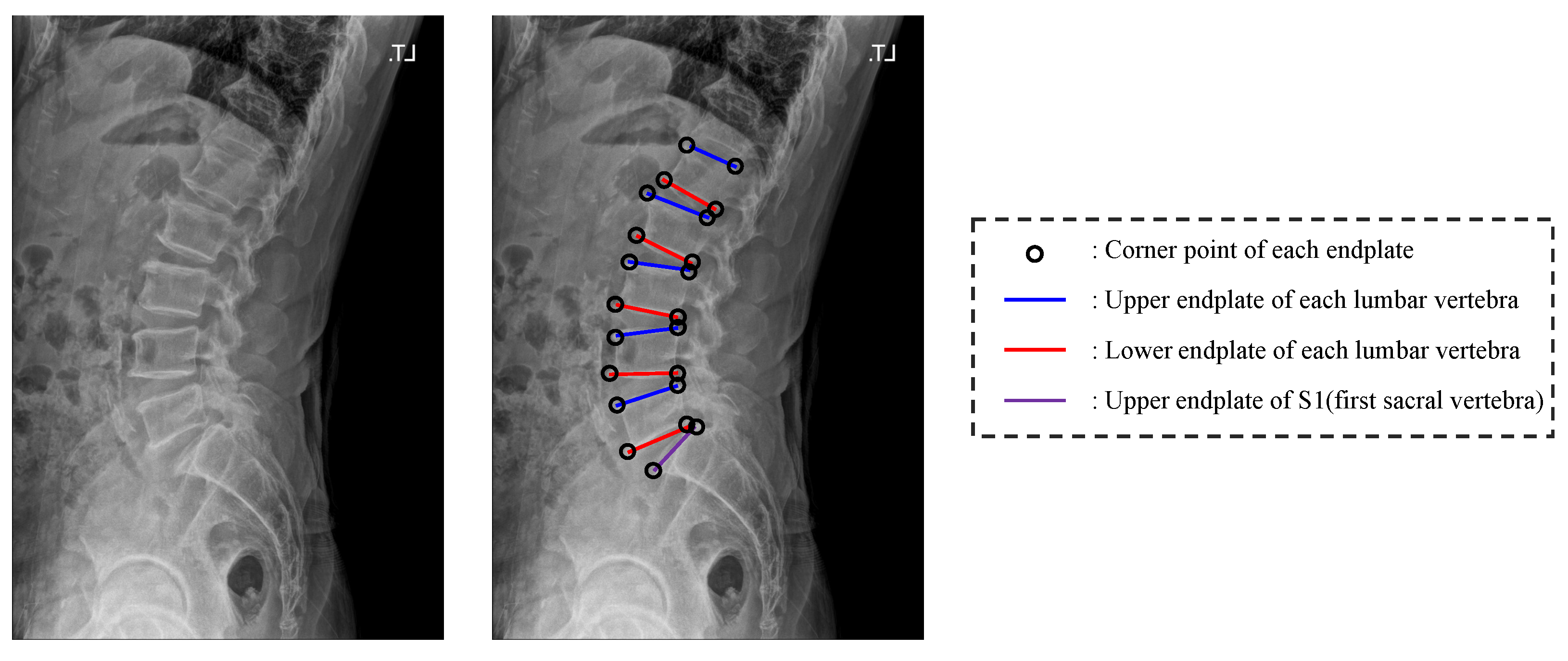
Figure 1.
Illustration for the corner point and upper and lower end plate of each vertebra.
Figure 1.
Illustration for the corner point and upper and lower end plate of each vertebra.
Figure 2.
An overview of the proposed network. The blue boxes denote the neural network and the green boxes denote the algorithmic process.
Figure 2.
An overview of the proposed network. The blue boxes denote the neural network and the green boxes denote the algorithmic process.
Figure 3.
Structure of our Pose-Net.
Figure 3.
Structure of our Pose-Net.
Figure 4.
Example of random spine cutout. (b,c) are results of the random spine cutout performed on (a).
Figure 4.
Example of random spine cutout. (b,c) are results of the random spine cutout performed on (a).
Figure 5.
Failure example. Blue circles and labels denote the ground truth of each vertebra. From left to right, the confidence maps for the centers of L1, L2, L3, L4, L5, and the sacrum are displayed with the input images, respectively.
Figure 5.
Failure example. Blue circles and labels denote the ground truth of each vertebra. From left to right, the confidence maps for the centers of L1, L2, L3, L4, L5, and the sacrum are displayed with the input images, respectively.
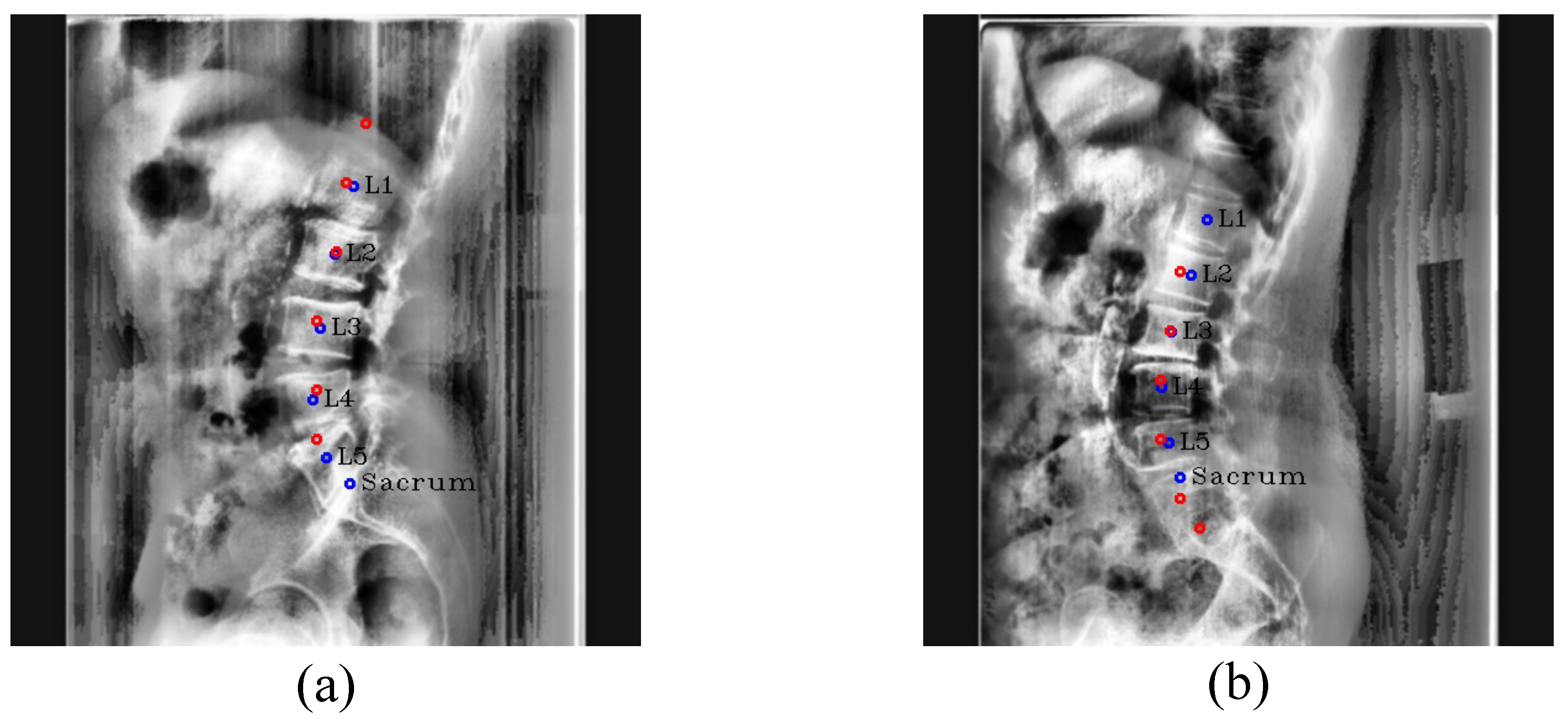
Figure 6.
