Design Strategies for Electrochemical Aptasensors for Cancer Diagnostic Devices
Abstract
1. Introduction
2. Electrochemical Techniques
3. Electrochemical Aptasensors
4. Electrochemical Assays for Cancer Biomarkers
4.1. Cancer Biomarkers
4.2. Human Epidermal Growth Factor Receptor-2
4.3. Urokinase Plasminogen Activator
4.4. Osteopontin
4.5. Mucin 1
| Strategy | Technique | LOD, M | Interference Studies | Ref. |
|---|---|---|---|---|
| POPhDA–AuNPs hybrid film/AuNPs silica/MWCNT C-Sh/Au | DPV | 1 × 10−12 | CEA, CA19-9, CA72-4, BSA | [141] |
| ALP-strp/Apt2/Biotin/Apt1/Strp-MBs/SPA | 7 × 10−11 | MUC4, MUC16 | [146] | |
| Apt-AuNPs/MCH/cDNA/Au | EIS | 10−10 | TNF-α, CEA | [142] |
| Exo I/MUC1/Apt-MB/CP/MCH/Au | SWV | 4 × 10−12 | Myo, BSA, CEA | [147] |
| Apt/EDC/MWCNT/SPCE | EIS | 0.02 U mL−1 | BSA, FBS, Lyz | [90] |
| Th/rGO-N′1,N′3 DHMIA/Apt/MUC1/Apt/Pdots/IL/Au | DPV | 6 × 10−11 | MUC4, Lys, Myo | [148] |
| SH-Apt-MB/Au | SWV | 4 × 10−9 | AA, UA, VEGF, BSA, PSA | [145] |
| MB-Apt/AuNPs/GCE | DPV, EIS | 24 × 10−9 | Lyz, BSA, Cyt C | [149] |
| Apt/ZrHCF NPs/ZrHCF/mFe3O4/mC/Au | EIS | 7.4 × 10−15 (0.9 pg mL−1) | CEA, IgG, BSA | [150] |
| AuNPs and GO doped PEDOT films APT/Strp/AuNPs-GO-PEDOT | DPV | 3.1 × 10−17 | MPT64, AChE, BSA | [144] |
| Exo I/Apt-MUC1/cDNA-MB/Naf/ITO | 3.3 × 10−15 M (0.4 pg mL−1) | CEA, GP73, HSA, ALP, AFP | [151] | |
| Metal ion electrochemical labels/Ru(NH3)63+ electronic wires | 3.33 × 10−15 | FBS, HCG, MUC16, CA19-9 | [152] | |
| AuNPs-DNA enzyme/H-2/MCH/c-DNA/Au | Amp | 3.3 × 10−16 (0.04 pg mL−1) | PSA, Thr, CEA, BSA | [153] |
| MXene probe/c-DNA-Fc/Apt + BSA/AuNPs/GCE | SWV | 3.3 × 10−13 | Not shown | [143] |
4.6. Carcinoma Antigen 125
4.7. Vascular Endothelial Growth Factor
| Strategy | Technique | LOD, M | Interference Studies | Ref. |
|---|---|---|---|---|
| Sandwich Apt/MBs-Ab/VEGF/Apt/AuIDE | EIS (capacitance) | 10.5 × 10−12 (401 pg mL−1) | BSA | [179] |
| NP/Strp-ALP/B-Apt/VEGF/MCH/SH-Apt/AuNPs/SPGE | DPV | 3 × 10−8 | HER2 | [180] |
| MB/Apt/GA/BSA-AuNCs/IL/GCE Apt/GA/BSA-AuNCs/IL/GCE | DPV EIS (Ferri/ferrocyanide) | signal-off: 0.32 × 10−12 signal-on: 0.48 × 10−12 | Not shown | [169] |
| SH-Apt/OMC–Aunano/SPCE | EIS (Ferri/ferrocyanide) | 26.2 × 10−15 (1 pg mL−1) | HIgG, HIgA, Lip, Lyz, HSA | [181] |
| Fc-Apt-alkyne/UDT + UDT-N3/Au | ACV | 6.2 × 10−9 | VEGF121, BSA, HSA, trypsin | [182] |
| DNA-Ag/Pt NCs/amino-Apt/GCE Peroxidase mimicking activity | Amp | 4.6 × 10−12 | Thr, HSA, HIgG | [183] |
| MCH/SH-Apt-MB/Au | SWV | 3.93 × 10−12 (0.15 ng mL−1) | AA, UA | [184] |
| Apt/Aunano/rGO-PAMAM-Th/SPCE | DPV | 0.7 × 10−12 | U, AA, D, Glu, HIgG, HIgA | [185] |
| BPEI-Fc-CB[7]-N3-GO/S1/MCH/S2/SH-Apt/Au | SWV | 0.21 × 10−15 (8 fg mL−1) | BSA, HSA, VEGFR1, VEGFR2, VEGF121 | [171] |
| MB/NH2-Apt/GA/AP/IL/GCE NH2-Apt/GA/AP/IL/GCE | DPV EIS (Ferri/ferrocyanide) | signal-off: 0.5 × 10−12 signal-on: 0.78 × 10−12 | Not shown | [172] |
| Hairpin DNA/AuNPs/GQD/Thi/MCH/SH-hairpin DNA/Au | SWV | 0.3 × 10−15 | PSA, BSA, Thr | [173] |
| Peptide Apt-based functionalised microneedles | EIS (capacitance) | 0.1 × 10−12 | HIgG, Con A, cholera toxin | [23] |
| SH-Apt/AuNAs@NC/Au | EIS (Ferri/ferrocyanide) | 0.18 × 10−12 (6.77 pg mL−1) | Lyz, HIgG, CEA, PSA | [174] |
