Application of Deep Convolutional Neural Networks in the Diagnosis of Osteoporosis
Abstract
:1. Introduction
2. Materials and Methods
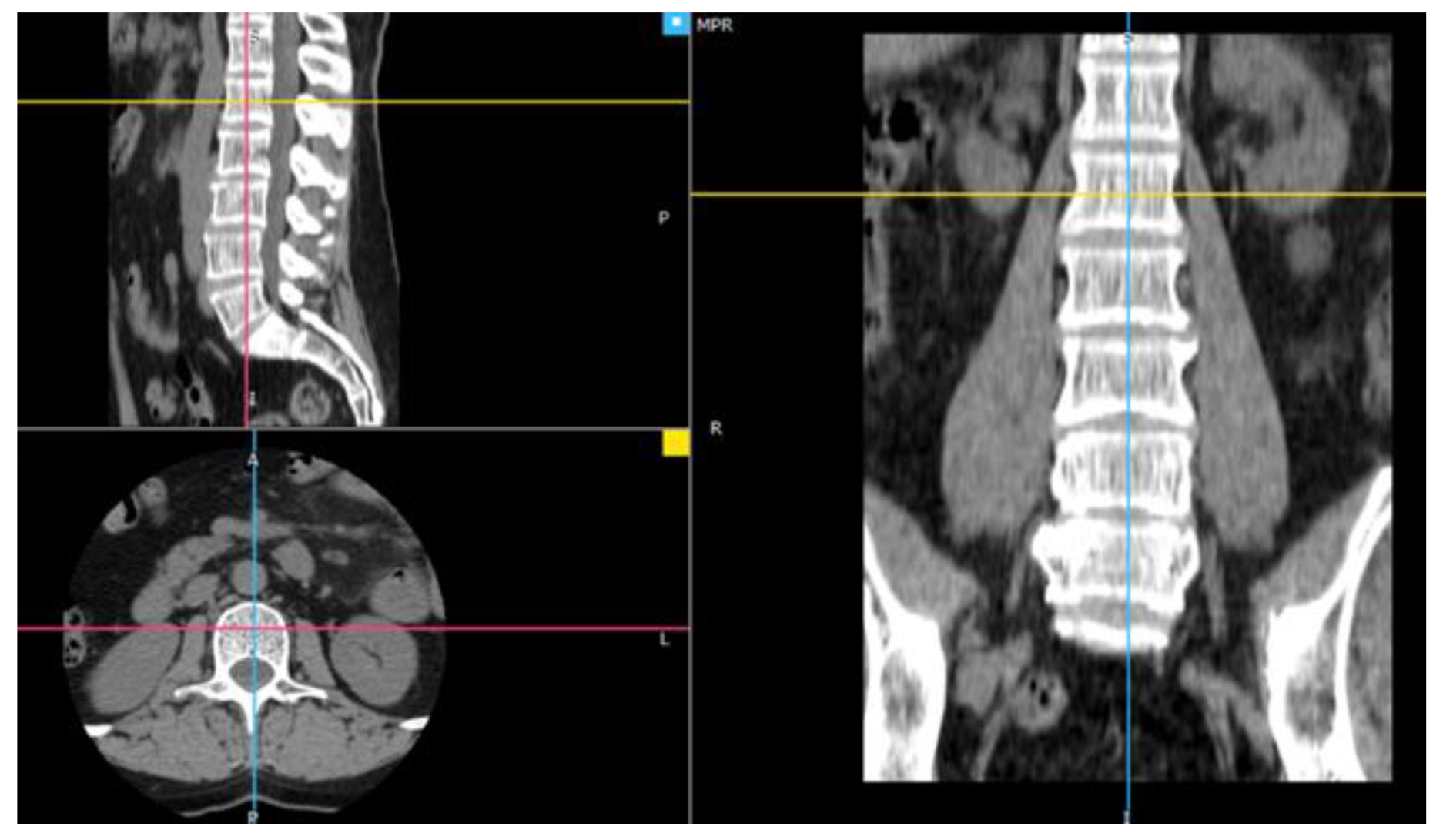
2.1. Material
2.2. Construction of the Classification Models
- Feature extraction—the convolution base has been frozen, and the added dense layers, creating a new classifier, were initiated randomly and trained over a period of 200 epochs using data augmentation. Layer freezing consists of preventing the update of their weights in the training process, so that the representations previously learned by the convolution base have not been modified during training.
