Emotion Recognition Using a Reduced Set of EEG Channels Based on Holographic Feature Maps
Abstract
:1. Introduction
2. Related Work
3. Materials
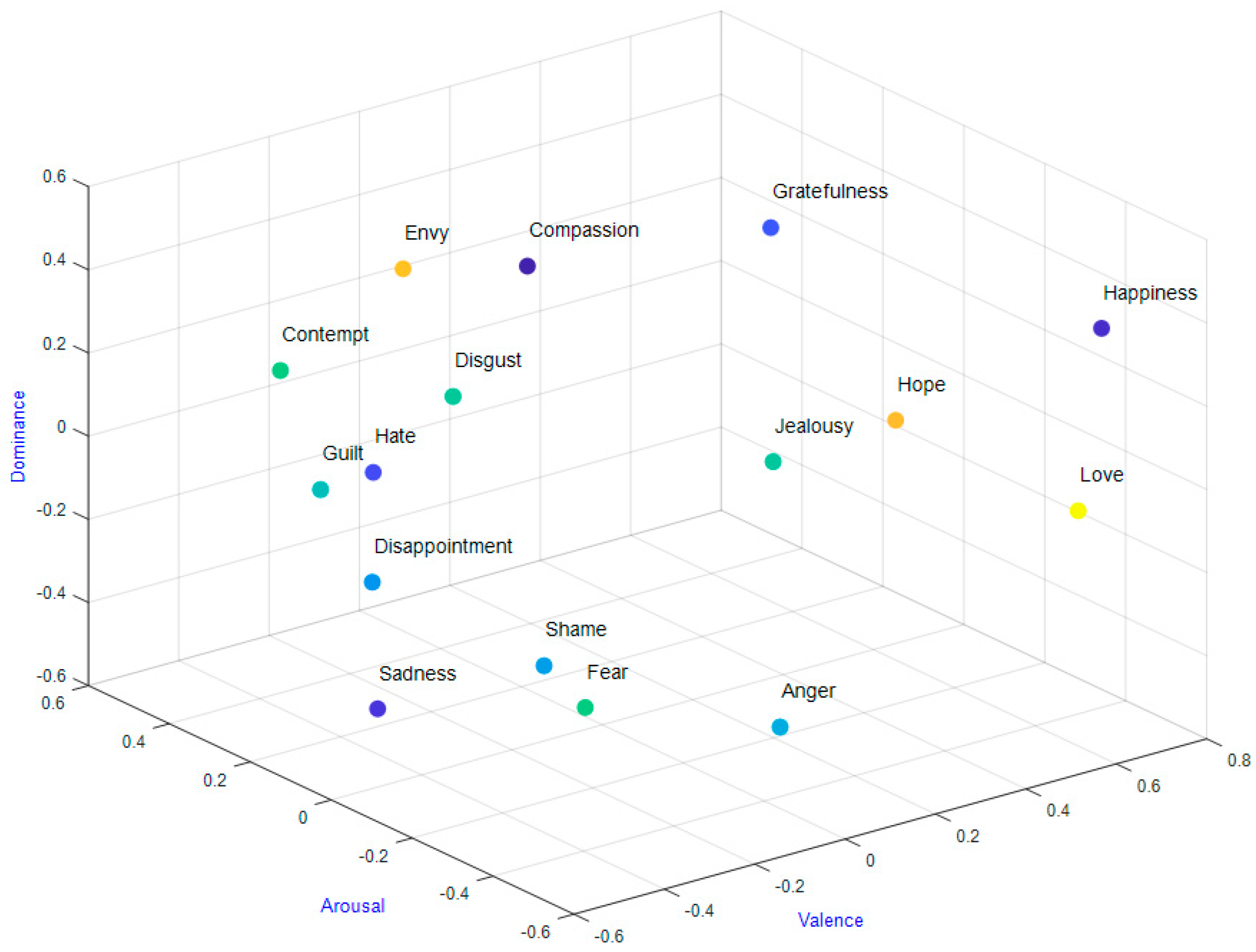
3.1. Selected Datasets
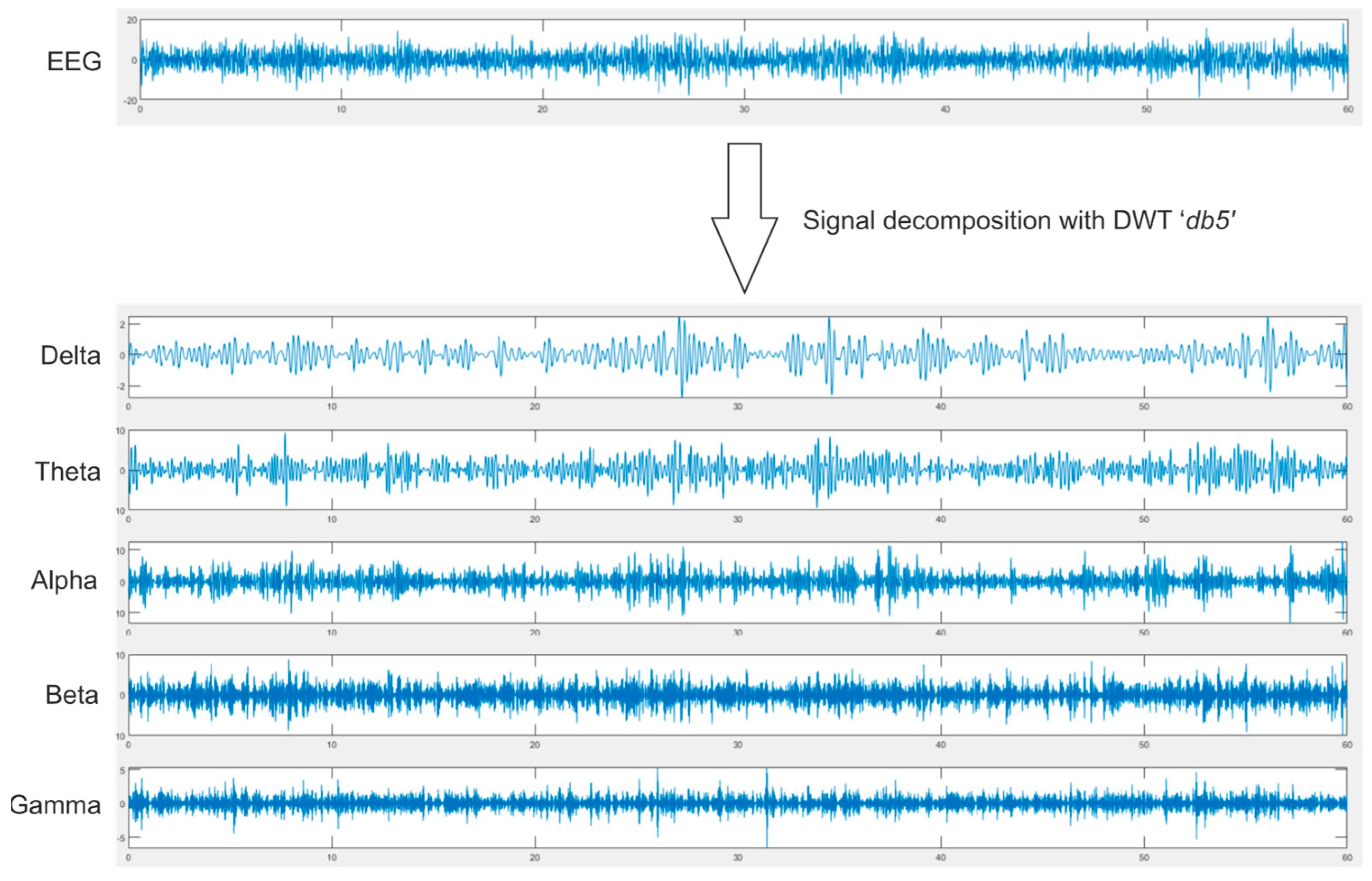
3.2. Selected Features
4. Methodology
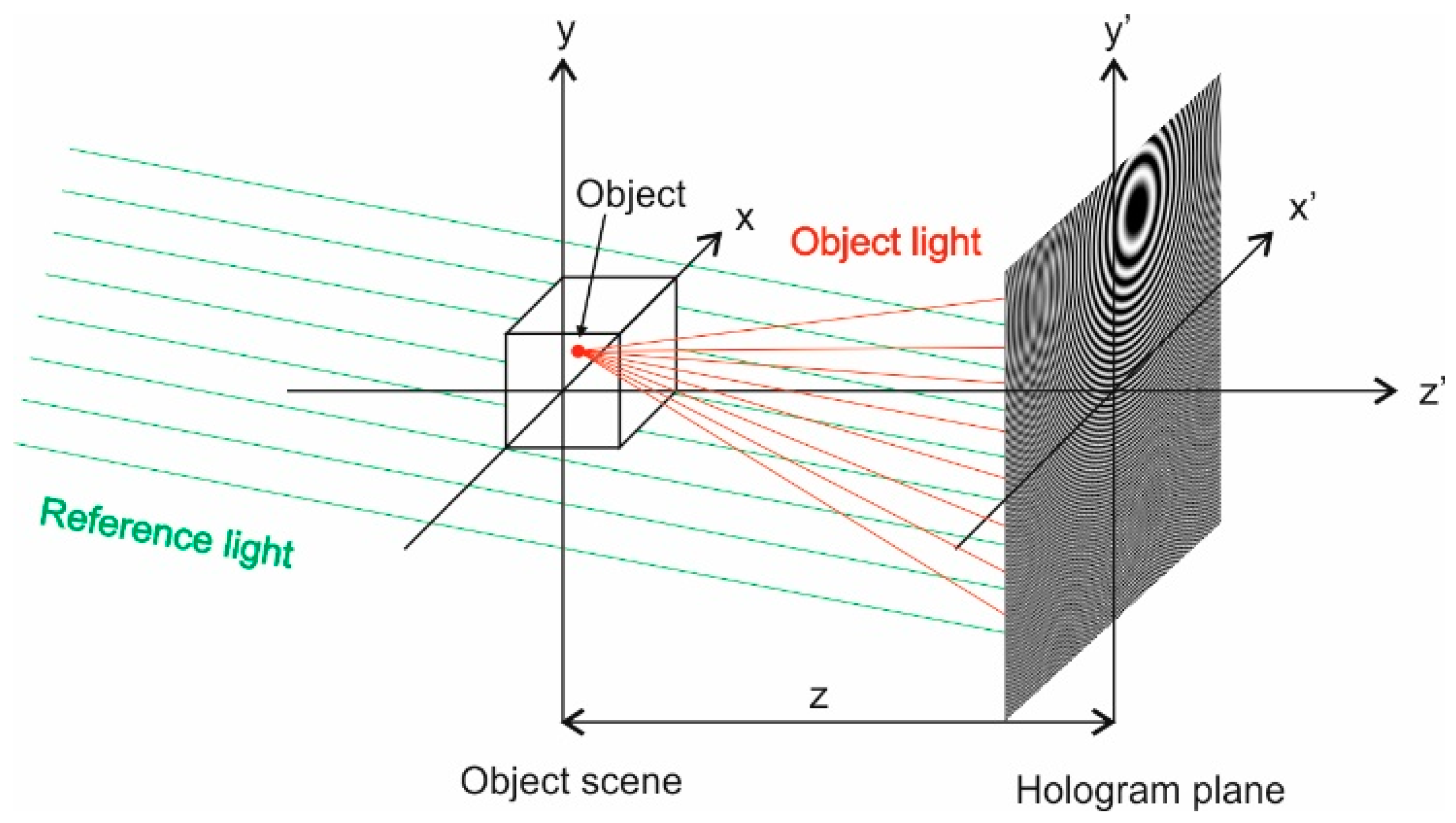
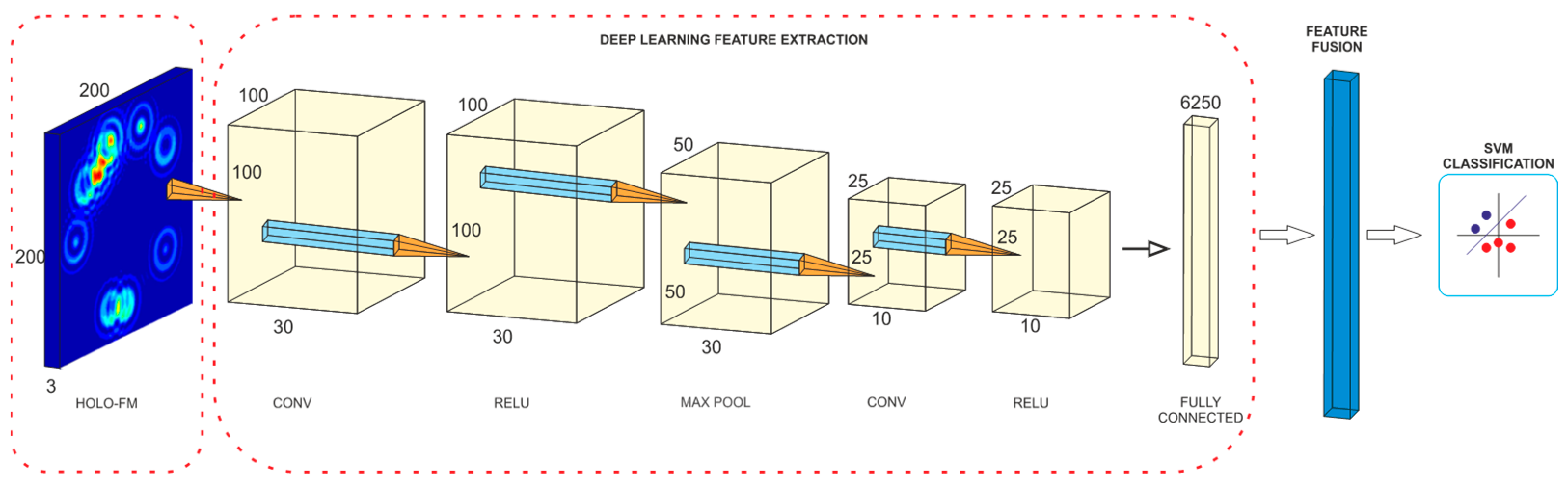
4.1. Feature Maps Creation
4.2. Channel Selection
| Algorithm 1. Pseudocode of ReliefF algorithm: |
| Inputs: Instance set S and the number of classes C Output: Weight vector w Step 1: For any feature fa, a = 1, 2, …, d, set the initial weight wa = 0 Step 2: for i = 1 to m do Randomly select xi from S; Select the k-nearest neighbors hj from the same class of x; Select the k-nearest neighbors mj(c) from different class from x for a = 1 to d do Update the weight by (9): End End |
4.3. Model Construction
5. Results and Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Khalil, R.A.; Jones, E.; Babar, M.I.; Jan, T.; Zafar, M.H.; Alhussain, T. Speech emotion recognition using deep learning techniques: A review. IEEE Access 2019, 7, 117327–117345. [Google Scholar] [CrossRef]
- Alreshidi, A.; Ullah, M. Facial Emotion Recognition Using Hybrid Features. Informatics 2020, 7, 6. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, F.; Bari, A.S.M.H.; Gavrilova, M.L. Emotion Recognition From Body Movement. IEEE Access 2020, 8, 11761–11781. [Google Scholar] [CrossRef]
- Chamola, V.; Vineet, A.; Nayyar, A.; Hossain, E. Brain-Computer Interface-Based Humanoid Control: A Review. Sensors 2020, 20, 3620. [Google Scholar] [CrossRef] [PubMed]
- Udovicic, G.; Topic, A.; Russo, M. Wearable technologies for smart environments: A review with emphasis on BCI. In Proceedings of the 24th International Conference on Software, Telecommunications and Computer Networks (SoftCOM), Split, Croatia, 22–24 September 2016; pp. 1–9. [Google Scholar]
- Gu, X.; Cao, Z.; Jolfaei, A.; Xu, P.; Wu, D.; Jung, T.P.; Lin, C.T. EEG-based brain-computer interfaces (BCIs): A survey of recent studies on signal sensing technologies and computational intelligence approaches and their applications. IEEE/ACM Trans. Comput. Biol. Bioinform. 2021, 8, 1645–1666. [Google Scholar] [CrossRef] [PubMed]
- Towle, V.L.; Bolanos, J.; Suarez, D.; Tan, K.; Grzeszczuk, R.; Levin, D.N.; Cakmur, R.; Frank, S.A.; Spire, J.-P. The spatial location of EEG electrodes: Locating the best-fitting sphere relative to cortical anatomy. Electroencephalogr. Clin. Neurophysiol. 1993, 86, 1–6. [Google Scholar] [CrossRef]
- Gui, Q.; Ruiz-Blondet, M.V.; Laszlo, S.; Jin, Z. A survey on brain biometrics. ACM Comput. Surv. (CSUR) 2019, 51, 1–38. [Google Scholar] [CrossRef]
- Alotaiby, T.; Abd El-Samie, F.E.; Alshebeili, S.A.; Ahmad, I. A review of channel selection algorithms for EEG signal processing. EURASIP J. Adv. Signal Process. 2015, 2015, 66. [Google Scholar] [CrossRef] [Green Version]
- Ansari-Asl, K.; Chanel, G.; Pun, T. A channel selection method for EEG classification in emotion assessment based on synchronization likelihood. In Proceedings of the 15th European Signal Processing Conference, Poznan, Poland, 3–7 September 2007; pp. 1241–1245. [Google Scholar]
