electromyographic signals: A taxonomy-based survey. Neurocomputing 2021, 452, 549–565. [Google Scholar] [CrossRef]
- Xiong, D.; Zhang, D.; Zhao, X.; Zhao, Y. Deep Learning for EMG-based Human-Machine Interaction: A Review. IEEE/CAA J. Autom. Sin. 2021, 8, 512–533. [Google Scholar] [CrossRef]
- Shehata, A.W.; Williams, H.E.; Hebert, J.S.; Pillarski, P.M. Machine Learning for the Control of Prosthetic Arms: Using Electromyographic Signals for Improved Performance. IEEE Signal Process. Mag. 2021, 38, 46–53. [Google Scholar] [CrossRef]
- Islam, J.; Ahmad, S.; Haque, F.; Bin, M.; Reaz, I. Myoelectric Pattern Recognition Performance Enhancement Using Nonlinear Features. arXiv 2021, 1–19. [Google Scholar] [CrossRef]
- Sarker, P.; Mirka, G. Effects of Sampling Frequency and Sample Window Size on Median Frequency of Surface EMG. Proc. Hum. Factors Ergon. Soc. Annu. Meet. 2019, 63, 1369–1372. [Google Scholar] [CrossRef]
- Tkach, D.; Huang, H.; Kuiken, T.A. Study of stability of time-domain features for electromyographic pattern recognition. J. Neuroeng. Rehabil. 2010, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Altın, C.; Er, O. Comparison of Different Time and Frequency Domain Feature Extraction Methods on Elbow Gesture’s EMG. Eur. J. Interdiscip. Stud. 2016, 5, 35. [Google Scholar] [CrossRef]
- Stergiou, N.; Giakas, G.; Byrne, J.E.; Pomeroy, V. Frequency domain characteristics of ground reaction forces during walking of young and elderly females. Clin. Biomech. 2002, 17, 615–617. [Google Scholar] [CrossRef]
- Ramzi, H. Blind Source Separation of Single-sensor Recordings: Application to Ground Reaction Force Signals. Ph.D. Thesis, Université de Lyon, Lyon, France, 2018. [Google Scholar]
- Soubra, R.; Diab, M.O.; Moslem, B. A mother wavelet selection study for vertical ground reaction force signals. In Proceedings of the BioSMART 2017–2017 2nd International Conference on Bio-engineering for Smart Technologies, Paris, France, 30 August–1 September 2017; pp. 1–5. [Google Scholar]
- Doulah, A.B.M.S.U.; Fattah, S.A.; Zhum, W.-P.; Ahmad, M.O. Wavelet Domain Feature Extraction Scheme Based on Dominant Motor Unit Action Potential of EMG Signal for Neuromuscular Disease Classification. IEEE Trans. Biomed. Circuits Syst. 2014, 8, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Spencer, R.; Thabtah, F.; Abdelhamid, N.; Thompson, M. Exploring feature selection and classification methods for predicting heart disease. Digit. Heal. 2020, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Radovic, M.; Ghalwash, M.; Filipovic, N.; Obradovic, Z. Minimum redundancy maximum relevance feature selection approach for temporal gene expression data. BMC Bioinformatics 2017, 18, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, J.L.; Lumsden, S.L. Locally linear embedding: Dimension reduction of massive protostellar spectra. Mon. Not. R. Astron. Soc. 2016, 461, 2250–2256. [Google Scholar] [CrossRef]