| SH-Apt/LPLE | EIS | 0.017 × 10−15 (0.64 fg mL−1) | Thr, PDGF-BB; VEGF121, HIgG | [178] |
| Sandwich Apt-GDH/VEGF/Biotin-Ab/ strp/DTBSU/AuWE | Amp | 105 × 10−12 | BSA | [177] |
| biotin-Apt-Fc/MB-strp/GO-GCE biotin-Apt-Fc/MB-strp/GO-PhA-GCE | SWV | 2.62 × 10−15 (1 pg mL−1) 0.18 × 10−12 (7 pg mL−1) | IL-6, CA-125, PSA, HIgG | [175] |
| SH-Apt-Fc + SH-PEG/AuNPs-MB/rGO-GCE | SWV | 2.62 × 10−15 (0.1 pg mL−1) | HIgG, IL-1β, PSA, IL-6 | [176] |
4.8. Prostate-Specific Antigen
4.9. Platelet-Derived Growth Factors
| Strategy | Technique | LOD, M | Interference Studies | Ref. |
|---|---|---|---|---|
| Sandwich assay based on MOS2/carbon aerogel composites | DPV | 0.3 × 10−12 | BSA, CEA, Hb, IgG | [208] |
| Structure-switching hairpin probe | CV | 2.67 × 10−12 (0.08 ng mL−1) | PDGF-AA, PDGF-AB, IgG, BSA | [215] |
| VS2-GR coupled with Exo ΙΙΙ-aided signal amplification leaf like VS2 nanosheets | DPV | 0.03 × 10−12 | BSA, IgE, Thr, CEA | [216] |
| DPV | 0.4 × 10−12 | Hb, Thr, BSA | [217] | |
| A background current-eliminated Apt sensing platform | ACV | 0.334 × 10−12 (10 pg mL−1) | PDGF-AA, PDGF-AB | [218] |
| Fe3O4@3D-rGO@plasma-polymerised (4-vinyl pyridine) nanocomposite | EIS | 0.98 × 10−12 (29.4 pg mL−1) | Thr, IgG, Lyz, BSA | [219] |
| Co3(PO4)2 BSA-based aptasensor | EIS | 0.123 × 10−12 (3.7 pg mL−1) | BSA, IgG, Lyz, Thr, PDFG-AA, PDGF-AB | [220] |
| Exo ΙΙΙ-aided signal amplification strategy | DPV | 20 × 10−15 | Hb, BSA, IgG, CEA, BSA | [221] |
| Apt-functionalised MHCPP | FET | 1.78 × 10−15 | ATP, BSA, Cal, PDGF-AA | [209] |
| Apt based EGFET sensor RCA | EGFET | 8.8 × 10−12 | Not shown | [222] |
| Apt based dual signal amplification strategy using hydroxyapatite NPs | SWV | 1.67 × 10−15 (50 fg mL−1) | AFP, CEA, IgG, HER2 | [223] |
| Sandwich Ab-Apt labelled ALP | SWV | 1.67 × 10−15 (50 fg mL−1) | AFP, CEA, IgG, p53, HER2 | [224] |
| EXPAR | DPV | 52 × 10−15 | PDGF-AA, PDGF-AB | [225] |
| Carbon-based nanocomposites with Apt-Ag-NCs | EIS | 26.5 × 10−15 (0.82 pg mL−1) | BSA, Thr, Lyz, IgG | [210] |
| Sandwich sensing system on 3D-IHC | Amp | 1.2 × 10−15 (0.03 pg mL−1) | AA, UA, Gly, Glu | [226] |
| Aptasensor based on new structure of GNPs containing α-CD | SWV | 0.52 × 10−9 | BSA, PSA, HSA, p53 | [227] |
| Se-doped MWCNTs-Gr, Hem/G-quadruplex and Y shaped DNA-aided target-triggered | DPV | 27 × 10−15 | Thr, IgG, PSA | [213] |
| Apt-Functionalised 3D CNWs | FET | 1.78 × 10−15 | Cal, ATP, BSA | [214] |
4.10. α-Fetoprotein (AFP)
| Strategy | Technique | LOD, g mL−1 | Interference Studies | Ref. |
|---|---|---|---|---|
| Apt/ZiP/Au | DPV | 3.1 × 10−15 | HSA, γ-globulin, Hb, ssDNA | [243] |
| Hairpin DNA-MB/Au | 8.76 × 10−12 | GP73, CEA | [240] | |
| Apt/Thi/rGO/AuNPs/SPCE | 5 × 10−8 | BSA, HSA, IgG, IgE | [232] | |
| Apt/GO/GCE | CV | 3 × 10−12 | PSA, CEA | [233] |
| Apt-PB NPs/GO/Au | DPV | 6.3 × 10−12 | NSE, CEA, MUC16, hCG, PSA | [234] |
| Apt/sAuNPs/Au | 0.23 × 10−12 | IgG, CEA, BSA, HSA | [235] | |
| Apt-3D NMCNMs/Au | 60.8 × 10−15 | BSA, PSA, CEA, IgG, EGFR, MUC1, VEGF | [236] | |
| Apt/Gs-Pyr BA-KCl HG/Au | EIS | 0.51 × 10−12 | Lyz, IgG, HSA, BSA, CEA | [237] |
| Apt/Cu MIL-96 OH/Au | 0.12 × 10−12 | CEA, IgG, Lyz, BSA, HSA | [238] | |
| HCR—two DNA hairpins-MB/DNA probe/AFP/Apt/Au | DPV | 0.041 × 10−12 | BSA, IgG, IgE | [242] |
