Data Fusion Approaches for the Characterization of Musts and Wines Based on Biogenic Amine and Elemental Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Solutions
2.2. Samples
2.3. Instruments
2.4. Analytical Procedures
2.4.1. Biogenic Amine Determination
2.4.2. Elemental Composition Determination
2.5. Data Analysis
3. Results and Discussion
3.1. Low-Level Data Fusion
3.2. Mid-Level Data Fusion
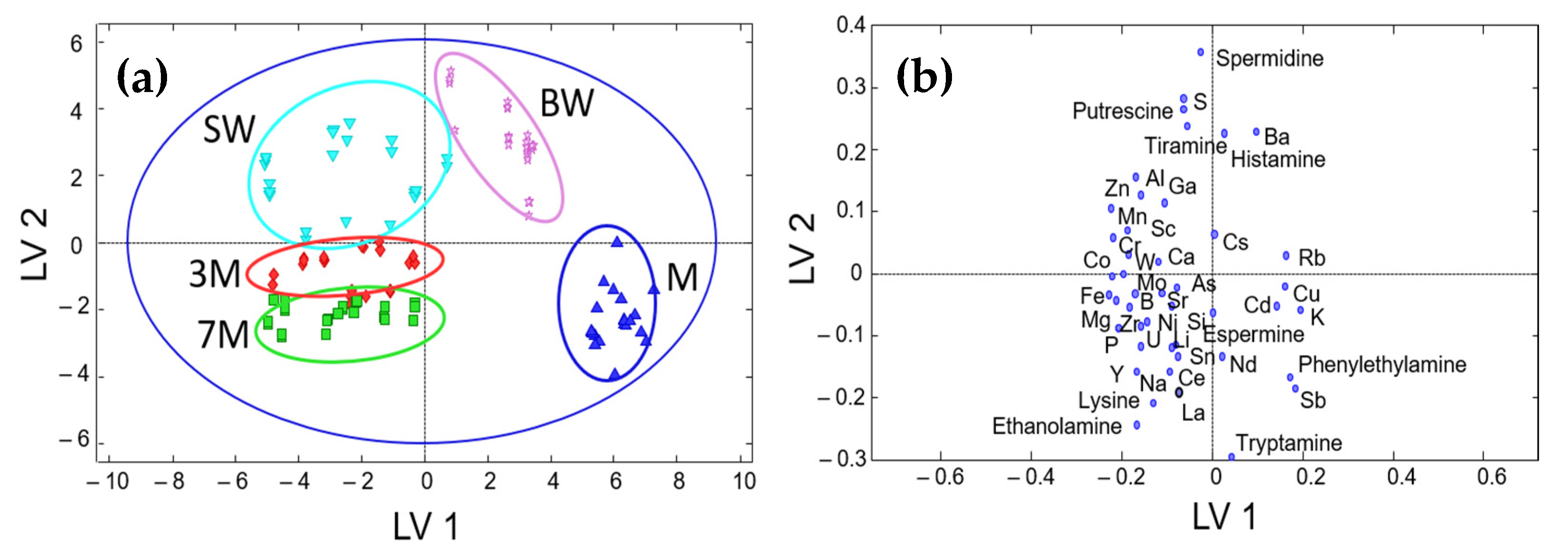
3.3. Sample Classification
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Borras, E.; Ferre, J.; Boque, R.; Mestres, M.; Acena, L.; Busto, O. Data fusion methodologies for food and beverage authentication and quality assessment—A review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bevilacqua, M.; Bro, R.; Marini, F.; Rinnan, A.; Rasmussen, M.A.; Skov, T. Recent chemometrics advances for foodomics. Trends Anal. Chem. 2017, 96, 42–51. [Google Scholar] [CrossRef]
- Callao, M.P.; Ruisanchez, I. An overview of multivariate qualitative methods for food fraud detection. Food Control 2018, 86, 283–293. [Google Scholar] [CrossRef]
- Biancolillo, A.; Marini, F.; Ruckebusch, C.; Vitale, R. Chemometric strategies for spectroscopy-based food authentication. App. Sci. 2020, 10, 6544. [Google Scholar] [CrossRef]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of electronic nose, electronic tongue and computer vision for animal source food authentication and quality assessment—A review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Ranaweera, R.K.R.; Capone, D.L.; Bastian, S.E.P.; Cozzolino, D.; Jeffery, D.W. A review of wine authentication using spectroscopic approaches in combination with chemometrics. Molecules 2021, 26, 4334. [Google Scholar] [CrossRef]
- Arslan, M.; Tahir, H.E.; Zareed, M.; Shi, J.Y.; Rakha, A.; Bilal, M.; Huang, X.W.; Li, Z.H.; Zou, X.B. Recent trends in quality control, discrimination and authentication of alcoholic beverages using nondestructive instrumental techniques—A review. Trends Food Sci. Technol. 2021, 107, 80–113. [Google Scholar] [CrossRef]
- Da Costa, N.L.; da Costa, M.S.; Barbosa, R. A Review on the application of chemometrics and machine learning algorithms to evaluate beer authentication. Food Anal. Methods 2021, 14, 136–155. [Google Scholar] [CrossRef]
- Schiavone, S.; Marchionni, B.; Bucci, R.; Marini, F.; Biancolillo, A. Authentication of Grappa (Italian grape marc spirit) by Mid and Near Infrared spectroscopies coupled with chemometrics. Vib. Spectrosc. 2020, 107, 103040. [Google Scholar] [CrossRef]
- Giannetti, V.; Mariani, M.B.; Marini, F.; Torrelli, P.; Biancolillo, A. Grappa and Italian spirits: Multi-platform investigation based on GC-MS, MIR and NIR spectroscopies for the authentication of the Geographical Indication. Microchem. J. 2020, 157, 104896. [Google Scholar] [CrossRef]