- Jatupaiboon, N.; Pan-ngum, S.; Israsena, P. Emotion classification using minimal EEG channels and frequency bands. In Proceedings of the 10th International Joint Conference on Computer Science and Software Engineering (JCSSE), Khon Kaen, Thailand, 29–31 May 2013; pp. 21–24. [Google Scholar]
- Keelawat, P.; Thammasan, N.; Numao, M.; Kijsirikul, B. A Comparative Study of Window Size and Channel Arrangement on EEG-Emotion Recognition Using Deep CNN. Sensors 2021, 21, 1678. [Google Scholar] [CrossRef] [PubMed]
- Ekman, P. An argument for basic emotions. Cogn. Emot. 1992, 6, 169–200. [Google Scholar] [CrossRef]
- Russell, J.A. A circumplex model of affect. J. Personal. Soc. Psychol. 1980, 38, 1161–1178. [Google Scholar] [CrossRef]
- Mehrabian, A. Comparison of the PAD and PANAS as models for describing emotions and for differentiating anxiety from depression. J. Psychopathol. Behav. Assess. 1997, 19, 331–357. [Google Scholar] [CrossRef]
- Gannouni, S.; Aledaily, A.; Belwafi, K.; Aboalsamh, H. Emotion detection using electroencephalography signals and a zero-time windowing-based epoch estimation and relevant electrode identification. Sci. Rep. 2021, 11, 7071. [Google Scholar] [CrossRef] [PubMed]
- Petrantonakis, P.C.; Hadjileontiadis, L.J. Emotion recognition from brain signals using hybrid adaptive filtering and higher order crossings analysis. IEEE Trans. Affect. Comput. 2010, 1, 81–97. [Google Scholar] [CrossRef]
- Valenzi, S.; Islam, T.; Jurica, P.; Cichocki, A. Individual classification of emotions using EEG. J. Biomed. Sci. Eng. 2014, 7, 604–620. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Chen, M.; Zhao, S.; Hu, S.; Shi, Z.; Cao, Y. ReliefF-based EEG sensor selection methods for emotion recognition. Sensors 2016, 16, 1558. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Liu, X.; Zhang, Y.; Cui, L.; Yu, X. A portable HCI system-oriented EEG feature extraction and channel selection for emotion recognition. Int. J. Intell. Syst. 2021, 36, 152–176. [Google Scholar] [CrossRef]
- Li, M.; Xu, H.; Liu, X.; Lu, S. Emotion recognition from multichannel EEG signals using K-nearest neighbor classification. Technol. Health Care 2018, 26, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Bazgir, O.; Mohammadi, Z.; Habibi, S.A.H. Emotion recognition with machine learning using EEG signals. In Proceedings of the 25th National and 3rd International Iranian Conference on Biomedical Engineering (ICBME), Qom, Iran, 29–30 November 2018; pp. 1–5. [Google Scholar]
- Mohammadi, Z.; Frounchi, J.; Amiri, M. Wavelet-based emotion recognition system using EEG signal. Neural Comput. Appl. 2017, 28, 1985–1990. [Google Scholar] [CrossRef]
- Özerdem, M.S.; Polat, H. Emotion recognition based on EEG features in movie clips with channel selection. Brain Inform. 2017, 4, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Hu, S.Y.; Song, H. Channel Selection Method for EEG Emotion Recognition Using Normalized Mutual Information. IEEE Access 2019, 7, 143303–143311. [Google Scholar] [CrossRef]
- Msonda, J.R.; He, Z.; Lu, C. Feature Reconstruction Based Channel Selection for Emotion Recognition Using EEG. In Proceedings of the IEEE Signal Processing in Medicine and Biology Symposium (SPMB), Philadelphia, PA, USA, 4 December 2021; pp. 1–7. [Google Scholar]
- Menon, A.; Natarajan, A.; Agashe, R.; Sun, D.; Aristio, M.; Liew, H.; Rabaey, J.M. Efficient emotion recognition using hyperdimensional computing with combinatorial channel encoding and cellular automata. arXiv 2021, arXiv:2104.02804. [Google Scholar]