Post-processing results. (a,c) are the resulting images before performing post-processing, and (b,d) are the resulting images after performing post-processing on (a,c), respectively. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and the blue circles and labels denote the ground truth of the center of each vertebra.
Figure 6.
Post-processing results. (a,c) are the resulting images before performing post-processing, and (b,d) are the resulting images after performing post-processing on (a,c), respectively. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and the blue circles and labels denote the ground truth of the center of each vertebra.
Figure 7.
Post-processing procedure. (a) denotes the boundary lines to make the value of the confidence map zero, (b) denotes the generated confidence map of the unextracted center by using the previous boundary lines, (c) denotes the process of removing the incorrectly extracted center point of T12 and inserting the correct center point, and (d) denotes the final center points after post-processing. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and the blue circles and labels denote the ground truth of the center of each vertebra.
Figure 7.
Post-processing procedure. (a) denotes the boundary lines to make the value of the confidence map zero, (b) denotes the generated confidence map of the unextracted center by using the previous boundary lines, (c) denotes the process of removing the incorrectly extracted center point of T12 and inserting the correct center point, and (d) denotes the final center points after post-processing. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and the blue circles and labels denote the ground truth of the center of each vertebra.
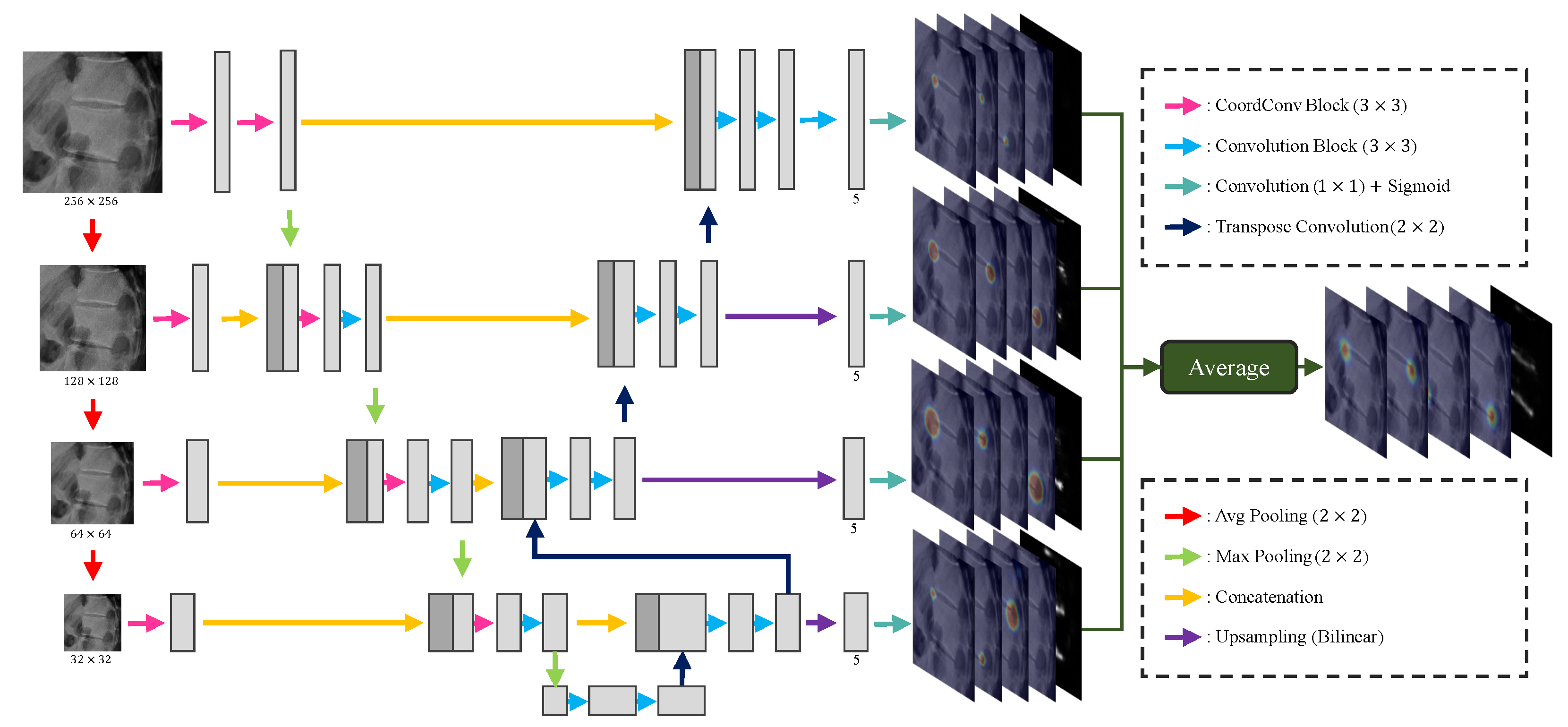
Figure 8.
An overview of our proposed M-Net. In each resolution, the channel of all features except the channel (5) of the last feature is the same as Kim et al. [
5].
Figure 8.
An overview of our proposed M-Net. In each resolution, the channel of all features except the channel (5) of the last feature is the same as Kim et al. [
5].