- Saeb, S.; Lonini, L.; Jayaraman, A.; Mohr, D.C.; Kording, K.P. The need to approximate the use-case in clinical machine learning. Gigascience 2017, 6, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Chaibub Neto, E.; Pratap, A.; Perumal, T.M.; Tummalacherla, M.; Snyder, P.; Bot, B.M.; Trister, A.D.; Friend, S.H.; Mangravite, L.; Omberg, L. Detecting the impact of subject characteristics on machine learning-based diagnostic applications. npj Digit. Med. 2019, 2, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chawla, N.V.; Bowyer, K.W.; Lawrence, O.; Hall, W. Philip Kegelmeyer SMOTE: Synthetic Minority Over-sampling Technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Kohavi, R.; Provost, F. Glossary of Terms. Mach. Learn. 1998, 30, 271–274. [Google Scholar] [CrossRef]
- Sinnreich, M.; Taylor, B.V.; Dyck, P.J.B. Diabetic neuropathies: Classification, clinical features, and pathophysiological basis. Neurologist 2005, 11, 63–79. [Google Scholar] [CrossRef]
- Hazari, A.; Maiya, A.G.; Shivashankara, K.N.; Agouris, I.; Monteiro, A.; Jadhav, R.; Kumar, S.; Shashi Kumar, C.G.; Mayya, S.S. Kinetics and kinematics of diabetic foot in type 2 diabetes mellitus with and without peripheral neuropathy: A systematic review and meta-analysis. Springerplus 2016, 5. [Google Scholar] [CrossRef] [Green Version]
- Alam, U.; Riley, D.R.; Jugdey, R.S.; Azmi, S.; Rajbhandari, S.; D’Août, K.; Malik, R.A. Diabetic Neuropathy and Gait: A Review. Diabetes Ther. 2017, 8, 1253–1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulton, A.J.M. The diabetic foot: From art to science. The 18th Camillo Golgi lecture. Diabetologia 2004, 47, 1343–1353. [Google Scholar] [CrossRef] [PubMed]
- Cruz-Vega, I.; Hernandez-Contreras, D.; Peregrina-Barreto, H.; Rangel-Magdaleno, J.D.J.; Ramirez-Cortes, J.M. Deep learning classification for diabetic foot thermograms. Sensors 2020, 20, 1762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäfer, Z.; Mathisen, A.; Svendsen, K.; Engberg, S.; Rolighed Thomsen, T.; Kirketerp-Møller, K. Toward Machine-Learning-Based Decision Support in Diabetes Care: A Risk Stratification Study on Diabetic Foot Ulcer and Amputation. Front. Med. 2021, 7, 1–13. [Google Scholar] [CrossRef]


| Control Group (n = 6) | Diabetic Group (n = 6) | Ulcer Diabetic Group (n = 9) | p | |
|---|---|---|---|---|
| Age(years) | 52.2 ± 6.9 | 56.0 ± 8.5 | 55.6 ± 6.0 | 0.7 a |
| Gender(male/female) | 3/3 | 2/4 | 1/8 | - |
| Weight (Kg) | 61.7 ± 5.3 | 66.7 ± 12.2 | 83.4 ± 10.5 | <0.001 a |
| Height (m) | 1.7 ± 0.09 | 1.6 ± 0.07 | 1.7 ± 0.07 | 0.008 a |
| BMI (kg/m2) | 22.1 ± 1.7 c | 25.1 ± 4.0 | 27.8 ± 3.4 | 0.029 a |
| Time of DM (years) | - | 15.2 ± 6.8 | 14.3 ± 5.8 | 0.80 a |
| Glycemia(mg/dL) | - | 142 ± 61.8 | 188.6 ± 61.2 | 0.17 a |
| MNSI Questionnaire d | - | 7 | 7.5 | 0.72 b |
| MNSI Clinical Examination d | - | 2.5 | 4 | 0.12 b |
| Combination | Muscles | No. of Features | After Removing Highly Correlated Features |
|---|---|---|---|
| One Channel | GL | 19 | 11 |
| VL | |||
| TA | |||
| Two Channel | GL-VL | 38 | 24 |
| GL-TA | 22 | ||
| TA-VL | 22 | ||
| Three Channel | GL-VL-Ta | 57 | 34 |
| Muscle | Feature Ranking Technique | Features | Accuracy (%) | Precision (%) | Sensitivity (%) | F1 Score (%) | AUC |
|---|---|---|---|---|---|---|---|