- Gupta, V.; Chopda, M.D.; Pachori, R.B. Cross-subject emotion recognition using flexible analytic wavelet transform from EEG signals. IEEE Sens. J. 2018, 19, 2266–2274. [Google Scholar] [CrossRef]
- Mert, A.; Akan, A. Emotion recognition from EEG signals by using multivariate empirical mode decomposition. Pattern Anal. Appl. 2018, 21, 81–89. [Google Scholar] [CrossRef]
- Liu, Y.; Fu, G. Emotion recognition by deeply learned multi-channel textual and EEG features. Future Gener. Comput. Syst. 2021, 119, 1–6. [Google Scholar] [CrossRef]
- Tong, L.; Zhao, J.; Fu, W. Emotion recognition and channel selection based on EEG Signal. In Proceedings of the 2018 11th International Conference on Intelligent Computation Technology and Automation (ICICTA), Changsha, China, 22–23 September 2018; pp. 101–105. [Google Scholar]
- Zhang, J.; Chen, M.; Hu, S.; Cao, Y.; Kozma, R. PNN for EEG-based Emotion Recognition. In Proceedings of the 2016 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Budapest, Hungary, 9–12 October 2016; pp. 2319–2323. [Google Scholar]
- Zheng, W.-L.; Lu, B.-L. Investigating critical frequency bands and channels for EEG-based emotion recognition with deep neural networks. IEEE Trans. Auton. Ment. Dev. 2015, 7, 162–175. [Google Scholar] [CrossRef]
- Pane, E.S.; Wibawa, A.D.; Pumomo, M.H. Channel selection of EEG emotion recognition using stepwise discriminant analysis. In Proceedings of the 2018 International Conference on Computer Engineering, Network and Intelligent Multimedia (CENIM), Surabaya, Indonesia, 26–27 November 2018; pp. 14–19. [Google Scholar]
- Cheah, K.H.; Nisar, H.; Yap, V.V.; Lee, C.Y.; Sinha, G.R. Optimizing residual networks and vgg for classification of eeg signals: Identifying ideal channels for emotion recognition. J. Healthc. Eng. 2021, 2021, 5599615. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W. Multichannel EEG-based emotion recognition via group sparse canonical correlation analysis. IEEE Trans. Cogn. Dev. Syst. 2016, 9, 281–290. [Google Scholar] [CrossRef]
- Sönmez, Y.Ü.; Varol, A. A Speech Emotion Recognition Model Based on Multi-Level Local Binary and Local Ternary Patterns. IEEE Access 2020, 8, 190784–190796. [Google Scholar] [CrossRef]
- Tuncer, T.; Dogan, S.; Acharya, U.R. Automated accurate speech emotion recognition system using twine shuffle pattern and iterative neighborhood component analysis techniques. Knowl.-Based Syst. 2021, 211, 106547. [Google Scholar] [CrossRef]
- Raghu, S.; Sriraam, N. Classification of focal and non-focal EEG signals using neighborhood component analysis and machine learning algorithms. Expert Syst. Appl. 2018, 113, 18–32. [Google Scholar] [CrossRef]
- Ferdinando, H.; Seppänen, T.; Alasaarela, E. Enhancing Emotion Recognition from ECG Signals using Supervised Dimensionality Reduction. In Proceedings of the 6th International Conference on Pattern Recognition Applications and Methods, ICPRAM 2017, Porto, Portugal, 24–26 February 2017; pp. 112–118. [Google Scholar]