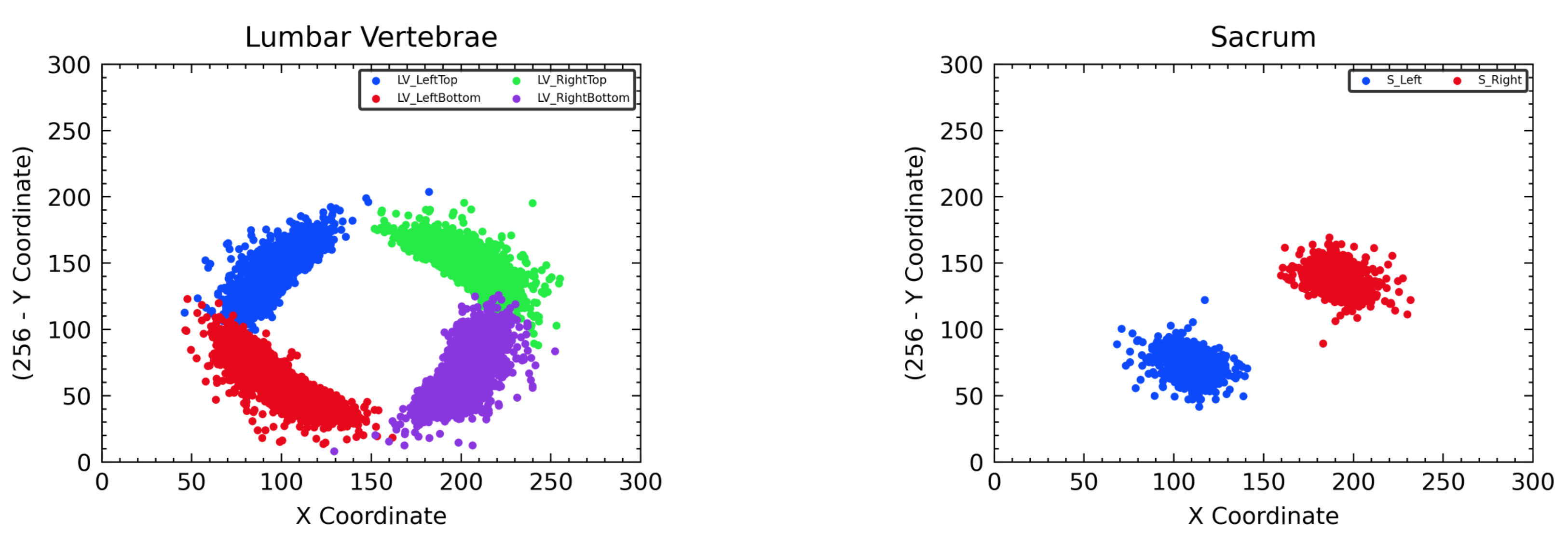
Figure 9.
Distribution of all landmark coordinates in the cropped vertebra images for training M-Net. The left side represents the case of lumbar vertebrae, and the right side represents the case of the sacrum.
Figure 9.
Distribution of all landmark coordinates in the cropped vertebra images for training M-Net. The left side represents the case of lumbar vertebrae, and the right side represents the case of the sacrum.
Figure 10.
Inference results between kernel size of Pose-Net. (b,d) are Pose-Net results with the kernel size of 13 × 13, while (a,c) are Pose-Net results with the kernel size of 7 × 7. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and blue circles and labels denote the ground truth of the centers.
Figure 10.
Inference results between kernel size of Pose-Net. (b,d) are Pose-Net results with the kernel size of 13 × 13, while (a,c) are Pose-Net results with the kernel size of 7 × 7. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and blue circles and labels denote the ground truth of the centers.
Figure 11.
Center detection failure cases. (a,b) are outlier cases caused by the similar appearance of L5 and the sacrum. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and blue circles and labels denote the ground truth of the centers.
Figure 11.
Center detection failure cases. (a,b) are outlier cases caused by the similar appearance of L5 and the sacrum. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and blue circles and labels denote the ground truth of the centers.
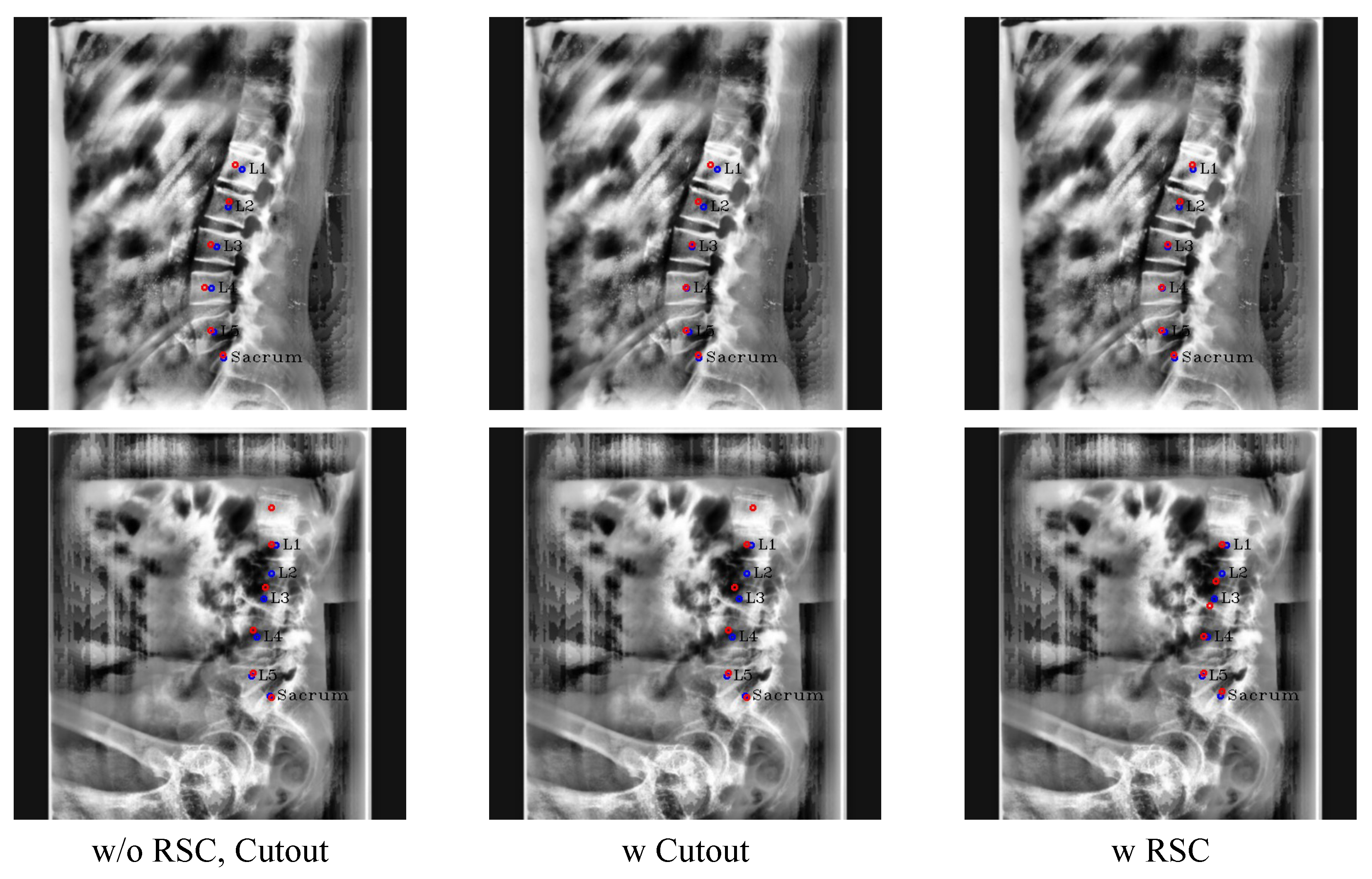
Figure 12.
Results of ablation study with the conventional cutout and RSC. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and blue circles and labels denote the ground truth of the centers.
Figure 12.
Results of ablation study with the conventional cutout and RSC. The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and blue circles and labels denote the ground truth of the centers.