- Fine tuning—the upper layers of the convolution base have been unfrozen and trained for a period of 300 epochs together with the new layers, also using data augmentation. At the end, the entire convolution base has been unfrozen.
3. Results
4. Discussion
| Item No. | Role | Research Material | The Method Used | Classifier Quality Assessment Parameter |
|---|---|---|---|---|
| Our research | Application of deep convolutional neural networks in the diagnosis of osteoporosis | CT images of L1 spongy tissue from 100 patients (50 healthy and 50 diagnosed with osteoporosis) | VGG16, VGG19, MobileNetV2, Xception, ResNet50, InceptionResNetV2 | ACC = 95%, TPR = 96%, TNR = 94% |
| [54] | Classification of osteoporosis on the basis of CR images of phalanges using DCNN | 101 computed radiography images of phalanges | An undefined convolutional network model from the Caffe package | TPR = 64.7%, FPR = 6.51% |
| [51] | Identification of vertebral compression fractures caused by osteoporosis | 3701 CT tests, 2681 (72%) were negative for the presence of VCF and 1020 (28%) were marked as positive for VCF, | Convolutional network and a classifier based on a recursive network | ACC = 89.1%, TPR = 83.9%, TNR = 93.8 % |
| [52] | Automatic detection of osteoporotic vertebral fractures on CT scans | 1432 CT images of the spine | (1) a function extraction module based on CNN ResNet34 and (2) an RNN module for aggregating the extracted features and making the final diagnosis. | ACC = 89.2%, F1 score = 90.8% |
| [55] | Metacarpal screening for osteoporosis | 4000 radiographs of the metacarpus | AlexNet | TPR = 82.4%, TNR = 95.7% |
| [53] | Diagnostic examination and prediction of the risk of osteoporosis in women | Age, weight, height, and T-score of the femoral neck of 1559 women | Radial basic function of artificial neural networks with the 2-4-1 architecture | ACC = 78.83%, AUC = 0.829, TPR = 51.0%, TNR = 90.12% |
| [56] | Prediction of osteoporosis from simple hip radiography | 1012 simple hip radiographs | VGG16 model | ACC = 81.2%, TPR = 91.1%, TNR = 68.9%, PPV = 78.5%, NPV = 86.1% |
| [16] | Predicting osteoporosis based on the mandibular cortical index on panoramic radiographs | Panoramic radiographs of mandibular 744 female patients | AlexNET, GoogleNET, ResNET-50, SqueezeNET, and ShuffleNET deep-learning models | ACC = 81.14%(AlexNET), ACC = 88.94% (GoogleNET), ACC = 98.56% (AlexNET), ACC = 92.79% (GoogleNET) |
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Osteoporosis Foundation. Key Statistics For Europe. Available online: https://www.osteoporosis.foundation/ (accessed on 18 September 2022).
- Camacho, P.M.; Petak, S.M.; Binkley, N.; Diab, D.L.; Eldeiry, L.S.; Farooki, A.; Harris, S.T.; Hurley, D.L.; Kelly, J.; Lewiecki, E.M.; et al. American College of Endocrinology Clinical Practice Guidelines for the Diagnosis and Treatment of Postmenopausal Osteoporosis—2020 Update. Endocr. Pract. 2020, 26, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Smets, J.; Shevroja, E.; Hügle, T.; Leslie, W.D.; Hans, D. Machine Learning Solutions for Osteoporosis—A Review. J. Bone Miner. Res. 2021, 36, 833–851. [Google Scholar] [CrossRef] [PubMed]
- Jolly, S.; Chaudhary, H.; Bhan, A.; Rana, H.; Goyal, A. Texture based Bone Radiograph Image Analysis for the assessment of Osteoporosis using Hybrid Domain. In Proceedings of the 2021 3rd International Conference on Signal Processing and Communication (ICPSC), Coimbatore, India, 13–14 May 2021; pp. 344–349. [Google Scholar] [CrossRef]