| GL | Chi-Square | Top 8 | 81.30 | 82.22 | 81.30 | 81.54 | 0.83 |
| mrmr | Top 11 | 80.92 | 81.59 | 80.92 | 81.11 | 0.82 | |
| Relieff | Top 7 | 84.73 | 84.89 | 84.73 | 84.76 | 0.85 | |
| fscnca | Top 10 | 81.30 | 82.27 | 81.30 | 81.54 | 0.83 | |
| TA | Chi-Square | Top 8 | 80.53 | 80.91 | 80.53 | 80.59 | 0.82 |
| mrmr | Top 9 | 84.35 | 85.06 | 84.35 | 84.48 | 0.87 | |
| Relieff | Top 9 | 81.30 | 81.58 | 81.30 | 81.40 | 0.82 | |
| fscnca | Top 11 | 83.59 | 83.93 | 83.59 | 83.67 | 0.86 | |
| VL | Chi-Square | Top 10 | 74.81 | 75.55 | 74.81 | 75.09 | 0.80 |
| mrmr | Top 11 | 75.95 | 76.77 | 75.95 | 76.24 | 0.82 | |
| Relieff | Top 8 | 77.10 | 77.96 | 77.10 | 77.39 | 0.83 | |
| fscnca | Top 11 | 76.72 | 77.25 | 76.72 | 76.84 | 0.83 |
| Muscles | Feature Ranking Technique | Features | Accuracy (%) | Precision (%) | Sensitivity (%) | F1 Score (%) | AUC |
|---|---|---|---|---|---|---|---|
| GL-TA | Chi-Square | Top 15 | 95.80 | 96.04 | 95.80 | 95.84 | 0.96 |
| mrmr | Top 17 | 94.27 | 94.38 | 94.27 | 94.30 | 0.95 | |
| Relieff | Top 17 | 95.04 | 95.16 | 95.04 | 95.05 | 0.96 | |
| fscnca | Top 22 | 93.51 | 93.67 | 93.51 | 93.55 | 0.94 | |
| TA-VL | Chi-Square | Top 18 | 92.37 | 92.71 | 92.37 | 92.44 | 0.94 |
| mrmr | Top 13 | 91.98 | 92.07 | 91.98 | 92.01 | 0.91 | |
| Relieff | Top 17 | 92.75 | 93.03 | 92.75 | 92.82 | 0.93 | |
| fscnca | Top 20 | 93.51 | 93.80 | 93.51 | 93.58 | 0.94 | |
| GL-VL | Chi-Square | Top 12 | 96.18 | 96.25 | 96.18 | 96.20 | 0.97 |
| mrmr | Top 14 | 93.51 | 93.59 | 93.51 | 93.52 | 0.95 | |
| Relieff | Top 16 | 95.80 | 95.84 | 95.80 | 95.81 | 0.97 | |
| fscnca | Top 21 | 92.75 | 93.12 | 92.75 | 92.82 | 0.96 |
| Muscles | Feature Ranking Technique | Features | Accuracy (%) | Precision (%) | Sensitivity (%) | F1 Score (%) | AUC |
|---|---|---|---|---|---|---|---|
| GL-TA-VL | Chi-Square | Top 27 | 95.80 | 95.86 | 95.80 | 95.78 | 0.99 |
| mrmr | Top 38 | 95.42 | 95.68 | 95.42 | 95.41 | 0.98 | |
| Relieff | Top 23 | 95.80 | 95.86 | 95.80 | 95.80 | 0.97 | |
| fscnca | Top 36 | 95.04 | 95.04 | 95.04 | 95.03 | 0.98 |
| Combination | Muscles | No. of Features | After Removing Highly Correlated Features |
|---|---|---|---|
| One Channel | GRFx | 195 | 56 |
| GRFy | 50 | ||
| GRFz | 37 | ||
| Two Channel | GRFx-GRFy | 390 | 102 |
| GRFx-GRFz | 87 | ||
| GRFy-GRFz | 86 | ||
| Three Channel | GRFx-GRFy-GRFz | 585 | 129 |
| GRF Component | Feature Ranking Technique | Features | Accuracy (%) | Precision (%) | Sensitivity (%) | F1 Score (%) | AUC |
|---|---|---|---|---|---|---|---|
| GRFx | Chi-Square | Top 38 | 97.37 | 97.41 | 97.37 | 97.36 | 0.98 |
| mrmr | Top 50 | 96.05 | 96.28 | 96.05 | 96.05 | 0.98 | |
| Relieff | Top 14 | 97.37 | 97.41 | 97.37 | 97.36 | 0.98 | |
| fscnca | Top 19 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 | |
| GRFy | Chi-Square | Top 15 | 92.11 | 92.28 | 92.11 | 92.11 | 0.95 |
| mrmr | Top 34 | 86.84 | 87.26 | 86.84 | 86.78 | 0.89 | |
| Relieff | Top 35 | 92.11 | 92.27 | 92.11 | 92.12 | 0.95 | |
| fscnca | Top 21 | 93.42 | 94.32 | 93.42 | 93.40 | 0.96 | |
| GRFz | Chi-Square | Top 24 | 94.74 | 94.75 | 94.74 | 94.72 | 0.95 |