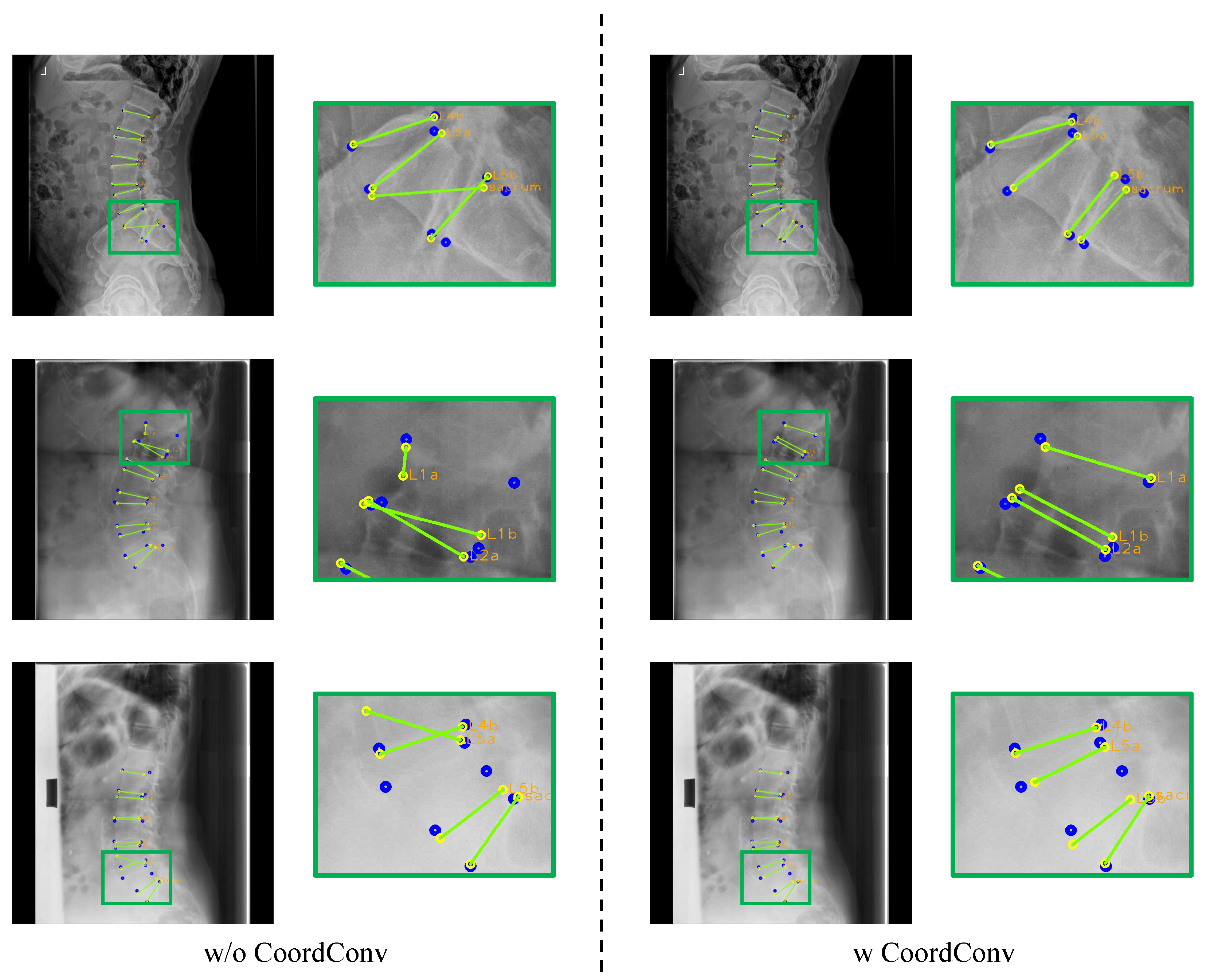
Figure 13.
Results of landmark detection depending on CoordConv. The blue circles represent the ground truth, while yellow circles with labels represent predicted results and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 13.
Results of landmark detection depending on CoordConv. The blue circles represent the ground truth, while yellow circles with labels represent predicted results and the green lines represent the line connecting the predicted landmarks of an endplate.
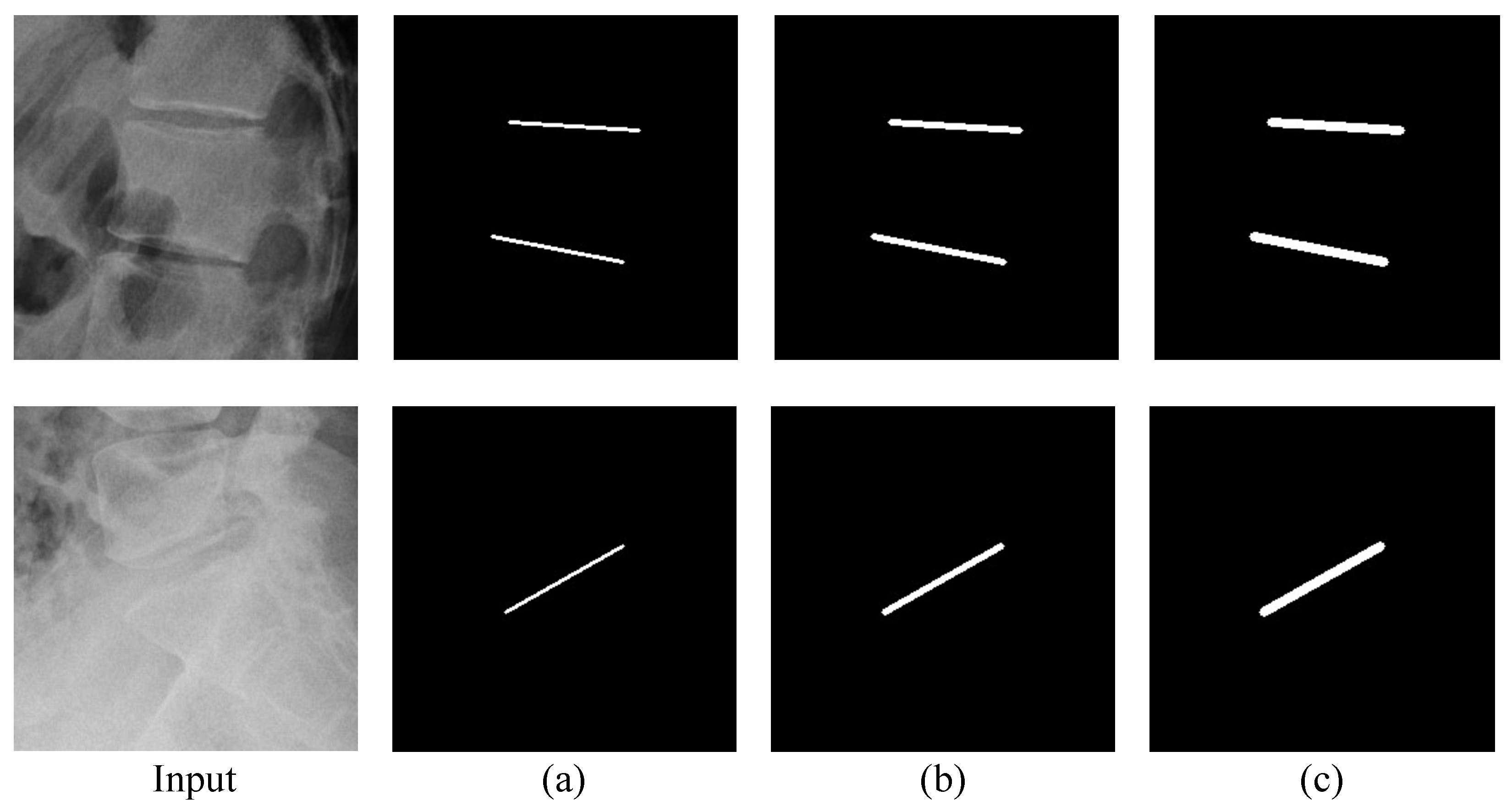
Figure 14.