| MB-DNA-AuNPs/AFP/Fc-capture probe/MCH/DNA1/Au | ACV | 0.27 × 10−15 | Thr, CEA, IgG, PSA | [241] |
4.11. Carcinoembryonic Antigen (CEA)
| Strategy | Technique | LOD, g mL−1 | Interference Studies | Ref. |
|---|---|---|---|---|
| Glu ox–Fc nanoporous AuNSs/CEA/MCH/Apt/Au | DPV | 0.45 × 10−12 | HSA, human IgG, mouse IgG | [262] |
| MCH/Apt/AuNPs-Hem/GCE | 40 × 10−15 | HSA, Thr, Lyz, Ins | [247] | |
| Apt-C-PPy MNTs/IDMA | FET | 1 × 10−15 | Thr, BSA, DP, AA, UA | [263] |
| AuNPs-Apt 2-CEA-Apt1/Au | Amp | 0.899 × 10−12 (5 fM) | PSA, BSA | [264] |
| H Apt +AuNRs Gr-strp NM/GCE | DPV | 1.5 × 10−12 | Myo, Fer, IgG | [248] |
| H1 + H2 + MnTMPyP/Pt-Pd–Apt2/CEA/BSA/Apt1/AuNPs/GCE | EIS | 0.03 × 10−12 | Thr, rabbit IgG, AFP, PSA | [265] |
| UiO-66-AgNCs-Apt/Au | EIS DPV | 8.88 × 10−12 4.93 × 10−12 | AA, MUC1, Thr, IgG, | [257] |
| BSA/Apt/AuNPs/Gr-MoSe2 hybrid/GCE | DPV | 0.03 × 10−12 | HSA, IgE, AFP, Thr, LDL | [249] |
| MoO42−/AuNRs-Apt/Ab-CEA/GO/GCE | SWV | 0.05 × 10−12 | AFP, MUC16, IgG | [250] |
| CuMOFs-PtNPs-Apt2-hemin-GOx/CEA/BSA/Apt1/AuNPs/GCE | EIS | 0.023 × 10−12 | Thr, IgG, AFP, PSA | [251] |
| Auxiliary DNA/Pb2+/Substrate chain-MB /DNAzyme: Hairpin-CEA/GrQD-IL-Naf/GCE | DPV | 0.34 × 10−15 | BSA, PSA, MUC1 | [252] |
| Compl. strands: DNA1 and DNA2/Hairpins: Apt1 + Apt2/SPAuE | 0.9 × 10−12 | IgE, Thr, HSA, Gly, Myo, PSA | [266] | |
| HRP/ConA/CEA/DNA Apt + MCH/Au | 3.4 × 10−9 | BSA, HSA, ɤ-globulin, AFP, CRP | [267] | |
| Apt/NiCoPBA/Au | EIS | 0.74 × 10−15 | BSA, HER2, IgG, PSA, VEGF, MUC1 | [268] |
| MSiNPs–MB–Av/IBNc /CEA/BSA/Apt/AuNPs/SPCE | DPV | 280 × 10−15 (buffer) 510 × 10−15 (HS) | Not shown | [258] |
| DMP DTPs HCR/Au | 18.2 × 10−15 | Hb, PSA, Thr | [269] | |
| EA/Apt/ Hem-GO-MWCNs/GCE | 0.82 × 10−15 | Ins, Ua, Glu, Arg, Gly, HSA | [253] | |
| Au-SiO2 Janus nanoparticles–Apt/Av-Fe3O4@SiO2 NanoCaptors/SPCE | Amp | 210 × 10−12 | Thr, IgG, HSA | [270] |
| HRP-Cu3(PO4)2 hybrid nanoflowers-AuNPs-Apt2-BSA/EA/BSA/Apt1/Hemin-rGO-AuNPs/GCE | DPV | 29 × 10−15 | Thr, Cys, Hb, PSA, | [271] |
| sDNA-Pb-MOF/H1 and H2-HCR/DNA/Apt/Au | SWV | 0.33 × 10−12 | Myo, BSA, MUC1, PSA | [254] |
| Apt/PDA + PSBMA/GCE | DPV | 3.3 × 10−15 | Lyz, ɤ-globulin, MB, IgG, BSA, HSA | [260] |
| MoO42−/CEA/Apt-Cu3(PO4)2 hybrid nanoflowers + GO composites/GCE | SWV | 2.4 × 10−15 | PSA, Thr, Hb | [255] |
| BSA/Apt/SA/APTES/Gr-PEDOT:PSS/PSE | EIS | 0.45 × 10−9 (buffer) 1.06 × 10−9 (HS) | BSA, PSA, Ins | [256] |
| DNA3-MB/CEA/Apt/MCH/DNA2/DNA1/SH-ssDNA/Au | SWV | 20 × 10−12 | BSA, PSA, CD86, EpCAM | [261] |
| AuNCs-Apt/CEA/ConA/NCMTs-Fe3O4-Cu silicate/Au | DPV | 5.38 × 10−12 | Hb, PDGF, Lyz, Thr, BSA, Cys, IgG, PSA | [259] |
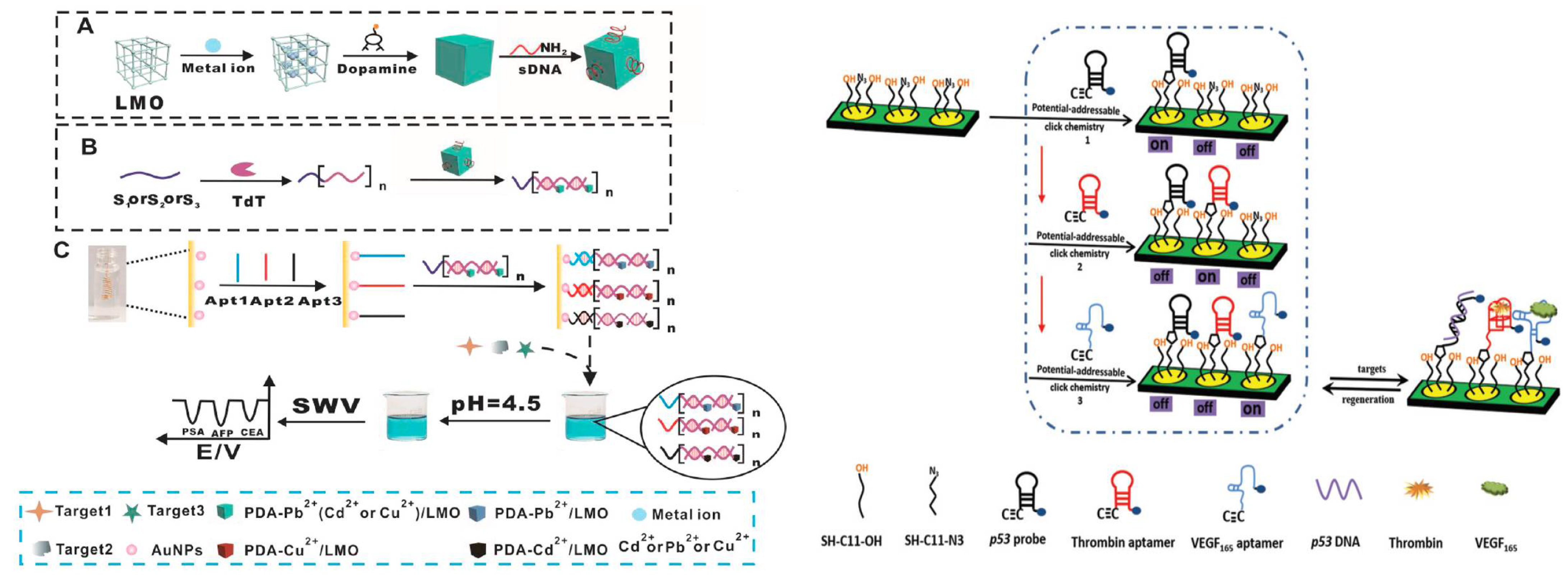
5. Multiplex Electrochemical Aptasensor Platforms for Several Cancer Biomarker Detection
6. Conclusions and Future Directions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. WHO Report on Cancer. Setting Priorities, Investing Wisely and Providing Care for All; World Health Organization: Geneva, Switzerland, 2020; ISBN 978-92-4-000129-9. [Google Scholar]
- Available online: http://www.statcan.gc.ca/cgi-bin/imdb/p2SV.pl?Function=getSurvey&SDDS=3207&lang=en&db=imdb&adm=8&dis=2 (accessed on 3 December 2020).