- Rios-Reina, R.; Callejon, R.M.; Savorani, F.; Amigo, J.M.; Cocchi, M. Data fusion approaches in spectroscopic characterization and classification of PDO wine vinegars. Talanta 2019, 98, 560–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez-Beltran, C.H.; Zuniga-Arroyo, V.M.; Andrade, J.M.; Cuadros-Rodriguez, L.; Perez-Caballero, G.; Jimenez-Carvelo, A.M. A sensor-based methodology to differentiate pure and mixed white tequilas based on fused infrared spectra and multivariate data treatment. Chemosensors 2021, 9, 47. [Google Scholar] [CrossRef]
- Biancolillo, A.; Bucci, R.; Magri, A.L.; Magri, A.D.; Marini, F. Data-fusion for multiplatform characterization of an Italian craft beer aimed at its authentication. Anal. Chim. Acta 2014, 820, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, N.; Savorani, F.; Bro, R.; Cocchi, M. Fused adjacency matrices to enhance information extraction: The beer benchmark. Anal. Chim. Acta 2019, 1061, 70–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios-Reina, R.; Azcarate, S.M.; Camina, J.M.; Goicoechea, H.C. Multi-level data fusion strategies for modeling three-way electrophoresis capillary and fluorescence arrays enhancing geographical and grape variety classification of wines. Anal Chim. Acta 2020, 1126, 52–62. [Google Scholar] [CrossRef]
- Tan, J.; Li, R.; Jiang, Z.T.; Zhang, Y.; Hou, Y.M.; Wang, Y.R.; Wu, X.; Gong, L. Geographical classification of Chinese Cabernet Sauvignon wines by data fusion of ultraviolet-visible and synchronous fluorescence spectroscopies: The combined use of multiple wavelength differences. Aust. J. Grape Wine Res. 2016, 22, 358–365. [Google Scholar] [CrossRef]
- Campmajo, G.; Saurina, J.; Nunez, O. FIA-HRMS fingerprinting subjected to chemometrics as a valuable tool to address food classification and authentication: Application to red wine, paprika, and vegetable oil samples. Food Chem. 2022, 373, 131491. [Google Scholar] [CrossRef]
- Ceto, X.; Capdevila, J.; Puig-Pujol, A.; del Valle, M. Cava wine authentication employing a voltammetric electronic tongue. Electroanalysis 2014, 26, 1504–1512. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Hernandez, C.; Salvo-Comino, C.; Martin-Pedrosa, F.; Garcia-Cabezon, C.; Rodriguez-Mendez, M.L. Analysis of red wines using an electronic tongue and infrared spectroscopy, Correlations with phenolic content and color parameters. LWT-Food Sci. Technol. 2020, 118, 108785. [Google Scholar] [CrossRef]
- Zhang, H.H.; Shao, W.Q.; Qiu, S.S.; Wang, J.; Wei, Z.B. Collaborative analysis on the marked ages of rice wines by electronic tongue and nose based on different feature data sets. Sensor 2020, 20, 1065. [Google Scholar] [CrossRef] [Green Version]
- Gutierrez, M.; Domingo, C.; Vila-Planas, J.; Ipatov, A.; Capdevila, F.; Demming, S.; Buttgenbach, S.; Llobera, A.; Jimenez-Jorquera, C. Hybrid electronic tongue for the characterization and quantification of grape variety in red wines. Sens. Actuators B Chem. 2011, 156, 695–702. [Google Scholar] [CrossRef]