Example of the ground truth of PAFs by width: (a) 2 pixels, (b) 4 pixels, and (c) 6 pixels.
Figure 14.
Example of the ground truth of PAFs by width: (a) 2 pixels, (b) 4 pixels, and (c) 6 pixels.
Figure 15.
Results of landmark detection with or without PAFs. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 15.
Results of landmark detection with or without PAFs. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 16.
Results of ablation study with CoordConv and PAFs. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 16.
Results of ablation study with CoordConv and PAFs. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 17.
Landmark detection failure cases. (a,b) are outlier cases caused by severe compression fracture and occlusion, respectively. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 17.
Landmark detection failure cases. (a,b) are outlier cases caused by severe compression fracture and occlusion, respectively. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
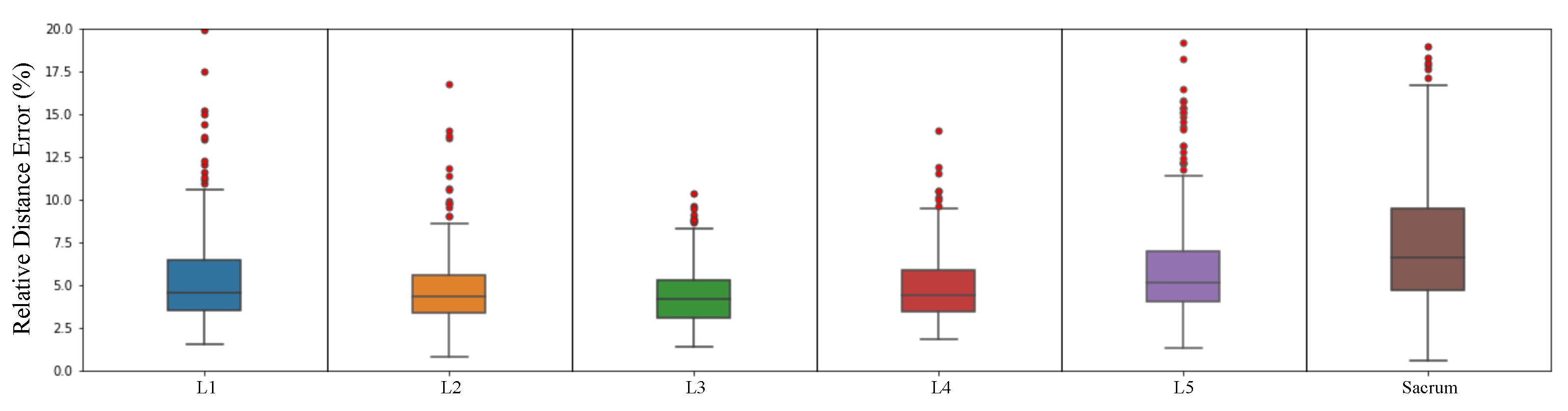
Figure 18.
Box plot of landmark relative distance error except for outliers.
Figure 18.
Box plot of landmark relative distance error except for outliers.
Figure 19.
Results of center detection. Our results are (
a,
c), and the results of Kim et al. [
5] are (
b,
d). The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and the blue circles and labels denote the ground truth of the center of each vertebra.
Figure 19.
Results of center detection. Our results are (
a,
c), and the results of Kim et al. [
5] are (
b,
d). The red circles denote the predicated centers of L1, L2, L3, L4, L5, and the sacrum in order from the top, and the blue circles and labels denote the ground truth of the center of each vertebra.
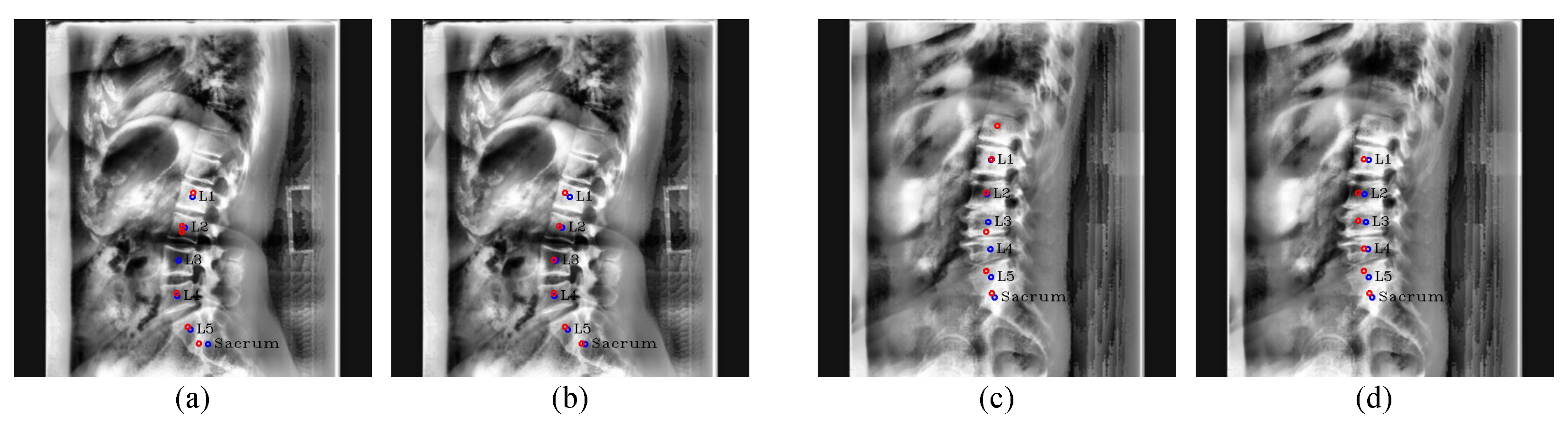
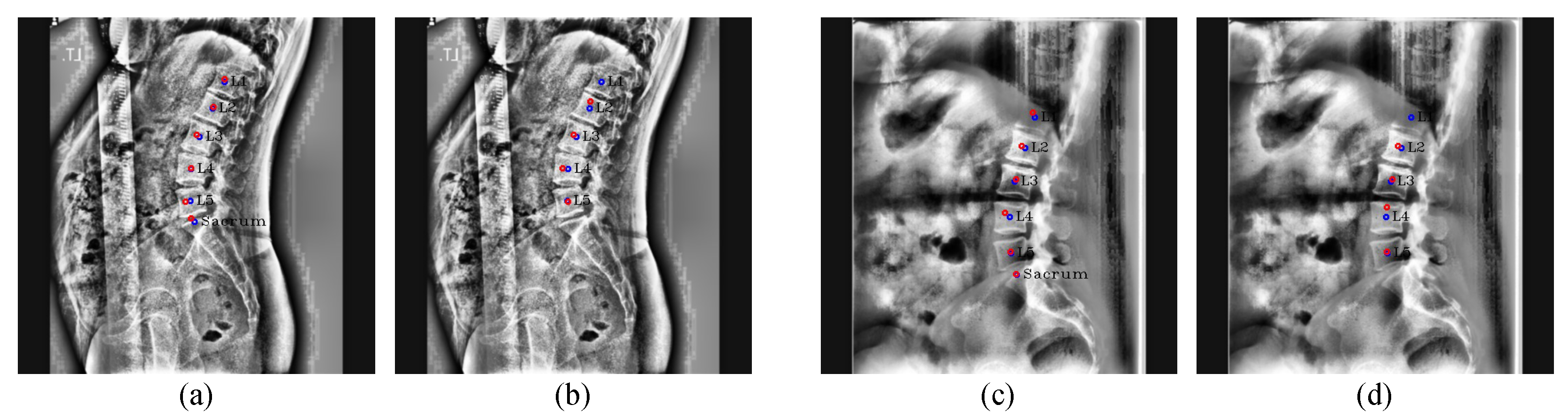
Figure 20.