- Available online: https://www.cancerresearchuk.org/ (accessed on 3 December 2020).
- Raab, S.S.; Grzybicki, D.M. Quality in cancer diagnosis. Cancer J. Clin. 2010, 60, 139–165. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.P.; Forman, D.; Bryant, H.; Butler, J.; Rachet, B.; Maringe, C.; Nur, U.; Tracey, E.; Coory, M.; Hatcher, J.; et al. Cancer survival in Australia, Canada, Denmark, Norway, Sweden, and the UK, 1995–2007 (the International Cancer Benchmarking Partnership): An analysis of population-based cancer registry data. Lancet 2011, 377, 127–138. [Google Scholar] [CrossRef]
- Crew, B. Worth the cost? A closer look at the da Vinci robot’s impact on prostate cancer surgery. Nature 2020, 580, S5–S7. [Google Scholar] [CrossRef]
- Available online: https://ourworldindata.org/cancer-death-rates-are-falling-five-year-survival-rates-are-rising (accessed on 2 December 2020).
- Soda, N.; Rehm, B.H.A.; Sonar, P.; Nguyen, N.-T.; Shiddiky, M.J.A. Advanced liquid biopsy technologies for circulating biomarker detection. J. Mater. Chem. B 2019, 7, 6670–6704. [Google Scholar]
- Hermann, T.; Patel, D.J. Adaptive recognition by nucleic acid aptamers. Science 2000, 287, 820–825. [Google Scholar] [CrossRef]
- Colas, P.; Cohen, B.; Jessen, T.; Grishina, I.; McCoy, J.; Brent, R. Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2. Nature 1996, 380, 548–550. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature 1992, 355, 850–852. [Google Scholar]
- Ferapontova, E.E.; Gothelf, K.V. Recent advances in electrochemical aptamer-based sensor. Curr. Org. Chem. 2011, 15, 498–505. [Google Scholar] [CrossRef]
- Díaz-Fernández, A.; Lorenzo-Gómez, R.; Miranda-Castro, R.; delos-Santos-Álvarez, N.; Lobo-Castañón, M.J. Electrochemical aptasensors for cancer diagnosis in biological fluids—A review. Anal. Chim. Acta 2020, 1124, 1–19. [Google Scholar]
- Hasegawa, H.; Taira, K.-I.; Sode, K.; Ikebukuro, K. Improvement of aptamer affinity by dimerization. Sensors 2008, 8, 1090–1098. [Google Scholar] [CrossRef]
- Riccardi, C.; Napolitano, E.; Musumeci, D.; Montesarchio, D. Dimeric and multimeric DNA aptamers for highly effective protein recognition. Molecules 2020, 25, 5227. [Google Scholar] [CrossRef] [PubMed]
- Kjems, J.; Ferapontova, E.E.; Gothelf, K.V. Nucleic Acid Nanotechnology; Springer: Berlin/Heidelberg, Germany, 2014; Volume 29. [Google Scholar]
- Kong, H.Y.; Byun, J. Nucleic acid aptamers: New methods for selection, stabilization, and application in biomedical science. Biomol. Ther. 2013, 21, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.rcsb.org/structure/2L5K (accessed on 2 December 2020).
- Available online: https://www.rcsb.org/structure/2M53 (accessed on 2 December 2020).
- Available online: https://www.rcsb.org/structure/4HQU (accessed on 2 December 2020).
- Li, J.; Tan, S.; Chen, X.; Zhang, C.-Y.; Zhang, Y. Peptide aptamers with biological and therapeutic applications. Curr. Med. Chem. 2011, 18, 4215–4222. [Google Scholar] [CrossRef] [PubMed]
- Mascini, M.; Palchetti, I.; Tombelli, S. Nucleic acid and peptide aptamers: Fundamentals and bioanalytical aspects. Angew. Chem. Int. Ed. 2012, 51, 1316–1332. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Na, J.; Jang, M.H.; Lee, H.; Lee, H.-S.; Lim, Y.-B.; Choi, H.; Chae, Y. A CMOS VEGF sensor for cancer diagnosis using a peptide aptamer-based functionalized microneedle. IEEE Trans. Biomed. Circuits Syst. 2019, 13, 1288–1299. [Google Scholar] [CrossRef]
- Zhang, Y.; Lai, B.S.; Juhas, M. Recent advances in aptamer discovery and applications. Molecules 2019, 24, 941. [Google Scholar] [CrossRef]
- Dunn, M.; Jimenez, R.; Chaput, J. Analysis of aptamer discovery and technology. Nat. Rev. Chem. 2017, 1, 0076. [Google Scholar] [CrossRef]
- Han, K.; Liang, Z.; Zhou, N. Design Strategies for Aptamer-Based Biosensors. Sensors 2010, 10, 4541–4557. [Google Scholar] [CrossRef]
- Wang, H.Q.; Wu, Z.; Tang, L.J.; Yu, R.Q.; Jiang, J.H. Fluorescence protection assay: A novel homogeneous assay platform toward development of aptamer sensors for protein detection. Nucleic Acids Res. 2011, 39, e122. [Google Scholar] [CrossRef]
- Cho, E.J.; Lee, J.W.; Ellington, A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef]
- Zhang, X.; Yadavalli, V.K. Surface immobilization of DNA aptamers for biosensing and protein interaction analysis. Biosens. Bioelectron. 2011, 26, 3142–3147. [Google Scholar] [CrossRef] [PubMed]
- Forouzanfar, S.; Alam, F.; Pala, N.; Wang, C. Review—A review of electrochemical aptasensors for label-free cancer diagnosis. J. Electrochem. Soc. 2020, 167, 067511. [Google Scholar] [CrossRef]
- Vasilescu, A.; Marty, J.-L. Electrochemical aptasensors for the assessment of food quality and safety. TrAC Trends Anal. Chem. 2016, 79, 60–70. [Google Scholar] [CrossRef]
- Labib, M.; Sargent, E.H.; Kelley, S.O. Electrochemical methods for the analysis of clinically relevant biomolecules. Chem. Rev. 2016, 116, 9001–9090. [Google Scholar] [CrossRef]