- Tang, C.; Zhang, W.; Li, H.; Li, L.; Li, Z.; Cai, A.; Wang, L.; Shi, D.; Yan, B. CNN-based qualitative detection of bone mineral density via diagnostic CT slices for osteoporosis screening. Osteoporos. Int. 2021, 32, 971–979. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Li, W.; Chen, X.; Chen, K.; Kang, H.; Yu, P.; Zhang, R.; Liao, J.; Hong, G.; Li, S. Opportunistic osteoporosis screening in multi-detector CT images using deep convolutional neural networks. Eur. Radiol. 2021, 31, 1831–1842. [Google Scholar] [CrossRef]
- Patil, K.A.; Prashanth, K.V.M.; Ramalingaiah, A. Texture Feature Extraction of Lumbar Spine Trabecular Bone Radiograph Image using Laplacian of Gaussian Filter with KNN Classification to Diagnose Osteoporosis. J. Phys. Conf. Ser. 2021, 2070, 012137. [Google Scholar] [CrossRef]
- Yamamoto, N.; Sukegawa, S.; Yamashita, K.; Manabe, M.; Nakano, K.; Takabatake, K.; Kawai, H.; Ozaki, T.; Kawasaki, K.; Nagatsuka, H.; et al. Effect of Patient Clinical Variables in Osteoporosis Classification Using Hip X-rays in Deep Learning Analysis. Medicina 2021, 57, 846. [Google Scholar] [CrossRef]
- Patil, K.A.; Prashant, K.V.M. Segmentation of Lumbar [L1-L4] AP Spine X-ray images using various Level Set methods to detect Osteoporosis. In Proceedings of the IEEE Bombay Section Signature Conference (IBSSC), Gwalior, India, 18–20 November 2021; pp. 1–6. [Google Scholar] [CrossRef]
- Janiesch, C.; Zschech, P.; Heinrich, K. Machine learning and deep learning. Electron. Mark. 2021, 31, 685–695. [Google Scholar] [CrossRef]
- Tang, D.; Zhou, J.; Wang, L.; Ni, M.; Chen, M.; Hassan, S.; Luo, R.; Chen, X.; He, X.; Zhang, L.; et al. A Novel Model Based on Deep Convolutional Neural Network Improves Diagnostic Accuracy of Intramucosal Gastric Cancer (With Video). Front. Oncol. 2021, 11, 622827. [Google Scholar] [CrossRef]
- Singh, V.; Asari, V.K.; Rajasekaran, R.A. Deep Neural Network for Early Detection and Prediction of Chronic Kidney Disease. Diagnostics 2022, 12, 116. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Belkacem, A.N.; Wang, X.; Sha, S.; Wang, C.; Chen, C. A convolutional neural network-based diagnostic method using resting-state electroencephalograph signals for major depressive and bipolar disorders. Biomed. Signal Process. Control. 2022, 72 Pt B, 103370. [Google Scholar] [CrossRef]
- Prudhvi Kumar, B.; Anithaashri, T.P. Novel Diagnostic System for COVID-19 Pneumonia Using Forward Propagation of Convolutional Neural Network Comparing with Artificial Neural Network. ECS Trans. 2022, 107, 13797. [Google Scholar] [CrossRef]
- Varalakshmi, P.; Sathyamoorthy, S.; Darshan, V.; Ramanujan, V.; Rajasekar, S.J.S. Detection of Osteoporosis with DEXA Scan Images using Deep Learning Models. In Proceedings of the International Conference on Advances in Computing, Communication and Applied Informatics (ACCAI), Chennai, India, 28–29 January 2022; pp. 1–6. [Google Scholar] [CrossRef]
- Tassoker, M.; Öziç, M.U.; Yuce, F. Comparison of five convolutional neural networks for predicting osteoporosis based on mandibular cortical index on panoramic radiographs. Dentomaxillofacial Radiol. 2022, 51, 20220108. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, T.; Taguchi, A.; Kakimoto, N. Osteoporosis screening support system from panoramic radiographs using deep learning by convolutional neural network. Dentomaxillofac Radiol. 2022, 51, 20220135. [Google Scholar] [CrossRef] [PubMed]
- Kendall, A.; Gal, Y. What uncertainties do we need in bayesian deep learning for computer vision? In Proceedings of the Annual Conference on Neural Information Processing Systems 2017, Long Beach, CA, USA, 4–9 December 2017; Volume 30.