Example of landmark detection result of ours and Kim et al. [
5]. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
Figure 20.
Example of landmark detection result of ours and Kim et al. [
5]. The blue circles represent the ground truth, while yellow circles with labels represent predicted results, and the green lines represent the line connecting the predicted landmarks of an endplate.
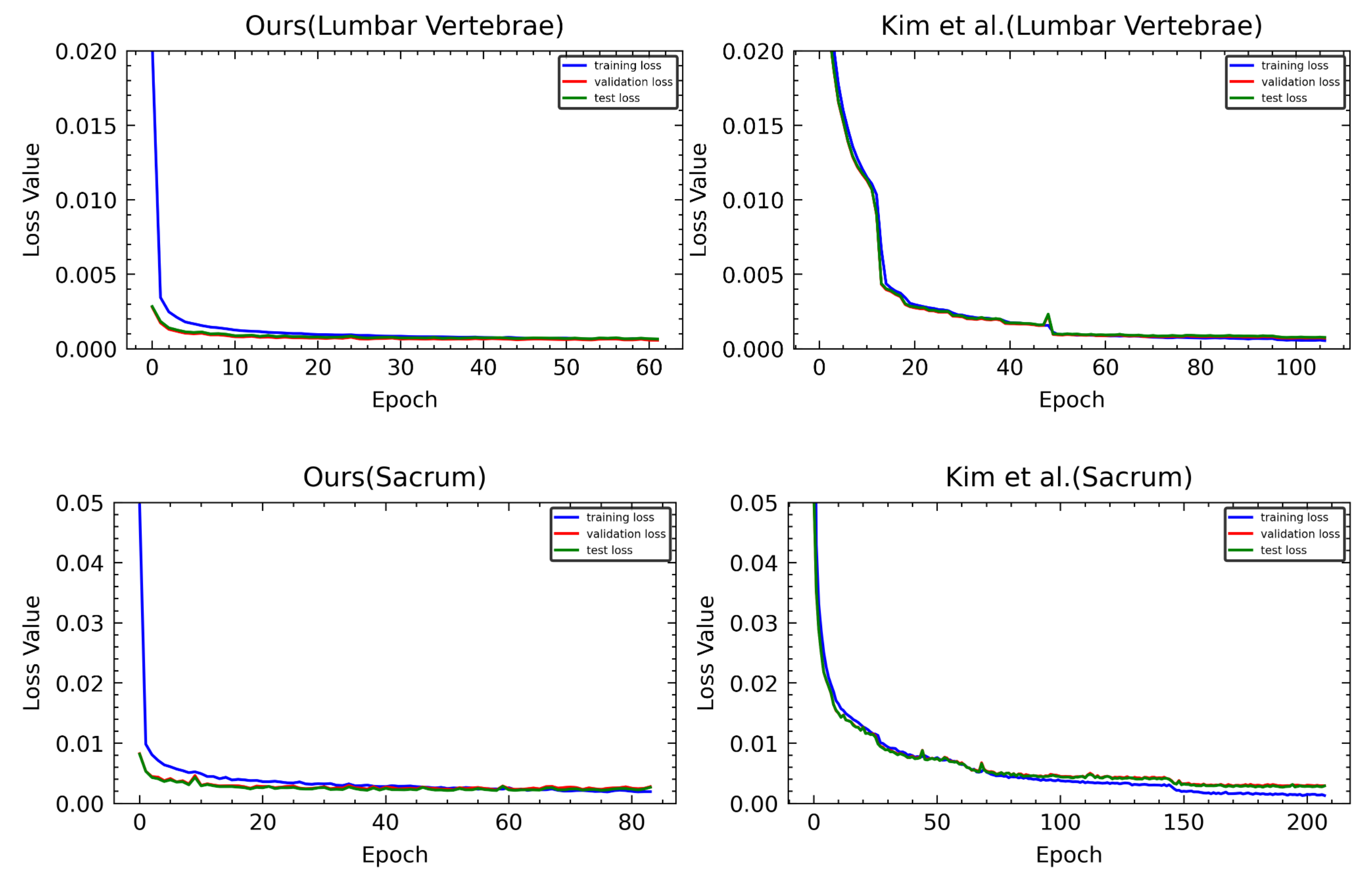
Figure 21.
Loss graphs of ours and Kim et al. [
5]. The first row is a loss graph of M-Net predicting the landmark of the lumbar vertebrae, and the second row is a loss graph of M-Net predicting the landmark of the sacrum.
Figure 21.
Loss graphs of ours and Kim et al. [
5]. The first row is a loss graph of M-Net predicting the landmark of the lumbar vertebrae, and the second row is a loss graph of M-Net predicting the landmark of the sacrum.
Table 1.
Average pixel values inside each vertebra (and the standard deviation).
Table 1.
Average pixel values inside each vertebra (and the standard deviation).
| Vertebra | Pixel Value |
|---|
| L1 | 137.11(30.99) |
| L2 | 132.20(29.65) |
| L3 | 130.83(28.02) |
| L4 | 140.09(25.58) |
| L5 | 163.02(25.62) |
Table 2.
Comparison of center distance error of Pose-Net according to kernel size change. The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
Table 2.
Comparison of center distance error of Pose-Net according to kernel size change. The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
| Kernel Size | L1 | L2 | L3 | L4 | L5 | Sacrum | Total | Outlier (%) |
|---|
| 7 (Inlier) | 6.2048(3.7175) | 5.6754(2.5657) | 5.8699(2.4392) | 5.9250(2.6028) | 6.5041(2.9050) | 6.5863(3.8569) | 6.1276(3.0809) | 3.6184 |
| 7 (All) | 7.3140(7.6024) | 6.9445(7.9221) | 7.1655(8.1824) | 6.9841(7.5247) | 7.4520(7.3884) | 7.5333(7.5562) | 7.2322(7.6934) | |
| 7->13 (Inlier) | 6.1645(2.6868) | 6.7569(2.6437) | 6.7818(2.5824) | 6.6531(2.5095) | 6.3572(2.7250) | 6.3670(3.6241) | 6.5134(2.8260) | 2.6316 |
| 7->13 (All) | 7.2846(7.3810) | 7.9211(7.6721) | 7.9006(7.6783) | 7.7393(7.5274) | 7.4110(7.5285) | 7.2813(7.3648) | 7.5896(7.5210) | |
Table 3.