- Malan, N.S.; Sharma, S. Feature selection using regularized neighbourhood component analysis to enhance the classification performance of motor imagery signals. Comput. Biol. Med. 2019, 107, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Canli, T.; Desmond, J.E.; Zhao, Z.; Gabrieli, J.D. Sex differences in the neural basis of emotional memories. Proc. Natl. Acad. Sci. USA 2002, 99, 10789–10794. [Google Scholar] [CrossRef] [Green Version]
- Cahill, L. Sex-related influences on the neurobiology of emotionally influenced memory. Ann. N. Y. Acad. Sci. 2003, 985, 163–173. [Google Scholar] [CrossRef]
- Bradley, M.M.; Codispoti, M.; Sabatinelli, D.; Lang, P.J. Emotion and motivation II: Sex differences in picture processing. Emotion 2001, 1, 300. [Google Scholar] [CrossRef] [PubMed]
- Chentsova-Dutton, Y.E.; Tsai, J.L. Gender differences in emotional response among European Americans and among Americans. Cogn. Emot. 2007, 21, 162–181. [Google Scholar] [CrossRef]
- Dadebayev, D.; Goh, W.W.; Tan, E.X. EEG-based emotion recognition: Review of commercial EEG devices and machine learning techniques. J. King Saud Univ. Comput. Inf. Sci. 2021; in press. [Google Scholar]
- Alarcão, S.M.; Fonseca, M.J. Emotions Recognition Using EEG Signals: A Survey. IEEE Trans. Affect. Comput. 2019, 10, 374–393. [Google Scholar] [CrossRef]
- Fraedrich, E.M.; Lakatos, K.; Spangler, G. Brain activity during emotion perception: The role of attachment representation. Attach. Hum. Dev. 2010, 12, 231–248. [Google Scholar] [CrossRef] [PubMed]
- Lomas, T.; Edginton, T.; Cartwright, T.; Ridge, D. Men developing emotional intelligence through meditation? Integrating narrative, cognitive and electroencephalography (EEG) evidence. Psychol. Men Masc. 2014, 15, 213. [Google Scholar] [CrossRef]
- Zhang, J.; Xu, H.; Zhu, L.; Kong, W.; Ma, Z. Gender recognition in emotion perception using eeg features. In Proceedings of the IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 2883–2887. [Google Scholar]
- Hu, J. An approach to EEG-based gender recognition using entropy measurement methods. Knowl.-Based Syst. 2018, 140, 134–141. [Google Scholar] [CrossRef]
- Van Putten, M.J.; Olbrich, S.; Arns, M. Predicting sex from brain rhythms with deep learning. Sci. Rep. 2018, 8, 3069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrieri, A.; Braccili, E.; Sgrò, F.; Meldolesi, G.N. Gender Identification in a Two-Level Hierarchical Speech Emotion Recognition System for an Italian Social Robot. Sensors 2022, 22, 1714. [Google Scholar] [CrossRef] [PubMed]
- Koelstra, S.; Muhl, C.; Soleymani, M.; Lee, J.-S.; Yazdani, A.; Ebrahimi, T.; Pun, T.; Nijholt, A.; Patras, I. DEAP: A database for emotion analysis; using physiological signals. IEEE Trans. Affect. Comput. 2012, 3, 18–31. [Google Scholar] [CrossRef] [Green Version]
- Katsigiannis, S.; Ramzan, N. DREAMER: A Database for Emotion Recognition Through EEG and ECG Signals from Wireless Low-cost Off-the-Shelf Devices. IEEE J. Biomed. Health Inform. 2018, 22, 98–107. [Google Scholar] [CrossRef] [Green Version]
- Miranda-Correa, J.A.; Abadi, M.K.; Sebe, N.; Patras, I. AMIGOS: A Dataset for Affect, Personality and Mood Research on Individuals and Groups. IEEE Trans. Affect. Comput. 2018, 12, 479–493. [Google Scholar] [CrossRef] [Green Version]
- Trnka, R.; Lačev, A.; Balcar, K.; Kuška, M.; Tavel, P. Modeling semantic emotion space using a 3D hypercube-projection: An innovative analytical approach for the psychology of emotions. Front. Psychol. 2016, 7, 522. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.D. Observations: Sam: The self-assessment manikin; an efficient cross-cultural measurement of emotional response. J. Advert. Res. 1995, 35, 63–68. [Google Scholar]