- Zhang, C.; Bailey, D.P.; Suslick, K.S. Colorimetric sensor arrays for the analysis of beers: A feasibility study. J. Agric. Food Chem. 2006, 54, 4925–4931. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Fang, M.; LaGasse, M.K.; Askim, J.R.; Suslick, K.S. Colorimetric recognition of aldehydes and ketones. Angew. Chem. Int. Ed. 2017, 56, 9860–9863. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Suslick, K.S. A hand-held optoelectronic nose for the identification of liquors. ACS Sens. 2018, 3, 121–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Suslick, K.S. The optoelectronic nose. Acc. Chem. Res. 2021, 54, 950–960. [Google Scholar] [CrossRef]
- Silvestri, M.; Elia, A.; Bertelli, D.; Salvatore, E.; Durante, C.; Vigni, M.L.; Marchetti, A.; Cocchi, M. A mid level data fusion strategy for the Varietal Classification of Lambrusco PDO wines. Chemom. Intell. Lab. Sys. 2014, 137, 181–189. [Google Scholar] [CrossRef] [Green Version]
- Belmonte-Sanchez, J.R.; Romero-Gonzalez, R.; Vidal, J.L.M.; Arrebola, F.J.; Garrido Frenich, A. H-1 NMR and multi-technique data fusion as metabolomic tool for the classification of golden rums by multivariate statistical analysis. Food Chem. 2020, 317, 126363. [Google Scholar] [CrossRef]
- Izquierdo-Llopart, A.; Saurina, J. Multi-sensor characterization of sparkling wines based on data fusion. Chemosensors 2021, 9, 200. [Google Scholar] [CrossRef]
- Cavdaroglu, C.; Ozen, B. Prediction of vinegar processing parameters with chemometric modelling of spectroscopic data. Microchem. J. 2021, 171, 106886. [Google Scholar] [CrossRef]
- Simon Sarkadi, L. Amino acids and biogenic amines as food quality factors. Pure Appl. Chem. 2019, 91, 289–300. [Google Scholar] [CrossRef]
- Saurina, J. Characterization of wines using compositional profiles and chemometrics. TrAC Trends Anal. Chem. 2010, 29, 234–245. [Google Scholar] [CrossRef]
- Sentellas, S.; Núñez, O.; Saurina, J. Recent advances in the determination of biogenic amines in food samples by (U)HPLC. J. Agric. Food Chem. 2016, 64, 7667–7678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mir-Cerda, A.; Izquierdo-Llopart, A.; Saurina, J.; Sentellas, S. Oenological processes and product qualities in the elaboration of sparkling wines determine the biogenic amine content. Fermentation 2021, 7, 144. [Google Scholar] [CrossRef]
- Mitar, I.; Ljubenkov, I.; Rohtek, N.; Prkic, A.; Andelic, I.; Vuletic, N. The content of biogenic amines in Croatian wines of different geographical origins. Molecules 2018, 23, 2570. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Han, F.L.; Liu, Y.J.; Wang, W.N. Determination of biogenic amines in wine using modified liquid-liquid extraction with high performance liquid chromatography-fluorescence detector. Food Anal. Methods 2020, 13, 911–922. [Google Scholar] [CrossRef]
- Palomino-Vasco, M.; Rodriguez-Caceres, M.I.; Mora-Diez, N.; Pardo-Botello, R.; Acedo-Valenzuela, M.I. Biogenic amines profile in red wines regarding aging and storage conditions. J. Food Compos. Anal. 2019, 83, 103295. [Google Scholar] [CrossRef]
- Perez-Magarino, S.; Ortega-Heras, M.; Martinez-Lapuente, L.; Guadalupe, Z.; Ayestara, B. Multivariate analysis for the differentiation of sparkling wines elaborated from autochthonous Spanish grape varieties: Volatile compounds, amino acids and biogenic amines. Eur. Food Res. Technol. 2013, 236, 827–841. [Google Scholar] [CrossRef]