- De Sousa Ribeiro, F.; Calivá, F.; Swainson, M.; Gudmundsson, K.; Leontidis, G.; Kollias, S. Deep bayesian self-training. Neural Comput. Appl. 2020, 32, 4275–4291. [Google Scholar] [CrossRef] [Green Version]
- Gal, Y.; Ghahramani, Z. Bayesian Convolutional Neural Networks with Bernoulli Approximate Variational Inference. arXiv 2015, arXiv:1506.02158 [stat.ML]. [Google Scholar]
- Ibrahim, M.; Louie, M.; Modarres, C.; Paisley, J.W. Global Explanations of Neural Networks: Mapping the Landscape of Predictions. In Proceedings of the 2019 AAAI/ACM Conference on AI, Ethics, and Society, Honolulu, HI, USA, 27–28 January 2019. [Google Scholar]
- Kästner, C. Interpretability and Explainability. Available online: https://ckaestne.medium.com/interpretability-andexplainability-a80131467856 (accessed on 8 October 2022).
- Loyola-González, O. Black-Box vs. White-Box: Understanding Their Advantages and Weaknesses from a Practical Point of View. IEEE Access 2019, 7, 154096–154113. [Google Scholar] [CrossRef]
- Cao, N.; Yan, X.; Shi, Y.; Chen, C. AI-Sketcher: A Deep Generative Model for Producing High-Quality Sketches. In Proceedings of the AAAI Conference on Artificial Intelligence, Honolulu, HI, USA, 27 January–1 February 2019; Volume 33, pp. 2564–2571. [Google Scholar]
- Shouling, J.; Jinfeng, L.; Tianyu, D.; Bo, L. Survey on Techniques, Applications and Security of Machine Learning Interpretability. J. Comput. Res. Dev. 2019, 56, 2071–2096. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Hinton, G.E.; Srivastava, N.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R.R. Improving neural networks by preventing co-adaptation of feature detectors. arXiv 2012, arXiv:1207.0580v1 [cs.NE]. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Xie, L.; Wang, J.; Wei, Z.; Wang, M.; Tian, Q. DisturbLabel: Regularizing CNN on the loss layer. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 4753–4762. [CrossRef]
- Ioffe, S.; Szegedy, C. Batch normalization: Accelerating deep network training by reducing internal covariate shift. In Proceedings of the International Conference on Machine Learning, Lille, France, 7–9 July 2015; pp. 448–456. [Google Scholar]
- Zheng, Q.; Yang, M.; Yang, J.; Zhang, Q.; Zhang, X. Improvement of generalization ability of deep CNN via implicit regularization in two-stage training process. IEEE Access 2018, 6, 15844–15869. [Google Scholar] [CrossRef]
- Emohare, O.; Cagan, A.; Morgan, R.; Davis, R.; Asis, M.; Switzer, J.; Polly, D.W. The Use of Computed Tomography Attenuation to Evaluate Osteoporosis Following Acute Fractures of the Thoracic and Lumbar Vertebra. Geriatr. Orthop. Surg. Rehabil. 2014, 5, 50–55. [Google Scholar] [CrossRef]
- Dzierżak, R.; Omiotek, Z.; Tkacz, E.; Uhlig, S. Comparison of the Classification Results Accuracy for CT Soft Tissue and Bone Reconstructions in Detecting the Porosity of a Spongy Tissue. J. Clin. Med. 2022, 11, 4526. [Google Scholar] [CrossRef] [PubMed]
- Dzierżak, R.; Omiotek, Z.; Tkacz, E.; Kępa, A. The Influence of the Normalisation of Spinal CT Images on the Significance of Textural Features in the Identification of Defects in the Spongy Tissue Structure. In Innovations in Biomedical Engineering, Proceedings of the Conference on Innovations in Biomedical Engineering, Katowice, Poland, 18–20 October 2018; Springer: Cham, Switzerland, 2019; pp. 55–66. [Google Scholar]
- Huang, G.; Liu, Z.; Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. arXiv 2018, arXiv:1608.06993v5 [cs.CV]. [Google Scholar]
- Keras. Keras library. Available online: https://keras.io/ (accessed on 10 February 2020).