- Wang, S.-H.; Li, H.-T.; Chang, E.-J.; Wu, A.-Y. Entropy-Assisted Emotion Recognition of Valence and Arousal Using XGBoost Classifier. In Proceedings of the Artificial Intelligence Applications and Innovations (AIAI), Rhodes, Greece, 25–27 May 2018; pp. 249–260. [Google Scholar]
- Topic, A.; Russo, M. Emotion recognition based on EEG feature maps through deep learning network. Eng. Sci. Technol. Int. J. 2021, 24, 1442–1454. [Google Scholar] [CrossRef]
- Lan, Z.; Sourina, O.; Wang, L.; Scherer, R.; Müller-Putz, G.R. Domain adaptation techniques for EEG-based emotion recognition: A comparative study on two public datasets. IEEE Trans. Cogn. Dev. Syst. 2018, 11, 85–94. [Google Scholar] [CrossRef]
- Li, X.; Song, D.; Zhang, P.; Zhang, Y.; Hou, Y.; Hu, B. Exploring EEG features in cross-subject emotion recognition. Front. Neurosci. 2018, 12, 162. [Google Scholar] [CrossRef] [Green Version]
- Pandey, P.; Seeja, K.R. Subject independent emotion recognition system for people with facial deformity: An EEG based approach. J. Ambient. Intell. Humaniz. Comput. 2020, 12, 2311–2320. [Google Scholar] [CrossRef]
- Cimtay, Y.; Ekmekcioglu, E. Investigating the Use of Pretrained Convolutional Neural Network on Cross-Subject and Cross-Dataset EEG Emotion Recognition. Sensors 2020, 20, 2034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Hu, B.; Moore, P.; Zhang, X.; Ma, X. Electroencephalogram-based emotion assessment system using ontology and data mining techniques. Appl. Soft Comput. 2015, 30, 663–674. [Google Scholar] [CrossRef]
- Murugappan, M.; Ramachandran, N.; Sazali, Y. Classification of human emotion from EEG using discrete wavelet transform. J. Biomed. Sci. Eng. 2010, 3, 390. [Google Scholar] [CrossRef] [Green Version]
- Atkinson, J.; Campos, D. Improving BCI-based emotion recognition by combining EEG feature selection and kernel classifiers. Expert Syst. Appl. 2016, 47, 35–41. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, X.; Sun, Y.; Zhang, J. Emotion Feature Analysis and Recognition Based on Reconstructed EEG Sources. IEEE Access 2020, 8, 11907–11916. [Google Scholar] [CrossRef]
- Zheng, W.; Liu, W.; Lu, Y.; Lu, B.; Cichocki, A. EmotionMeter: A Multimodal Framework for Recognizing Human Emotions. IEEE Trans. Cybern. 2019, 49, 1110–1122. [Google Scholar] [CrossRef]
- Chai, X.; Wang, Q.; Zhao, Y.; Li, Y.; Liu, D.; Liu, X.; Bai, O. A Fast, Efficient Domain Adaptation Technique for Cross-Domain Electroencephalography (EEG)-Based Emotion Recognition. Sensors 2017, 17, 1014. [Google Scholar] [CrossRef] [Green Version]
- Khosrowabadi, R.; Rahman, A.W.b.A. Classification of EEG correlates on emotion using features from Gaussian mixtures of EEG spectrogram. In Proceedings of the 3rd International Conference on Information and Communication Technology for the Moslem World (ICT4M), Jakarta, Indonesia, 13–14 December 2010; pp. E102–E107. [Google Scholar]
- Sourina, O.; Liu, Y. A fractal-based algorithm of emotion recognition from EEG using arousal-valence model. In Proceedings of the International Conference on Bio-Inspired Systems and Signal Processing, Rome, Italy, 26–29 January 2011; pp. 209–214. [Google Scholar]