- Scozzari, A. Electrochemical sensing methods: A brief review. In Algal Toxins: Nature, Occurrence, Effect and Detection; Evangelista, V., Ed.; Springer: Berlin/Heidelberg, Germany, 2008. [Google Scholar]
- Salek-Maghsoudi, A.; Vakhshiteh, F.; Torabi, R. Recent advances in biosensor technology in assessment of early diabetes biomarkers. Biosens. Bioelectron. 2018, 99, 122–135. [Google Scholar] [CrossRef]
- Karunakaran, C.; Bhargava, K.; Benjamin, R. (Eds.) Biosensors and Bioelectronics; Elsevier: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Bettazzi, F.; Marraza, G.; Minunii, M. Biosensors and related bioanalytical tools. Compr. Anal. Chem. 2017, 77, 1–33. [Google Scholar]
- Caygill, R.L.; Blair, G.E.; Millner, P.A. A review on viral biosensors to detect human pathogens. Anal. Chim. Acta 2010, 681, 8–15. [Google Scholar] [CrossRef]
- Ramaley, L.; Krause, M. Theory of square wave voltammetry. Anal. Chem. 1969, 41, 1362–1365. [Google Scholar] [CrossRef]
- Gupta, V.; Jain, R.; Radhapyari, K.; Jadon, N.; Agarwal, S. Voltammetric techniques for the assay of pharmaceuticals—A review. Anal. Biochem. 2011, 408, 179–196. [Google Scholar] [CrossRef]
- Laborda, E.; Molina, A.; Martinez-Ortiz, F.; Compton, R. Electrode modification using porous layers. Maximising the analytical response by choosing the most suitable voltammetry: Differential pulse vs. square wave vs. linear sweep voltammetry. Electrochim. Acta 2012, 73, 3–9. [Google Scholar]
- Chen, A.; Shah, B. Electrochemical sensing and biosensing based on square wave voltammetry. Anal. Methods 2013, 5, 2158–2173. [Google Scholar]
- Yang, L.; Bashir, R. Electrical/electrochemical impedance for rapid detection of foodborne pathogenic bacteria. Biotechnol. Adv. 2008, 26, 135–150. [Google Scholar] [PubMed]
- Vu, C.-A.; Chen, W.-Y. Field-effect transistor biosensors for biomedical applications: Recent advances and future prospects. Sensors 2019, 19, 4214. [Google Scholar]
- Shui, B.; Tao, D.; Florea, A.; Cheng, J.; Zhao, Q.; Gu, Y.; Li, W.; Jaffrezic-Renault, N.; Mei, Y.; Guo, Z. Biosensors for Alzheimer’s disease biomarker detection: A review. Biochimie 2018, 147, 13–24. [Google Scholar] [PubMed]
- Radecki, J.; Radecka, H.; Cieśla, J.; Tudek, B. Chemical sensors and biosensors in genetically modified food control. BioTechnologia 2006, 3, 67–78. [Google Scholar]
- Kwak, B.S.; Kim, H.O.; Kim, J.H.; Lee, S.; Jung, H.-I. Quantitative analysis of sialic acid on erythrocyte membranes using a photothermal biosensor. Biosens. Bioelectron. 2012, 35, 484–488. [Google Scholar]
- Fogel, R.; Limson, J.; Seshia, A.A. Acoustic biosensors. Essays Biochem. 2016, 60, 101–110. [Google Scholar]
- Zangar, R.C.; Daly, D.S.; White, A.M. ELISA microarray technology as a high-throughput system for cancer biomarker validation. Expert Rev. Proteom. 2006, 3, 37–44. [Google Scholar]
- Arya, S.K.; Estrela, P. Recent advances in enhancement strategies for electrochemical ELISA-based immunoassays for cancer biomarker detection. Sensors 2018, 18, 2010. [Google Scholar]
- Celine, I.L.; Duarte, J.A.C.; Rocha-Santos, T.A.P. Detection of ErbB2: Nanotechnological solutions for clinical diagnostics; in a book I send: Immunosensors in Clinical Laboratory Diagnostics. RSC Adv. 2014, 4, 3422. [Google Scholar]
- Cooper, M. Optical biosensors in drug discovery. Nat. Rev. Drug Discov. 2002, 1, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Kulasingam, V.; Jung, B.P.; Blasutig, I.M.; Baradaran, S.; Chan, M.K.; Aytekin, M.; Colantonio, D.A.; Adeli, K. Pediatric reference intervals for 28 chemistries and immunoassays on the Roche cobas® 6000 analyzer—A CALIPER pilot study. Clin. Biochem. 2010, 43, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Wang, J. Analytical Electrochemistry, 3rd ed.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2006. [Google Scholar]
- Rusling, J.F.; Kumar, C.V.; Gutkind, J.S.; Patel, V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst 2010, 135, 2496. [Google Scholar] [CrossRef] [PubMed]
- Baird, G.S. Where are all the aptamers? Am. J. Clin. Pathol. 2010, 134, 529. [Google Scholar] [CrossRef] [PubMed]
- Binte Jamal, R.; Shipovskov, S.; Ferapontova, E.E. Electrochemical immuno- and aptamer-based assays for bacteria: Pros and cons over traditional detection schemes. Sensors 2020, 20, 5561. [Google Scholar] [CrossRef]
- Alvarez-Martos, I.; Campos, R.; Ferapontova, E.E. Surface state of the dopamine RNA aptamer affects specific recognition and binding of dopamine by the aptamer-modified electrodes. Analyst 2015, 140, 4089–4096. [Google Scholar] [CrossRef]
- Bogomolova, A.; Komarova, E.; Reber, K.; Gerasimov, T.; Yavuz, O.; Bhatt, S.; Aldissi, M. Changes in electrochemical impedance spectroscopy in protein biosensing. Anal. Chem. 2009, 81, 3944–3949. [Google Scholar] [CrossRef]
- Pividori, M.I.; Merkoci, A.; Alegret, S. Electrochemical genosensor design: Immobilisation of oligonucleotides onto transducer surfaces and detection methods. Biosens. Bioelectron. 2000, 15, 291–303. [Google Scholar] [CrossRef]
- Ferapontova, E.E. DNA electrochemistry and electrochemical sensors for nucleic acids. Annu. Rev. Anal. Chem. 2018, 11, 197–218. [Google Scholar] [CrossRef]