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. arXiv 2017, arXiv:1610.02357v3 [cs.CV]. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the International Conference on Learning Representations (ICLR 2015), San Diego, CA, USA, 7–9 May 2015. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep Residual Learning for Image Recognition. arXiv 2015, arXiv:1512.03385v1 [cs.CV]. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Identity Mappings in Deep Residual Networks. arXiv 2016, arXiv:1603.05027v3 [cs.CV]. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the Inception Architecture for Computer Vision. arXiv 2015, arXiv:1512.00567v3 [cs.CV]. [Google Scholar]
- Szegedy, C.; Ioffe, S.; Vanhoucke, V.; Alemi, A. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning. arXiv 2016, arXiv:1602.07261v2 [cs.CV]. [Google Scholar] [CrossRef]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Hartwig, A. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. arXiv 2017, arXiv:1704.04861v1 [cs.CV]. [Google Scholar]
- Sandler, M.; Howard, A.; Zhu, M.; Zhmoginov, A.; Chen, L.-C. MobileNetV2: Inverted Residuals and Linear Bottlenecks. arXiv 2019, arXiv:1801.04381v4 [cs.CV]. [Google Scholar]
- Zoph, B.; Vasudevan, V.; Shlens, J.; Le, Q.V. Learning Transferable Architectures for Scalable Image Recognition. arXiv 2018, arXiv:1707.07012v4 [cs.CV]. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.-J.; Li, K.; Fei-Fei, L. ImageNet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009. [Google Scholar]
- Mahdianpari, M.; Salehi, B.; Rezaee, M.; Mohammadimanesh, F.; Zhang, Y. Very Deep Convolutional Neural Networks for Complex Land Cover Mapping Using Multispectral Remote Sensing Imagery. Remote Sens. 2018, 10, 1119. [Google Scholar] [CrossRef] [Green Version]
- Dong, K.; Zhou, C.; Ruan, Y.; Li, Y. MobileNetV2 Model for Image Classification. In Proceedings of the 2020 2nd International Conference on Information Technology and Computer Application (ITCA), Guangzhou, China, 18–20 December 2020; pp. 476–480. [Google Scholar] [CrossRef]
- Wu, Z.; Shen, C.; Hengel, A. Wider or Deeper: Revisiting the ResNet Model for Visual Recognition. Pattern Recognit. 2019, 90, 119–133. [Google Scholar] [CrossRef] [Green Version]
- Peng, C.; Liu, Y.; Yuan, X.; Chen, Q. Research of image recognition method based on enhanced inception-ResNet-V2. Multimed. Tools Appl. 2022, 81, 34345–34365. [Google Scholar] [CrossRef]
- Bar, A.; Wolf, L.; Amitai, O.B.; Toledano, E.; Elnekave, E. Compression Fractures Detection on CT. In Proceedings of the SPIE Medical Imaging, Orlando, FL, USA, 12–14 February 2017. [Google Scholar]
- Tomita, N.; Cheung, Y.Y.; Hassanpour, S. Deep neural networks for automatic detection of osteoporotic vertebral fractures on CT scans. Comput. Biol. Med. 2018, 98, 8–15. [Google Scholar] [CrossRef]
- Meng, J.; Sun, N.; Chen, Y.; Li, Z.; Cui, X.; Fan, J.; Cao, H.; Zheng, W.; Jin, O.; Jiang, L.; et al. Artificial neural networkoptimizes self-examination of osteoporosis risk in women. J. Int. Med. Res. 2019, 47, 3088–3098. [Google Scholar] [CrossRef]