- Higuchi, T. Approach to an irregular time series on the basis of the fractal theory. Phys. D Nonlinear Phenom. 1988, 31, 277–283. [Google Scholar] [CrossRef]
- Hjorth, B. EEG analysis based on time domain properties. Electroencephalogr. Clin. Neurophysiol. 1970, 29, 306–310. [Google Scholar] [CrossRef]
- Jenke, R.; Peer, A.; Buss, M. Feature extraction and selection for emotion recognition from EEG. IEEE Trans. Affect. Comput. 2014, 5, 327–339. [Google Scholar] [CrossRef]
- Oh, S.-H.; Lee, Y.-R.; Kim, H.-N. A novel EEG feature extraction method using Hjorth parameter. Int. J. Electron. Electr. Eng. 2014, 2, 106–110. [Google Scholar] [CrossRef] [Green Version]
- Abdul-Latif, A.A.; Cosic, I.; Kumar, D.K.; Polus, B.; Da Costa, C. Power changes of EEG signals associated with muscle fatigue: The root mean square analysis of EEG bands. In Proceedings of the 2004 Intelligent Sensors, Sensor Networks and Information Processing Conference, Melbourne, VIC, Australia, 14–17 December 2004; pp. 531–534. [Google Scholar]
- Murugappan, M.; Rizon, M.; Nagarajan, R.; Yaacob, S. Inferring of human emotional states using multichannel EEG. Eur. J. Sci. Res. 2010, 48, 281–299. [Google Scholar]
- Nakisa, B.; Rastgoo, M.N.; Tjondronegoro, D.; Chandran, V. Evolutionary computation algorithms for feature selection of EEG-based emotion recognition using mobile sensors. Expert Syst. Appl. 2018, 93, 143–155. [Google Scholar] [CrossRef] [Green Version]
- Dzedzickis, A.; Kaklauskas, A.; Bucinskas, V. Human Emotion Recognition: Review of Sensors and Methods. Sensors 2020, 20, 592. [Google Scholar] [CrossRef] [Green Version]
- Zheng, W.; Zhu, J.; Lu, B. Identifying Stable Patterns over Time for Emotion Recognition from EEG. IEEE Trans. Affect. Comput. 2019, 10, 417–429. [Google Scholar] [CrossRef] [Green Version]
- Song, T.; Zheng, W.; Song, P.; Cui, Z. EEG Emotion Recognition Using Dynamical Graph Convolutional Neural Networks. IEEE Trans. Affect. Comput. 2018, 11, 532–541. [Google Scholar] [CrossRef] [Green Version]
- Musha, T.; Terasaki, Y.; Haque, H.A.; Ivamitsky, G.A. Feature extraction from EEGs associated with emotions. Artif. Life Robot. 1997, 1, 15–19. [Google Scholar] [CrossRef]
- Bos, D.O. EEG-based emotion recognition. Influ. Vis. Audit. Stimuli 2006, 56, 1–17. [Google Scholar]
- Gabor, D. A New Microscopic Principle. Nature 1948, 161, 777–778. [Google Scholar] [CrossRef]
- Lobaz, P. CGDH Tools: Getting started in computer generated display holography. In Proceedings of the 11th International Symposium on Display Holography—ISDH 2018, Aveiro, Portugal, 25–29 June 2018; pp. 159–165. [Google Scholar]
- Tsang, P.W.M.; Poon, T. Review on the State-of-the-Art Technologies for Acquisition and Display of Digital Holograms. IEEE Trans. Ind. Inform. 2016, 12, 886–901. [Google Scholar] [CrossRef]
- Kononenko, I. Estimating attributes: Analysis and extensions of RELIEF. In Proceedings of the European Conference on Machine Learning, Berlin, Germany, 6–8 April 1994; pp. 171–182. [Google Scholar]
- Zhang, J.; Yin, Z.; Chen, P.; Nichele, S. Emotion recognition using multi-modal data and machine learning techniques: A tutorial and review. Inf. Fusion 2020, 59, 103–126. [Google Scholar] [CrossRef]