| mrmr | Top 18 | 93.42 | 93.47 | 93.42 | 93.42 | 0.94 | |
| Relieff | Top 32 | 93.42 | 93.71 | 93.42 | 93.41 | 0.93 | |
| fscnca | Top 13 | 93.42 | 93.65 | 93.42 | 93.45 | 0.95 |
| GRF Component | Feature Ranking Technique | Features | Accuracy (%) | Precision (%) | Sensitivity (%) | F1 Score (%) | AUC |
|---|---|---|---|---|---|---|---|
| GRFy-GRFz | Chi-Square | Top 3 | 94.74 | 94.83 | 94.74 | 94.67 | 0.92 |
| mrmr | Top 42 | 96.05 | 96.05 | 96.05 | 96.05 | 0.97 | |
| Relieff | Top 45 | 94.74 | 94.82 | 94.74 | 94.75 | 0.96 | |
| fscnca | Top 20 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 | |
| GRFx-GRFz | Chi-Square | Top 24 | 94.74 | 94.93 | 94.74 | 94.75 | 0.96 |
| mrmr | Top 19 | 97.37 | 97.43 | 97.37 | 97.37 | 0.99 | |
| Relieff | Top 41 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 | |
| fscnca | Top 7 | 98.68 | 98.72 | 98.68 | 98.68 | 0.98 | |
| GRFx-GRFy | Chi-Square | Top 17 | 96.05 | 96.24 | 96.05 | 96.07 | 0.98 |
| mrmr | Top 87 | 97.37 | 97.43 | 97.37 | 97.37 | 0.97 | |
| Relieff | Top 17 | 96.05 | 96.07 | 96.05 | 96.04 | 0.99 | |
| fscnca | Top 24 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 |
| GRF Component | Feature Ranking Technique | Accuracy (%) | Precision (%) | Sensitivity (%) | F1 Score (%) | AUC | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| GRFx-GRFy-GRFz | Chi-Square | Top 111 | 97.37 | 97.41 | 97.37 | 97.36 | 1.00 |
| mrmr | Top 64 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 | |
| Relieff | Top 106 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 | |
| fscnca | Top 21 | 98.68 | 98.72 | 98.68 | 98.68 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haque, F.; Reaz, M.B.I.; Chowdhury, M.E.H.; Ezeddin, M.; Kiranyaz, S.; Alhatou, M.; Ali, S.H.M.; Bakar, A.A.A.; Srivastava, G. Machine Learning-Based Diabetic Neuropathy and Previous Foot Ulceration Patients Detection Using Electromyography and Ground Reaction Forces during Gait. Sensors 2022, 22, 3507. https://doi.org/10.3390/s22093507
Haque F, Reaz MBI, Chowdhury MEH, Ezeddin M, Kiranyaz S, Alhatou M, Ali SHM, Bakar AAA, Srivastava G. Machine Learning-Based Diabetic Neuropathy and Previous Foot Ulceration Patients Detection Using Electromyography and Ground Reaction Forces during Gait. Sensors. 2022; 22(9):3507. https://doi.org/10.3390/s22093507
Chicago/Turabian StyleHaque, Fahmida, Mamun Bin Ibne Reaz, Muhammad Enamul Hoque Chowdhury, Maymouna Ezeddin, Serkan Kiranyaz, Mohammed Alhatou, Sawal Hamid Md Ali, Ahmad Ashrif A Bakar, and Geetika Srivastava. 2022. "Machine Learning-Based Diabetic Neuropathy and Previous Foot Ulceration Patients Detection Using Electromyography and Ground Reaction Forces during Gait" Sensors 22, no. 9: 3507. https://doi.org/10.3390/s22093507
APA StyleHaque, F., Reaz, M. B. I., Chowdhury, M. E. H., Ezeddin, M., Kiranyaz, S., Alhatou, M., Ali, S. H. M., Bakar, A. A. A., & Srivastava, G. (2022). Machine Learning-Based Diabetic Neuropathy and Previous Foot Ulceration Patients Detection Using Electromyography and Ground Reaction Forces during Gait. Sensors, 22(9), 3507. https://doi.org/10.3390/s22093507