- Hatano, K.; Murakami, S.; Lu, H.; Tan, J.K.; Kim, H.; Aoki, T. Classification of Osteoporosis from Phalanges CR Images Based on DCNN. In Proceedings of the 2017 17th International Conference on Control, Automation and Systems (ICCAS 2017), Jeju, Korea, 18–21 October 2017. [Google Scholar]
- Nahom, N.; Teitel, J.; Morris, M.R.; Sani, N.; Mitten, D.; Hammert, W.C. Convolutional Neural Network for Second Metacarpal Radiographic Osteoporosis Screening. J. Hand Surg. Am. 2020, 45, 175–181. [Google Scholar]
- Jang, R.; Choi, J.H.; Kim, N.; Chang, J.S.; Yoon, P.W.; Kim, C.-H. Prediction of osteoporosis from simple hip radiography using deep learning algorithm. Sci. Rep. 2021, 11, 19997. [Google Scholar] [CrossRef]



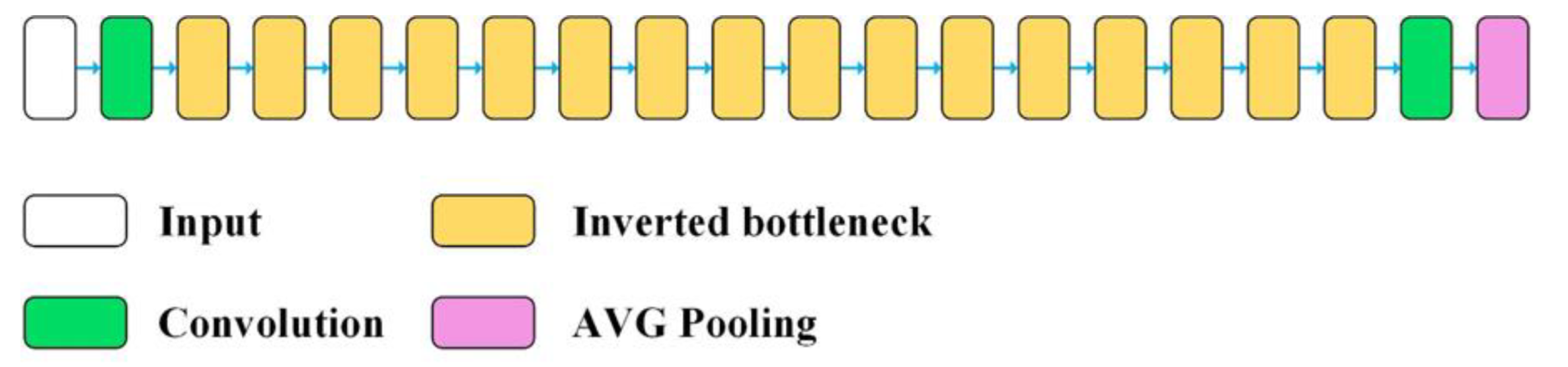











| No. | Model | The Number of Layers of the CB | Fine-Tuned Layers of the CB | Type of Fine-Tuned Layers |
|---|---|---|---|---|
| 1 | VGG16 | 19 | 12–19 | 2D convolution |
| 2 | VGG19 | 22 | 13–22 | 2D convolution |
| 3 | Xception | 132 | 117–132 | depth-wise separable 1D and 2D convolution, batch normalization |
| 4 | MobileNetV2 | 155 | 136–155 | 2D convolution, depth-wise convolution, batch normalization |
| 5 | ResNet50 | 175 | 150–175 | 2D convolution, batch normalization |
| 6 | InceptionResNetV2 | 780 | 631–780 | 2D convolution, batch normalization |
| Parameter | Value | |
|---|---|---|
| Training | epochs | 200 (feature extraction), 300 (fine tuning) |
| batch size | 5 | |
| Model | loss function | binary cross entropy |
| optimizer | root mean square propagation (RMSprop) | |
| metrics | accuracy | |
| Optimizer | learning rate | 2 × 10−5 (feature extraction), 10−5 (fine tuning) |
| rho | 0.9 | |
| momentum | 0.0 | |
| epsilon | 10−7 | |
| centred | False | |
| Data augmentation | rotation range | 40° |
| width shift range | 0.2 | |
| height shift range | 0.2 | |
| shear range | 0.2 | |
| zoom range | 0.2 | |
| horizontal flip | True |
| Model | GPU Inference Time [ms] | CPU Inference Time [ms] | Latency Increasing (tCPU / tGPU) |
|---|---|---|---|
| VGG16 | 5.6 | 20.6 | 3.7 |
| VGG19 | 7.0 | 26.1 | 3.7 |
| Xception | 25.2 | 58.0 | 2.3 |
| ResNet50 | 22.0 | 33.9 | 1.5 |
| MobileNetV2 | 21.1 | 65.0 | 3.1 |
| InceptionResNetV2 | 93.8 | 115.0 | 1.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dzierżak, R.; Omiotek, Z. Application of Deep Convolutional Neural Networks in the Diagnosis of Osteoporosis. Sensors 2022, 22, 8189. https://doi.org/10.3390/s22218189
Dzierżak R, Omiotek Z. Application of Deep Convolutional Neural Networks in the Diagnosis of Osteoporosis. Sensors. 2022; 22(21):8189. https://doi.org/10.3390/s22218189
Chicago/Turabian StyleDzierżak, Róża, and Zbigniew Omiotek. 2022. "Application of Deep Convolutional Neural Networks in the Diagnosis of Osteoporosis" Sensors 22, no. 21: 8189. https://doi.org/10.3390/s22218189
APA StyleDzierżak, R., & Omiotek, Z. (2022). Application of Deep Convolutional Neural Networks in the Diagnosis of Osteoporosis. Sensors, 22(21), 8189. https://doi.org/10.3390/s22218189