- Goldberger, J.; Hinton, G.E.; Roweis, S.; Salakhutdinov, R.R. Neighbourhood components analysis. In Proceedings of the 17th International Conference on Neural Information Processing Systems, Vancouver, BC, Canada, 1 December 2004; Volume 17, pp. 513–520. [Google Scholar]
- Siddharth, S.; Jung, T.P.; Sejnowski, T.J. Utilizing deep learning towards multi-modal bio-sensing and vision-based affective computing. IEEE Trans. Affect. Comput. 2022, 13, 96–107. [Google Scholar] [CrossRef] [Green Version]
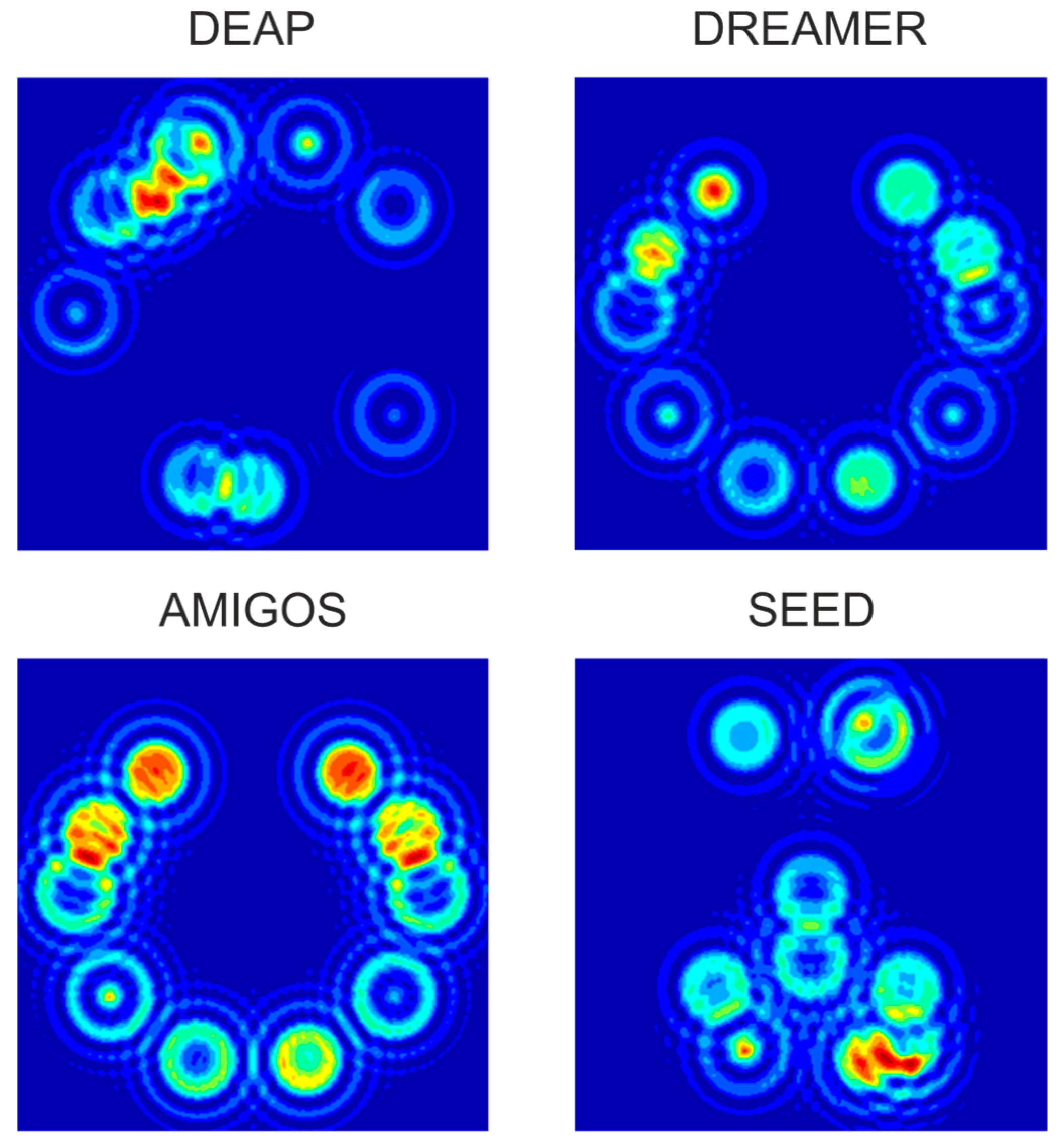
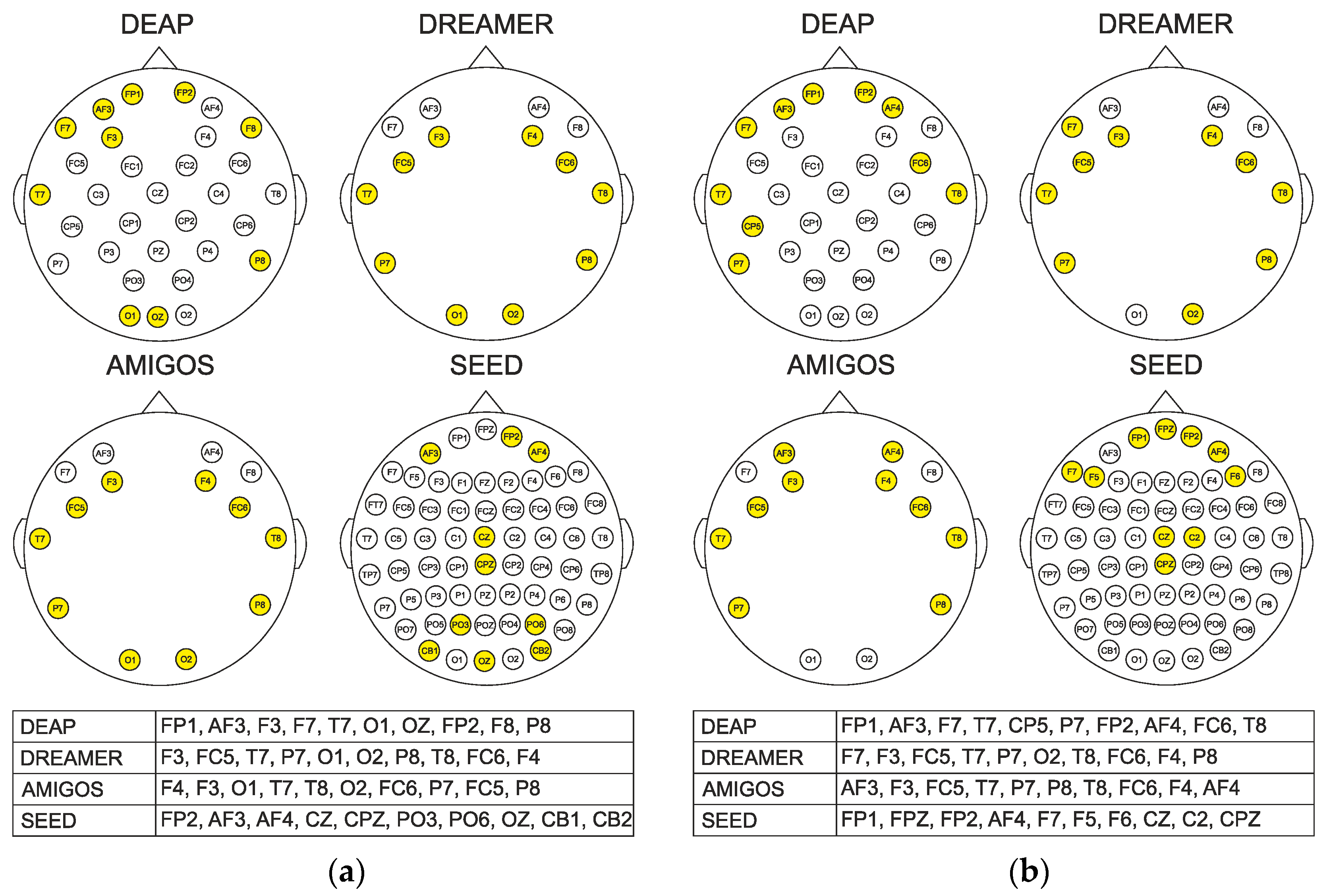
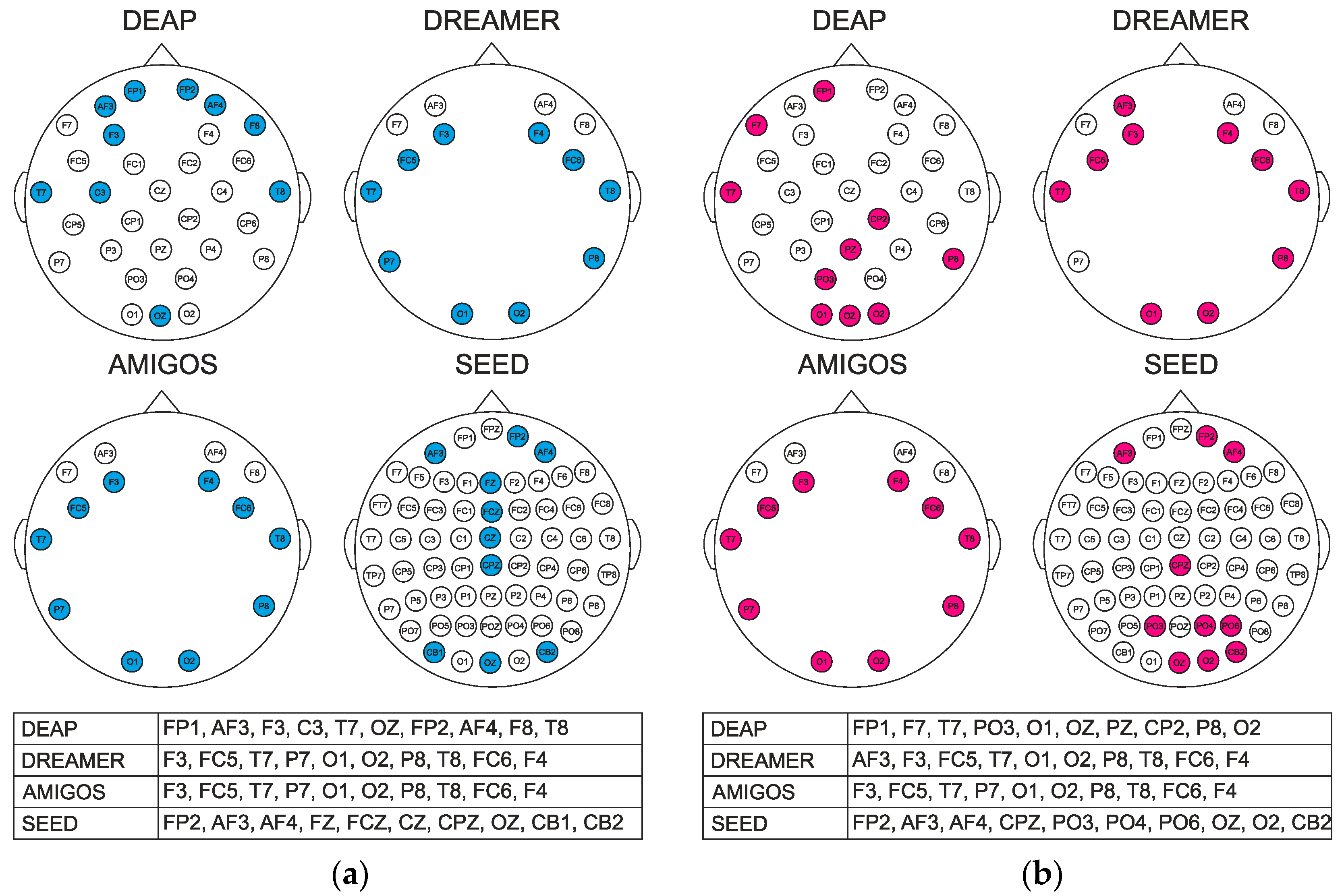
| Dataset | DEAP | DREAMER | AMIGOS | SEED |
|---|---|---|---|---|
| Participants | 32 | 23 | 40 | 15 |
| Trials | 40 | 18 | 16 | 10 |
| Channels | 32 | 14 | 14 | 62 |
| Affective states | Valence Arousal Dominance | Valence Arousal Dominance | Valence Arousal Dominance | Valence |
| Rating scale range (threshold) | 1–9 (4.5) | 1–5 (2.5) | 1–9 (4.5) | N/A |
| Percentage | Channels |
|---|---|
| >75% | F4, F3 |
| 60–75% | T7, FP1, FP2, T8, F7, F8 |
| 45–60% | O1, P7, P8, O2 |
| 30–45% | FC5, FC6, C4, C3, AF3, AF4 |
| <30% | P3, P4, Pz |
| DEAP | DREAMER | AMIGOS | SEED | |||
|---|---|---|---|---|---|---|
| R-HOLO-FM | Valence | Accuracy | 83.26 | 90.76 | 88.54 | 88.19 |
| F1-score | 87.13 | 89.09 | 89.79 | 88.51 | ||
| Arousal | Accuracy | 83.85 | 92.92 | 91.51 | N/A | |
| F1-score | 86.80 | 89.25 | 87.77 | N/A | ||
| Dominance | Accuracy | 88.58 | 92.97 | 90.34 | N/A | |
| F1-score | 86.56 | 89.47 | 87.83 | N/A | ||
| N-HOLO-FM | Valence | Accuracy | 81.88 | 86.12 | 88.53 | 88.31 |
| F1-score | 86.16 | 88.48 | 87.90 | 88.59 | ||
| Arousal | Accuracy | 82.45 | 89.07 | 91.32 | N/A | |
| F1-score | 85.76 | 88.58 | 87.58 | N/A | ||
| Dominance | Accuracy | 88.35 | 89.82 | 86.10 | N/A | |
| F1-score | 89.95 | 89.04 | 87.89 | N/A | ||
| Random | Valence | Accuracy | 51.33 | 48.79 | 49.57 | 43.33 |
| F1-score | 50.42 | 48.21 | 49.43 | 43.21 | ||
| Arousal | Accuracy | 49.06 | 51.45 | 49.78 | N/A | |
| F1-score | 48.13 | 49.21 | 46.96 | N/A | ||
| Dominance | Accuracy | 50.70 | 51.21 | 57.33 | N/A | |
| F1-score | 49.35 | 45.87 | 57.33 | N/A | ||
| Majority | Valence | Accuracy | 63.13 | 61.11 | 56.47 | 50.00 |
| F1-score | 38.70 | 37.93 | 36.09 | 33.33 | ||
| Arousal | Accuracy | 63.75 | 72.43 | 65.95 | N/A | |
| F1-score | 38.93 | 42.02 | 39.74 | N/A | ||
| Dominance | Accuracy | 66.72 | 77.05 | 54.74 | N/A | |
| F1-score | 40.02 | 43.52 | 35.38 | N/A | ||
| Class ratio | Valence | Accuracy | 45.94 | 48.79 | 51.72 | 50.67 |
| F1-score | 47.66 | 48.79 | 49.14 | 49.33 | ||
| Arousal | Accuracy | 45.94 | 39.61 | 45.69 | N/A | |
| F1-score | 42.03 | 40.10 | 43.97 | N/A | ||
| Dominance | Accuracy | 42.03 | 35.75 | 51.72 | N/A | |
| F1-score | 38.59 | 30.92 | 51.72 | N/A |
| Study | Used Feature(s) | Classification Method(s) | Number of Channels | Best Accuracy |
|---|---|---|---|---|
| Koelstra et al. [54] | PSD | NB | 32 | V: 57.60 A: 62.00 |
| Li et al. [21] | Entropy and energy | kNN | 18 | V: 85.74 A: 87.90 |
| Bazgir et al. [22] * | Entropy and energy | SVM | 10 | V: 91.10 A: 91.30 |
| Mohammadi et al. [23] | Entropy and energy | kNN | 10 | V: 86.75 A: 84.05 |
| Özerdem et al. [24] | Various time and frequency domain features | MLPNN | 5 | V: 77.14 |
| Wang et al. [25] | Band energy (spectrogram) | SVM | 8 for V 10 for A | V: 74.41 A: 73.64 |