Comparison of center distance error of Pose-Net according to the size of the random spine cutout (RSC) region. The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
Table 3.
Comparison of center distance error of Pose-Net according to the size of the random spine cutout (RSC) region. The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
| Ratio | L1 | L2 | L3 | L4 | L5 | Sacrum | Total | Outlier (%) |
|---|
| 1.0 (Inlier) | 5.9408(2.6590) | 5.8152(2.3992) | 5.6638(2.4129) | 6.0802(2.5584) | 5.7691(2.5759) | 6.5375(3.4117) | 5.9677(2.7031) | 2.9605 |
| 1.0 (All) | 7.0061(7.3041) | 6.9253(7.6269) | 6.8778(8.1331) | 7.2323(7.9045) | 6.8937(7.9268) | 7.6288(7.9528) | 7.0940(7.8065) | |
| 0.8 (Inlier) | 5.9864(2.6269) | 6.2773(2.4904) | 5.6182(2.3728) | 6.0225(2.4082) | 6.0451(2.5293) | 6.1910(3.5926) | 6.0234(2.7071) | 2.3026 |
| 0.8 (All) | 6.8910(6.9462) | 7.2230(7.0324) | 6.6239(7.5476) | 6.9955(7.3794) | 7.0123(7.2798) | 7.1463(7.7933) | 6.9820(7.3280) | |
| 0.6 (Inlier) | 5.9070(2.7222) | 6.1465(2.4507) | 5.6808(2.3847) | 5.9585(2.5786) | 6.1200(2.6225) | 6.8137(3.5138) | 6.1044(2.7567) | 1.9737 |
| 0.6 (All) | 6.7998(6.9070) | 7.0464(6.8417) | 6.6200(7.1450) | 6.9024(7.2187) | 7.0595(7.1900) | 7.7733(7.8280) | 7.0336(7.1948) | |
| 0.4 (Inlier) | 6.1318(2.8215) | 6.3584(2.5233) | 5.8741(2.4999) | 5.9819(2.6023) | 6.0978(2.6394) | 6.2191(3.6614) | 6.1105(2.8205) | 3.6184 |
| 0.4 (All) | 7.4260(8.0499) | 7.6042(7.6288) | 6.9495(7.4526) | 6.9735(7.2562) | 7.0888(7.3214) | 7.2202(7.8299) | 7.2104(7.5883) | |
Table 4.
Ablation study of the conventional cutout (CO) and random spine cutout (RSC). The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
Table 4.
Ablation study of the conventional cutout (CO) and random spine cutout (RSC). The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
| | L1 | L2 | L3 | L4 | L5 | Sacrum | Total | Outlier (%) |
|---|
| w/o (RSC, CO) (Inlier) | 6.1645(2.6868) | 6.7569(2.6437) | 6.7818(2.5824) | 6.6531(2.5095) | 6.3572(2.7250) | 6.3670(3.6241) | 6.5134(2.8260) | 2.6316 |
| w/o (RSC, CO) (All) | 7.2846(7.3810) | 7.9211(7.6721) | 7.9006(7.6783) | 7.7393(7.5274) | 7.4110(7.5285) | 7.2813(7.3648) | 7.5896(7.5210) | |
| w CO (Inlier) | 6.3759(3.0089) | 6.3484(2.9113) | 6.1966(2.5607) | 5.8308(2.5506) | 6.2939(2.7560) | 6.2061(4.4343) | 6.2086(3.1060) | 2.9605 |
| w CO (All) | 7.5616(7.6095) | 7.5371(7.8315) | 7.3814(7.9178) | 7.0085(8.1010) | 7.4556(8.1073) | 7.3054(8.6486) | 7.3749(8.0335) | |
| w RSC (Inlier) | 5.9070(2.7222) | 6.1465(2.4507) | 5.6808(2.3847) | 5.9585(2.5786) | 6.1200(2.6225) | 6.8137(3.5138) | 6.1044(2.7567) | 1.9737 |
| w RSC (All) | 6.7998(6.9070) | 7.0464(6.8417) | 6.6200(7.1450) | 6.9024(7.2187) | 7.0595(7.1900) | 7.7733(7.8280) | 7.0336(7.1948) | |
Table 5.
Ablation study of CoordConv(CC). CC(1) means using CoordConv in the first encoding layer of M-Net, and CC(2) means using CoordConv in the first two encoding layers, as shown in
Figure 8. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
Table 5.
Ablation study of CoordConv(CC). CC(1) means using CoordConv in the first encoding layer of M-Net, and CC(2) means using CoordConv in the first two encoding layers, as shown in
Figure 8. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
| | L1 | L2 | L3 | L4 | L5 | Sacrum | Total |
|---|
| w/o CC (D) | 13.4629(14.5039) | 10.8182(6.1662) | 10.2746(5.6941) | 10.6966(4.8587) | 13.2534(8.1749) | 22.7648(20.0812) | 13.5451(12.1363) |
| w/o CC (RD) | 6.4143(7.0429) | 4.9426(2.9314) | 4.6324(2.6764) | 4.9216(2.4185) | 6.3256(4.2349) | 9.5112(8.7448) | 6.1246(5.5049) |
| w CC(1) (D) | 13.8476(15.5072) | 10.7795(4.8753) | 10.5213(5.4359) | 10.9129(4.7082) | 13.5546(7.9351) | 21.3077(17.3570) | 13.4873(11.2722) |
| w CC(1) (RD) | 6.5605(7.5749) | 4.9090(2.3354) | 4.7562(2.9093) | 4.9928(2.2583) | 6.4696(4.0365) | 8.8381(7.7252) | 6.0877(5.2320) |
| w CC(2) (D) | 13.2129(13.9569) | 10.4010(5.0925) | 10.1377(4.6663) | 10.6879(5.3652) | 12.8991(6.6441) | 22.1391(16.5678) | 13.2463(10.7381) |
| w CC(2) (RD) | 6.3072(7.0595) | 4.7426(2.4540) | 4.5529(2.1768) | 4.9061(2.4465) | 6.1400(3.3254) | 9.1456(7.3295) | 5.9657(4.9308) |
Table 6.
Performance according to the width of the ground truth of PAFs. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
Table 6.