- Ferapontova, E.E. Hybridization biosensors relying on electrical properties of nucleic acids. Electroanalysis 2017, 29, 6–13. [Google Scholar] [CrossRef]
- Farjami, E.; Campos, R.; Nielsen, J.; Gothelf, K.; Kjems, J.; Ferapontova, E.E. RNA aptamer-based electrochemical biosensor for selective and label-free analysis of dopamine. Anal. Chem. 2013, 85, 121–128. [Google Scholar] [CrossRef]
- Álvarez-Martos, I.; Ferapontova, E.E. Electrochemical label-free aptasensor for specific analysis of dopamine in serum in the presence of structurally related neurotransmitters. Anal. Chem. 2016, 88, 3608–3616. [Google Scholar] [CrossRef]
- Oberhaus, F.V.; Frense, D.; Beckmann, D. Immobilization techniques for aptamers on gold electrodes for the electrochemical detection of proteins: A Review. Biosensors 2020, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Zamay, G.S.; Zamay, T.N.; Kolovskii, V.A.; Shabanov, A.V.; Glazyrin, Y.E.; Veprintsev, D.V.; Krat, A.V.; Zamay, S.S.; Kolovskaya, O.S.; Gargaun, A.; et al. Electrochemical aptasensor for lung cancer-related protein detection in crude blood plasma samples. Sci. Rep. 2016, 6, 34350. [Google Scholar] [CrossRef] [PubMed]
- Mazaafrianto, D.N.; Ishida, A.; Maeki, M.; Tani, H.; Tokeshi, M. Label-free electrochemical sensor for ochratoxin a using a microfabricated electrode with immobilized aptamer. ACS Omega 2018, 3, 16823–16830. [Google Scholar] [CrossRef]
- Ding, S.; Mosher, C.; Lee, X.Y.; Das, S.R.; Cargill, A.A.; Tang, X.; Chen, B.; McLamore, E.S.; Gomes, C.L.; Hostetter, J.M.; et al. Rapid and label-free detection of interferon gamma via an electrochemical aptasensor comprised of a ternary surface monolayer on a gold interdigitated electrode array. ACS Sens. 2017, 2, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Su, Z.; Xu, H.; Xu, X.; Zhang, Y.; Ma, Y.; Li, C.; Xi, Q. Effective covalent immobilization of quinone and aptamer onto a gold electrode via thiol addition for sensitive and selective protein biosensing. Talanta 2017, 164, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Jolly, P.; Formisano, N.; Tkáč, J.; Kasák, P.; Frost, C.G.; Estrela, P. Label-free impedimetric aptasensor with antifouling surface chemistry: A prostate specific antigen case study. Sens. Actuators B Chem. 2015, 209, 306–312. [Google Scholar] [CrossRef]
- Liu, N.; Song, J.; Lu, Y.; Davis, J.J.; Gao, F.; Luo, X. Electrochemical aptasensor for ultralow fouling cancer cell quantification in complex biological media based on designed branched peptides. Anal. Chem. 2019, 91, 8334–8340. [Google Scholar] [CrossRef]
- Jain, A.; Cheng, K. The principles and applications of avidin-based nanoparticles in drug delivery and diagnosis. J. Control. Release 2017, 245, 27–40. [Google Scholar] [CrossRef]
- Pandey, A.K.; Rajput, Y.S.; Sharma, R.; Singh, D. Immobilized aptamer on gold electrode senses trace amount of aflatoxin M1. Appl. Nanosci. 2017, 7, 893–903. [Google Scholar] [PubMed]
- Karapetis, S.; Nikolelis, D.; Hianik, T. Label-free and redox markers-based electrochemical aptasensors for Aflatoxin M1 detection. Sensors 2018, 18, 4218. [Google Scholar]
- Tahiri-Alaoui, A.; Frigotto, L.; Manville, N.; Ibrahim, J.; Romby, P.; James, W. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucleic Acids Res. 2002, 30, 10. [Google Scholar]
- Li, Y.; Lu, Y. Functional Nucleic Acids for Analytical Applications, 3rd ed.; Springer: New York, NY, USA, 2009; pp. 179–199. [Google Scholar]
- De-los-Santos-Álvarez, P.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Current strategies for electrochemical detection of DNA with solid electrodes. Anal. Bioanal. Chem. 2004, 378, 104–118. [Google Scholar] [PubMed]
- Jolly, P.; Batistuti, M.R.; Ustuner, S.; Mulato, M.; Arya, S.K.; Estrela, P. Chapter 9 Nucleic acid-based aptasensors for cancer diagnostics: An insight into immobilisation strategies. In Next Generation Point-of-Care Biomedical Sensors Technologies for Cancer Diagnosis; Chandra, P., Tan, Y.N., Singh, S.P., Eds.; Springer: Singapore, 2017; pp. 205–231. [Google Scholar]
- Gan, X.; Zhao, H. Understanding signal amplification strategies of nanostructured electrochemical sensors for environmental pollutants. Curr. Opin. Electrochem. 2019, 17, 56–64. [Google Scholar]
- Liu, J.; Morris, M.D.; Macazo, F.C.; Schoukroun-Barnes, L.R.; White, R.J. The current and future role of aptamers in electroanalysis. J. Electrochem. Soc. 2014, 161, H301–H313. [Google Scholar]
- Wang, Z.; Yu, J.; Gui, R.; Jin, H.; Xia, Y. Carbon nanomaterials-based electrochemical aptasensors. Biosens. Bioelectron. 2016, 79, 136–149. [Google Scholar]
- George, J.M.; Antony, A.; Mathew, B. Metal oxide nanoparticles in electrochemical sensing and biosensing: A review. Microchim. Acta 2018, 185, 358. [Google Scholar]
- Gui, R.; Jin, H.; Guo, H.; Wang, Z. Recent advances and future prospects in molecularly imprinted polymers-based electrochemical biosensors. Biosens. Bioelectron. 2018, 100, 56–70. [Google Scholar]