- Gil, R.L.; Amorim, C.G.; Montenegro, M.C.B.S.M.; Araujo, A.N. HPLC-potentiometric method for determination of biogenic amines in alcoholic beverages: A reliable approach for food quality control. Food Chem. 2022, 372, 131288. [Google Scholar] [CrossRef]
- Bozic, J.T.; Butinar, L.; Marusic, M.B.; Korte, D.; Vodopivec, B.M. Determination of biogenic amines formation by autochthonous lactic acid bacteria from ’Refosk’ grapes using different analytical methods. LWT Food Sci. Technol. 2022, 156, 112908. [Google Scholar] [CrossRef]
- Han, S.Y.; Hao, L.L.; Shi, X.; Niu, J.M.; Zhang, B. Development and application of a new QuEChERS method in UHPLC-QqQ-MS/MS to detect seven biogenic amines in Chinese wines. Foods 2019, 8, 552. [Google Scholar] [CrossRef] [Green Version]
- Gonzalvez, A.; Llorens, A.; Cervera, M.L.; Armenta, S.; de la Guardia, M. Elemental fingerprint of wines from the protected designation of origin Valencia. Food Chem. 2009, 112, 26–34. [Google Scholar] [CrossRef]
- Katerinopoulou, K.; Kontogeorgos, A.; Salmas, C.E.; Patakas, A.; Ladavos, A. Geographical origin authentication of agri-food products: A review. Foods 2020, 9, 489. [Google Scholar] [CrossRef] [PubMed]
- Granell, B.; Izquierdo-Llopart, A.; Sahuquillo, A.; López-Sánchez, J.F.; Saurina, J. Characterization of musts, wines, and sparkling wines based on their elemental composition determined by ICP-OES and ICP-MS. Beverages 2022, 8, 3. [Google Scholar] [CrossRef]
- Kozak, M.; Scaman, C.H. Unsupervised classification methods in food sciences: Discussion and outlook. J. Sci. Food Agric. 2008, 88, 1115–1127. [Google Scholar] [CrossRef]
- Granato, D.; Putnik, P.; Kovacevic, D.B.; Santos, J.S.; Calado, V.; Rocha, R.S.; Da Cruz, A.G.; Jarvis, B.; Rodionova, O.Y.; Pomerantsev, A. Trends in chemometrics: Food authentication, microbiology, and effects of processing. Compr. Rev. Food Sci. Food Saf. 2018, 17, 663–677. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Analytes | Sample Type | Method | Remarks | Ref. |
|---|---|---|---|---|
| Putrescine, ethanolamine, histamine, tyramine, cadaverine, phenylethylamine, agmatine, tryptamine, spermine, and spermidine | Musts, base wines, and sparkling wine; Xarel·lo and Pinot Noir varieties | HPLC-FLD with precolumn derivatization using dansyl-Cl | Putrescine, ethanolamine, tyramine, and histamine are the most important in wine quality | [33] |
| Isopenthylamine, ethanolamine, methylamine, ethylamine, spermidine, spermine, putrescine, tyramine, histamine, cadaverine, and tryptamine | Red and white Croatian wines from Hrvatsko zagorje and Dalmatia regions | HPLC-UV with precolumn derivatization using dansyl-Cl | BAs are a discriminating factor for a grape variety and geographical origin for red wines | [34] |
| Putrescine, histamine, tyramine, cadaverine, phenylethylamine, tryptamine, spermine, and spermidine | Red and white wines from Chinese markets | HPLC-FLD with precolumn derivatization using dansyl-Cl; liquid–liquid extraction of derivatives | Predominant BAs were putrescine, tyramine, and 2-phenylethylamine | [35] |