Performance according to the width of the ground truth of PAFs. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
| | L1 | L2 | L3 | L4 | L5 | Sacrum | Total |
|---|
| w/o PAFs (D) | 13.4629(14.5039) | 10.8182(6.1662) | 10.2746(5.6941) | 10.6966(4.8587) | 13.2534(8.1749) | 22.7648(20.0812) | 13.5451(12.1363) |
| w/o PAFs (RD) | 6.4143(7.0429) | 4.9426(2.9314) | 4.6324(2.6764) | 4.9216(2.4185) | 6.3256(4.2349) | 9.5112(8.7448) | 6.1246(5.5049) |
| w PAFs-width 2 (D) | 13.9451(14.8208) | 10.6166(5.6704) | 9.9683(3.9839) | 10.9204(5.0152) | 13.4877(8.9793) | 22.3652(18.6160) | 13.5505(11.7209) |
| w PAFs-width 2 (RD) | 6.6628(7.4286) | 4.8374(2.6447) | 4.4580(1.9171) | 5.0171(2.5283) | 6.4009(4.4703) | 9.2942(8.2189) | 6.1117(5.4070) |
| w PAFs-width 4 (D) | 12.2740(9.0398) | 10.4992(5.8889) | 9.8556(5.0278) | 10.7250(4.8108) | 13.2953(7.5819) | 21.6285(17.7035) | 13.0463(10.2522) |
| w PAFs-width 4 (RD) | 5.8339(4.6423) | 4.8065(3.0228) | 4.4455(2.7755) | 4.9670(2.4390) | 6.3455(3.8456) | 8.9756(7.7470) | 5.8957(4.7022) |
| w PAFs-width 6 (D) | 13.7257(17.3613) | 10.6346(6.0329) | 9.9886(3.9369) | 10.9204(5.9805) | 13.3778(9.0107) | 21.7235(17.0877) | 13.3951(11.9373) |
| w PAFs-width 6 (RD) | 6.5436(8.4218) | 4.8677(3.0381) | 4.4686(1.8798) | 5.0277(2.8025) | 6.3426(4.4093) | 9.0349(7.7065) | 6.0475(5.5391) |
Table 7.
Ablation study of CoordConv(CC) and PAFs. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
Table 7.
Ablation study of CoordConv(CC) and PAFs. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
| | L1 | L2 | L3 | L4 | L5 | Sacrum | Total |
|---|
| w/o (CC, PAFs) (D) | 13.4629(14.5039) | 10.8182(6.1662) | 10.2746(5.6941) | 10.6966(4.8587) | 13.2534(8.1749) | 22.7648(20.0812) | 13.5451(12.1363) |
| w/o (CC, PAFs) (RD) | 6.4143(7.0429) | 4.9426(2.9314) | 4.6324(2.6764) | 4.9216(2.4185) | 6.3256(4.2349) | 9.5112(8.7448) | 6.1246(5.5049) |
| w CC (D) | 13.2129(13.9569) | 10.4010(5.0925) | 10.1377(4.6663) | 10.6879(5.3652) | 12.8991(6.6441) | 22.1391(16.5678) | 13.2463(10.7381) |
| w CC (RD) | 6.3072(7.0595) | 4.7426(2.4540) | 4.5529(2.1768) | 4.9061(2.4465) | 6.1400(3.3254) | 9.1456(7.3295) | 5.9657(4.9308) |
| w PAFs (D) | 12.2740(9.0398) | 10.4992(5.8889) | 9.8556(5.0278) | 10.7250(4.8108) | 13.2953(7.5819) | 21.6285(17.7035) | 13.0463(10.2522) |
| w PAFs (RD) | 5.8339(4.6423) | 4.8065(3.0228) | 4.4455(2.7755) | 4.9670(2.4390) | 6.3455(3.8456) | 8.9756(7.7470) | 5.8957(4.7022) |
| w (CC, PAFs) (D) | 12.9856(12.2581) | 10.2726(4.3833) | 9.9563(4.1797) | 10.6939(4.4190) | 12.9161(6.7633) | 21.3219(16.1635) | 13.0244(10.0280) |
| w (CC, PAFs) (RD) | 6.1385(5.8977) | 4.7145(2.1368) | 4.4771(2.2258) | 4.9122(2.1481) | 6.1483(3.4313) | 8.8486(7.2449) | 5.8732(4.5841) |
Table 8.
Average landmark detection time according to the usage of CoordConv and PAFs. Increase Rate denotes the relative increment in inference time compared to w/o (CoordConv, PAFs).
Table 8.
Average landmark detection time according to the usage of CoordConv and PAFs. Increase Rate denotes the relative increment in inference time compared to w/o (CoordConv, PAFs).
| | w/o (CoordConv, PAFs) | w CoordConv | w PAFs | w (CoordConv, PAFs) |
|---|
| Average Inference Time (ms) | 12.98(0.24) | 21.71(0.37) | 13.01(0.27) | 21.87(0.53) |
| Increase Rate (%) | 0 | 67.3 | 0.2 | 68.5 |
Table 9.
Average outlier ratio for each vertebra.
Table 9.
Average outlier ratio for each vertebra.
| | L1 | L2 | L3 | L4 | L5 | Sacrum |
|---|
| Outlier (%) | 2.68 | 0 | 0.34 | 0.34 | 0.67 | 5.70 |
Table 10.
Comparison of center detection performance between ours and Kim et al. [
5]. The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
Table 10.
Comparison of center detection performance between ours and Kim et al. [
5]. The value having the lowest distance error for each column in (Inlier) is marked in bold and underlined for (All).
| | L1 | L2 | L3 | L4 | L5 | Total | Outlier (%) |
|---|
| Ours (Inlier) | 5.9070(2.7222) | 6.1465(2.4507) | 5.6808(2.3847) | 5.9585(2.5786) | 6.1200(2.6225) | 5.9626(2.5567) | 1.9737 |
| Ours (All) | 6.7998(6.9070) | 7.0464(6.8417) | 6.6200(7.1450) | 6.9024(7.2187) | 7.0595(7.1900) | 6.8856(7.0548) | |
| Kim et al. [5] (Inlier) | 11.1426(31.2408) | 5.8518(2.8318) | 6.2710(2.6000) | 6.2763(2.6771) | 5.7273(2.5778) | 7.0538(14.3036) | 4.6053 |
| Kim et al. [5] (All) | 18.8371(50.7964) | 6.4124(5.5082) | 6.7307(5.2587) | 6.7293(5.1520) | 6.0839(5.1002) | 8.9587(23.6900) | |
Table 11.
Comparison of landmark detection performance between our method and Kim et al. [
5]. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
Table 11.
Comparison of landmark detection performance between our method and Kim et al. [
5]. The lowest value for each column in distance error (D) is marked in bold and underlined in relative distance error (RD).
| | L1 | L2 | L3 | L4 | L5 | Total |
|---|
| Ours (D) | 12.9856(12.2581) | 10.2726(4.3833) | 9.9563(4.1797) | 10.6939(4.4190) | 12.9161(6.7633) | 11.3649(7.2139) |
| Ours (RD) | 6.1385(5.8977) | 4.7145(2.1368) | 4.4771(2.2258) | 4.9122(2.1481) | 6.1483(3.4313) | 5.2781(3.5530) |
| Kim et al. [5] (D) | 29.3115(105.0534) | 20.6214(84.7178) | 9.6515(6.8754) | 11.1020(11.0167) | 16.4756(21.1937) | 17.4324(61.6963) |
| Kim et al. [5] (RD) | 14.6237(54.6018) | 9.8399(42.7799) | 4.2791(2.7548) | 4.9899(4.3484) | 7.9765(10.7887) | 8.3418(31.6554) |