- Maduraiveeran, G.; Sasidharan, M.; Ganesan, V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. 2018, 103, 113–129. [Google Scholar]
- Walcarius, A. Silica-based electrochemical sensors and biosensors: Recent trends. Curr. Opin. Electrochem. 2018, 10, 88–97. [Google Scholar] [CrossRef]
- Pietrantonio, F.D.; Cannatà, D.; Benetti, M. Biosensor technologies based on nanomaterials. In Functional Nanostructured Interfaces for Environmental and Biomedical Applications, Micro and Nano Technologies, 3rd ed.; Dca, V., Suchea, M.P., Eds.; Elsevier: Oxford, UK, 2009; pp. 181–242. [Google Scholar]
- Cho, I.H.; Kim, D.H.; Park, S. Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis. Biomater. Res. 2020, 24, 6. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.O.; Mamba, B.; Feleni, U. Poly (propylene imine) dendrimer: A potential nanomaterial for electrochemical application. Mater. Chem. Phys. 2020, 244, 122641. [Google Scholar] [CrossRef]
- Negahdary, M. Electrochemical aptasensors based on the gold nanostructures. Talanta 2020, 216, 120999. [Google Scholar] [CrossRef]
- Pan, M.; Yang, J.; Liu, K.; Yin, Z.; Ma, T.; Liu, S.; Xu, L.; Wang, S. Noble Metal Nanostructured Materials for Chemical and Biosensing Systems. Nanomaterials 2020, 10, 209. [Google Scholar] [CrossRef]
- Nawaz, M.A.H.; Rauf, S.; Catanante, G.; Nawaz, M.H.; Nunes, G.; Marty, J.L.; Hayat, A. One step assembly of thin films of carbon nanotubes on screen printed interface for electrochemical aptasensing of breast cancer biomarker. Sensors 2016, 16, 1651. [Google Scholar] [CrossRef]
- Dauphin-Ducharme, P.; Yang, K.; Arroyo-Curras, N.; Ploense, K.L.; Zhang, Y.; Gerson, J.; Kurnik, M.; Kippin, T.E.; Stojanovic, M.N.; Plaxco, K.W. Electrochemical aptamer-based sensors for improved therapeutic drug monitoring and high-precision, feedback-controlled drug delivery. ACS Sens. 2019, 4, 2832–2837. [Google Scholar] [CrossRef]
- Zhang, X.; Duan, Y.F.S.; Su, L.; Zhang, J.; He, F. Mycobacterium tuberculosis strain H37Rv electrochemical sensor mediated by aptamer and AuNPs−DNA. ACS Sens. 2019, 4, 849–855. [Google Scholar] [CrossRef]
- Amor-Gutiérrez, O.; Selvolini, G.; Fernández-Abedul, M.T.; Escosura-Muñiz, A.; Marrazza, G. Folding-based electrochemical aptasensor for the determination of β-lactoglobulin on poly-L-lysine modified graphite electrodes. Sensors 2020, 20, 2349. [Google Scholar] [CrossRef]
- Villalonga, A.; Pérez-Calabuig, A.M.; Villalonga, R. Electrochemical biosensors based on nucleic acid aptamers. Anal. Bioanal. Chem. 2020, 412, 55–72. [Google Scholar] [CrossRef]
- Song, S.; Wang, L.; Li, J.; Fan, C.; Zhao, J. Aptamer-based biosensors. Trends Anal. Chem. 2008, 27, 108–117. [Google Scholar] [CrossRef]
- Plaxco, K.W.; Soh, H.T. Switch-based biosensors: A new approach towards real-time, in vivo molecular detection. Trends. Biotechnol. 2011, 29, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Kaur, H.; Bruno, J.G.; Kumar, A.; Sharma, T.K. Aptamers in the therapeutics and diagnostics pipelines. Theranostics 2018, 8, 4016–4032. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute at the National Institutes of Health, NCI Dictionary of Cancer Terms. Available online: https://www.cancer.gov/publications/dictionaries/cancer-terms/def/biomarker (accessed on 2 December 2020).
- Duffy, M.J. Tumor Markers in Clinical Practice: A Review Focusing on Common Solid Cancers. Med. Princ. Pract. 2013, 22, 4–11. [Google Scholar] [CrossRef]
- Yarden, S.; Sliwkowski, M.X. Untangling the ErbB signalling network. Nat. Rev. Mol. Cell Biol. 2001, 2, 127–137. [Google Scholar] [CrossRef]
- Perez, E.A.; Cortés, J.; Gonzalez-Angulo, A.M.; Bartlett, J.M.S. HER2 testing: Current status and future directions. Cancer Treat. Rev. 2014, 40, 276–284. [Google Scholar] [CrossRef]
- Ross, J.S.; Slodkowska, E.A.; Symmans, W.F.; Pusztai, L.; Ravdin, P.M.; Hortobagyi, G.N. The HER-2 receptor and breast cancer: Ten years of targeted anti-HER-2 therapy and personalized medicine. Oncologist 2009, 14, 320–368. [Google Scholar] [CrossRef]
- Salimian, R.; Kékedy-Nagy, L.; Ferapontova, E.E. Specific picomolar detection of a breast cancer biomarker HER-2/neu protein in serum: Electrocatalytically amplified electroanalysis by the aptamer/PEG-modified electrode. ChemElectroChem 2017, 4, 872–879. [Google Scholar] [CrossRef]
- Emami, M.; Shamsipur, M.; Saber, R.; Irajirad, R. An electrochemical immunosensor for detection of a breast cancer biomarker based on antiHER2-iron oxide nanoparticle bioconjugates. Analyst 2014, 139, 2858–2866. [Google Scholar] [CrossRef]
- Zhu, Y.; Chandra, P.; Shim, Y.B. Ultrasensitive and selective electrochemical diagnosis of breast cancer based on a hydrazine–Au nanoparticle–aptamer bioconjugate. Anal. Chem. 2013, 85, 1058–1064. [Google Scholar] [CrossRef]
- Al-Khafaji, Q.A.M.; Harris, M.; Tombelli, S.; Laschi, S.; Turner, A.P.F.; Mascini, M.; Marrazza, G. An electrochemical immunoassay for HER2 detection. Electroanalysis 2012, 24, 735–742. [Google Scholar] [CrossRef]