| Putrescine, ethanolamine, histamine, tyramine, cadaverine, phenylethylamine, tryptamine, and agmatine | Red Spanish wines; monovarietal ‘Tempranillo’ wines (young, oak, and aged) | UHPLC-FLD with precolumn derivatization using OPA | Storage time, temperature, and bottle closing influence BA levels. Cork stopper and refrigeration are the best conditions to prevent the increase in histamine and tyramine | [36] |
| Volatile compounds, amino acids, and amines; agmatine, histamine, spermidine, tyrosine, phenylethylamine, isoamylamine, putrescine, tyramine, and tryptamine | Spanish Sparkling wines from Verdejo, Viura, Malvasia, Albarin, Godello, Prieto Picudo, and Garnacha; “Champenoise” method | HPLC-UV with precolumn derivatization using diethyl ethoxymethylenemalonate | Albarin and Prieto Picudo wines showed the highest BA content | [37] |
| Methylamine, ethylamine, putrescine, cadaverine, histamine, spermidine, spermine, phenylethylamine, tyramine, and tryptamine | Alcoholic beverages including red and white wine | Ion-pair chromatography with butane-sulfonic acid; HPLC-potentiometric detection; multi-walled carbon nanotube sensing membrane | Tyramine and tryptamine are the most abundant in red wine; spermidine, spermine, and tryptamine are the most abundant in white wine | [38] |
| Histamine, putrescine, cadaverine, and tyramine | “Refosk” wine from Slovenian-Italian Karst region | HPLC-UV with precolumn derivatization using dansyl-Cl | Some strains of Lactobacillus have the ability to produce BAs | [39] |
| Cadaverine, hexylamine, histamine, phenylethylamine, putrescine, and tyramine | Chinese wines | Direct separation and detection by UHPLC-QqQ-MS/MS; QuEChERS for sample treatment | Histidine is correlated with alcoholic degree and grape variety; phenylethylamine is correlated with pH and storage time | [40] |
| Grape Variety | Quality | Must | Base Wine | Stabilized Wine | 3-Month Sparkling Wine | 7-Month Sparkling Wine |
|---|---|---|---|---|---|---|
| Pinot Noir | A | MPA | BWPA | SWPA | C3PA | C7PA |
| B | MPB | BWPB | SWPB | C3PB | C7PB | |
| C | MPC | BWPC | SWPC | C3PC | C7PC | |
| D | MPD | BWPD | SWPD | C3PD | C7PD | |
| Xarel·lo | A | MXA | BWXA | SWXA | C3XA | C7XA |
| B | MXB | BWXB | SWXB | C3XB | C7XB | |
| C | MXC | BWXC | SWXC | C3XC | C7XC | |
| D | MXD | BWXD | SWXD | C3XD | C7XD |
| Sample | Ethanolamine | Putrescine | Histamine | S | K | Na |
|---|---|---|---|---|---|---|
| MPA | 2.99 | 2.52 | 0.16 | 2.68 | 93.3 | 1.22 |
| MPB | 2.70 | 1.42 | 0.14 | 5.86 | 151.0 | 2.77 |
| MPC | 3.85 | 4.84 | 0.17 | 8.04 | 124.1 | 2.05 |
| MPD | 3.49 | 2.14 | 0.13 | 3.83 | 151.3 | 2.59 |
| MXA | 2.72 | 1.29 | 0.11 | 3.48 | 72.9 | 1.87 |
| MXB | 4.01 | 0.43 | 0.10 | 2.91 | 87.6 | 2.16 |
| MXC | 5.30 | 3.29 | 0.11 | 3.46 | 120.8 | 1.78 |
| MXD | 4.09 | 2.81 | 0.12 | 3.31 | 95.2 | 1.40 |
| BWPA | 3.14 | 4.01 | 0.19 | 32.5 | 47.4 | 0.59 |
| BWPB | 5.21 | 3.42 | 0.18 | 33.9 | 79.0 | 0.50 |