| Msonda et al. [26] | EMD IMFs | LSVC | 8 | V: 67.00 |
| Menon et al. [27] ** | Various time and frequency domain features | HDC | Feature channel vector set | V: 76.70 A: 74.20 |
| Gupta et al. [28] | IP | RF | 6 | V: 79.99 A: 79.95 |
| Mert et al. [29] | Various time and frequency domain features | MEMD + ANN | 18 | V: 72.87 A: 75.00 |
| Zhang et al. [32] | Band power | PNN | 9 for V 8 for A | V: 81.21 A: 81.76 |
| Our method | R-HOLO-FM | CNN + SVM | 10 | V: 83.26 A: 83.85 D: 88.58 |
| Our method | N-HOLO-FM | CNN + SVM | 10 | V: 81.88 A: 82.45 D: 88.35 |
| Study | Used Feature(s) | Classification Method(s) | Number of Channels | Best Accuracy |
|---|---|---|---|---|
| Katsigiannis et al. [55] | PSD | SVM | 14 | V: 62.49 A: 62.17 D: 61.84 |
| Msonda et al. [26] | EMD IMF | LSVC | 8 | V: 80.00 |
| Our method | R-HOLO-FM | CNN + SVM | 10 | V: 90.76 A: 92.92 D: 92.97 |
| Our method | N-HOLO-FM | CNN + SVM | 10 | V: 86.12 A: 89.07 D: 89.82 |
| Study | Used Feature(s) | Classification Method(s) | Number of Channels | Best Accuracy |
|---|---|---|---|---|
| Miranda et al. [56] | PSD, SPA | SVM | 14 | V: 57.60 A: 59.20 |
| Msonda et al. [26] | EMD IMF | LR | 8 | V: 78.00 |
| Menon et al. [27] * | Various time and frequency domain features | HDC | Feature channel vector set | V: 87.10 A: 80.50 |
| Our method | R-HOLO-FM | CNN + SVM | 10 | V: 88.54 A: 91.51 D: 90.34 |
| Our method | N-HOLO-FM | CNN + SVM | 10 | V: 88.53 A: 91.32 D: 86.10 |
| Study | Used Feature(s) | Classification Method(s) | Number of Channels | Best Accuracy |
|---|---|---|---|---|
| Zheng et al. [33] | Feature map from DE | DBN + SVM | 12 | V: 86.65 |
| Gupta et al. [28] | IP | RF | 12 | V: 90.48 |
| Pane et al. [34] | DE | SDA + LDA | 15 | V: 99.85 |
| Cheah et al. [35] | Extracted with VGG14 | VGG14 1D kernel (T-then-S) | 10 | V: 91.67 |
| Zheng [36] | Raw EEG features | GSCCA | 12 | V: 83.72 |
| Our method | R-HOLO-FM | CNN + SVM | 10 | V: 88.19 |
| Our method | N-HOLO-FM | CNN + SVM | 10 | V: 88.31 |
| Dataset | Valence | Arousal | Dominance |
|---|---|---|---|
| DEAP | 82.55 | 82.27 | 88.81 |
| DREAMER | 90.26 | 91.87 | 93.24 |
| AMIGOS | 83.63 | 87.84 | 90.40 |
| SEED | 82.07 | N/A | N/A |
| Dataset | Valence | Arousal | Dominance |
|---|---|---|---|
| DEAP | 87.82 | 89.26 | 82.12 |
| DREAMER | 89.58 | 93.27 | 89.42 |
| AMIGOS | 88.92 | 92.08 | 85.44 |
| SEED | 87.70 | N/A | N/A |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topic, A.; Russo, M.; Stella, M.; Saric, M. Emotion Recognition Using a Reduced Set of EEG Channels Based on Holographic Feature Maps. Sensors 2022, 22, 3248. https://doi.org/10.3390/s22093248
Topic A, Russo M, Stella M, Saric M. Emotion Recognition Using a Reduced Set of EEG Channels Based on Holographic Feature Maps. Sensors. 2022; 22(9):3248. https://doi.org/10.3390/s22093248
Chicago/Turabian StyleTopic, Ante, Mladen Russo, Maja Stella, and Matko Saric. 2022. "Emotion Recognition Using a Reduced Set of EEG Channels Based on Holographic Feature Maps" Sensors 22, no. 9: 3248. https://doi.org/10.3390/s22093248
APA StyleTopic, A., Russo, M., Stella, M., & Saric, M. (2022). Emotion Recognition Using a Reduced Set of EEG Channels Based on Holographic Feature Maps. Sensors, 22(9), 3248. https://doi.org/10.3390/s22093248