| BWPC | 5.35 | 24.10 | 4.00 | 32.7 | 96.6 | 2.02 |
| BWPD | 6.13 | 21.43 | 3.68 | 22.5 | 77.5 | 3.38 |
| BWXA | 3.86 | 1.81 | 0.11 | 17.7 | 38.8 | 0.50 |
| BWXB | 5.14 | 3.05 | 0.11 | 43.0 | 78.9 | 1.37 |
| BWXC | 5.75 | 10.79 | 1.76 | 57.9 | 63.7 | 3.27 |
| BWXD | 6.51 | 13.07 | 1.94 | 40.9 | 75.7 | 2.44 |
| SWPA | 3.43 | 3.77 | 0.20 | 37.5 | 34.6 | 1.12 |
| SWPB | 5.49 | 2.80 | 0.31 | 25.9 | 37.6 | 2.23 |
| SWPC | 4.75 | 10.81 | 1.20 | 24.3 | 46.0 | 2.82 |
| SWPD | 6.57 | 15.87 | 1.78 | 16.4 | 30.4 | 5.18 |
| SWXA | 3.22 | 0.95 | 0.11 | 16.9 | 34.0 | 0.70 |
| SWXB | 6.29 | 2.43 | 0.21 | 22.7 | 27.2 | 2.00 |
| SWXC | 5.94 | 14.37 | 2.32 | 21.6 | 28.8 | 3.98 |
| SWXD | 7.13 | 10.26 | 1.77 | 21.5 | 35.7 | 4.63 |
| C3PA | 2.89 | 2.11 | 0.18 | 14.2 | 26.2 | 2.48 |
| C3PB | 6.05 | 2.69 | 0.30 | 25.9 | 37.1 | 2.28 |
| C3PC | 4.94 | 12.00 | 1.40 | 20.8 | 44.7 | 3.43 |
| C3PD | 6.42 | 15.90 | 2.32 | 16.8 | 25.4 | 4.69 |
| C3XA | 3.41 | 1.11 | 0.13 | 11.7 | 30.7 | 2.05 |
| C3XB | 7.14 | 3.14 | 0.26 | 24.4 | 14.2 | 2.40 |
| C3XC | 7.25 | 17.83 | 2.64 | 21.2 | 25.9 | 5.23 |
| C3XD | 6.08 | 9.47 | 1.69 | 19.9 | 39.3 | 5.16 |
| C7PA | 2.73 | 1.39 | 0.14 | 14.5 | 30.6 | 2.42 |
| C7PB | 5.50 | 2.28 | 0.26 | 25.3 | 40.7 | 2.20 |
| C7PC | 5.04 | 11.47 | 1.38 | 21.5 | 45.5 | 3.46 |
| C7PD | 6.74 | 18.47 | 2.90 | 19.1 | 21.8 | 5.70 |
| C7XA | 3.44 | 0.94 | 0.12 | 12.2 | 32.4 | 1.99 |
| C7XB | 5.55 | 3.11 | 0.26 | 23.4 | 30.4 | 3.42 |
| C7XC | 5.88 | 10.84 | 1.87 | 20.8 | 39.7 | 5.41 |
| C7XD | 6.06 | 18.50 | 2.79 | 21.2 | 41.0 | 5.04 |
| Classification Rate | |||||
|---|---|---|---|---|---|
| Step | Must | Base Wine | Stabilized Wine | 3-Month Sparkling Wine | 7-Month Sparkling Wine |
| Calibration | 100% | 100% | 90% 1 | 87% 2 | 75% 3 |
| Validation | 100% | 70% 4 | 100% | 100% | 100% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mir-Cerdà, A.; Granell, B.; Izquierdo-Llopart, A.; Sahuquillo, À.; López-Sánchez, J.F.; Saurina, J.; Sentellas, S. Data Fusion Approaches for the Characterization of Musts and Wines Based on Biogenic Amine and Elemental Composition. Sensors 2022, 22, 2132. https://doi.org/10.3390/s22062132
Mir-Cerdà A, Granell B, Izquierdo-Llopart A, Sahuquillo À, López-Sánchez JF, Saurina J, Sentellas S. Data Fusion Approaches for the Characterization of Musts and Wines Based on Biogenic Amine and Elemental Composition. Sensors. 2022; 22(6):2132. https://doi.org/10.3390/s22062132
Chicago/Turabian StyleMir-Cerdà, Aina, Biel Granell, Anaïs Izquierdo-Llopart, Àngels Sahuquillo, José Fermín López-Sánchez, Javier Saurina, and Sonia Sentellas. 2022. "Data Fusion Approaches for the Characterization of Musts and Wines Based on Biogenic Amine and Elemental Composition" Sensors 22, no. 6: 2132. https://doi.org/10.3390/s22062132
APA StyleMir-Cerdà, A., Granell, B., Izquierdo-Llopart, A., Sahuquillo, À., López-Sánchez, J. F., Saurina, J., & Sentellas, S. (2022). Data Fusion Approaches for the Characterization of Musts and Wines Based on Biogenic Amine and Elemental Composition. Sensors, 22(6), 2132. https://doi.org/10.3390/s22062132