- Malecka, K.; Pankratov, D.; Ferapontova, E.E. Femtomolar electroanalysis of a breast cancer biomarker HER-2/neu protein in human serum. Anal. Chim. Acta 2019, 1077, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Biliran, H.; Sheng, S. Pleiotrophic inhibition of pericellular urokinase-type plasminogen activator system by endogenous tumor suppressive maspin. Cancer Res. 2001, 61, 8676–8682. [Google Scholar] [PubMed]
- Andreasen, P.A.; Egelund, R.; Petersen, H.H. The plasminogen activation system in tumor growth, invasion, and metastasis. Cell. Mol. Life Sci. 2000, 57, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Danø, K.; Behrendt, N.; Høyer-Hansen, G.; Johnsen, M.; Lund, L.R.; Ploug, M.; Rømer, J. Plasminogen activation and cancer. Thromb. Haemost. 2005, 93, 676–681. [Google Scholar]
- Lang, D.S.; Heilenkötter, U.; Schumm, W.; Behrens, O.; Simon, R.; Vollmer, E.; Goldmann, T. Optimized immunohistochemistry in combination with image analysis: A reliable alternative to quantitative ELISA determination of uPA and PAI-1 for routine risk group discrimination in breast cancer. Breast 2013, 22, 736–743. [Google Scholar] [CrossRef]
- Fowler, C.B.; Man, Y.G.; Mason, J.T. An Ultra-sensitive immunoassay for quantifying biomarkers in breast tumor Tissue. J. Cancer 2014, 5, 115–124. [Google Scholar] [CrossRef]
- Hofmann, R.; Lehmer, A.; Hartung, R.; Robrecht, C.; Buresch, M.; Grothe, F. Prognostic value of urokinase plasminogen activator and plasminogen activator inhibitor-1 in renal cell cancer. J. Urol. 1996, 155, 858–862. [Google Scholar] [CrossRef]
- Jarczewska, M.; Kekedy-Nagy, L.; Nielsen, J.S.; Campos, R.; Kjems, J.; Malinowska, E.; Ferapontova, E.E. Electroanalysis of pM-levels of urokinase plasminogen activator in serum by phosphorothioated RNA aptamer. Analyst 2015, 140, 3794–3802. [Google Scholar] [CrossRef]
- Campos, R.; Kotlyar, A.; Ferapontova, E.E. DNA-mediated electron transfer in DNA duplexes tethered to gold electrodes via phosphorothioated dA tags. Langmuir 2014, 30, 11853–11857. [Google Scholar] [CrossRef]
- Ferapontova, E.E.; Gothelf, K.V. Effect of serum on an RNA aptamer-based electrochemical sensor for theophylline. Langmuir 2009, 25, 4279–4283. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Cui, Y.; Owen, S.; Li, W.; Cheng, S.; Jiang, W.G. Human osteopontin: Potential clinical applications in cancer. Int. J. Mol. Med. 2017, 39, 1327–1337. [Google Scholar] [CrossRef]
- Wei, R.; Wong, J.P.C.; Kwok, H.F. Osteopontin—A promising biomarker for cancer therapy. J. Cancer 2017, 8, 2173–2183. [Google Scholar] [CrossRef] [PubMed]
- Anborgh, P.H.; Lee, D.J.; Stam, P.F.; Tuck, A.B.; Chambers, A.F. Role of osteopontin as a predictive biomarker for anti-EGFR therapy in triple-negative breast cancer. Expert Opin. Ther. Targets 2018, 22, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Lan, Z.; Fu, D.; Yu, X.; Xi, M. Diagnostic values of osteopontin combined with CA125 for ovarian cancer: A meta-analysis. Fam. Cancer 2016, 15, 221–230. [Google Scholar] [CrossRef]
- Wiśniewski, T.; Żyromska, A.; Makarewicz, R.; Żekanowska, E. Osteopontin and angiogenic factors as new biomarkers of prostate cancer. Urol. J. 2018, 16, 134–140. [Google Scholar]
- Chakraborty, D.; Viveka, T.S.; Arvind, K.; Shyamsundar, V.; Kanchan, M.; Alex, S.A.; Chandrasekaran, N.; Vijayalakshmi, R.; Mukherjee, A. A facile gold nanoparticle–based ELISA system for detection of osteopontin in saliva: Towards oral cancer diagnostics. Clin. Chim. Acta 2018, 477, 166–172. [Google Scholar] [CrossRef]
- Yan, C.H.; Lv, M.; Li, H.; Song, X.; Yan, F.; Cao, S.; Ren, X. Osteopontin is a novel prognostic biomarker in early-stage non-small cell lung cancer after surgical resection. J. Cancer Res. Clin. Oncol. 2015, 141, 1371–1378. [Google Scholar] [CrossRef]
- Bruha, R.; Vitek, L.; Smid, V. Osteopontin—A potential biomarker of advanced liver disease. Ann. Hepatol. 2020, 19, 344–352. [Google Scholar] [CrossRef]
- Gu, X.; Gao, X.S.; Ma, M.; Qin, S.; Qi, X.; Li, X.; Sun, S.; Yu, H.; Wang, W.; Zhou, D. Prognostic significance of osteopontin expression in gastric cancer: A meta-analysis. Oncotarget 2016, 7, 69666–69673. [Google Scholar] [CrossRef]
- Rychlíkováa, J.; Veckaa, M.; Jáchymováb, M.; Macášeka, J.; Hrabáka, P.; Zemana, M.; Vávrováa, L.; Šoupalc, J.; Krechlera, T.; Žák, A. Osteopontin as a discriminating marker for pancreatic cancer and chronic pancreatitis. Cancer Biomark. 2016, 17, 55–65. [Google Scholar]
- Assidi, M.; Gomaa, W.; Jafri, M.; Hanbazazh, M.; Al-Ahwal, M.; Pushparaj, P.; Al-Harbi, A.; Al-Qahtani, M.; Buhmeida, A.; Al-Maghrabi, J. Prognostic value of Osteopontin (SPP1) in colorectal carcinoma requires a personalized molecular approach. Tumor Biol. 2019, 41, 1–11. [Google Scholar]
- Zhao, M.; Xu, H.; Liang, F.; He, J.; Zhang, J. Association of osteopontin expression with the prognosis of glioma patient: A meta-analysis. Tumor Biol. 2015, 36, 429–436. [Google Scholar]
- Guarino, V.; Faviana, P.; Salvatore, G.; Castellone, M.D.; Cirafici, A.M.; De Falco, V.; Celetti, A.; Giannini, R.; Basolo, F.; Melillo, R.M.; et al. Osteopontin Is Overexpressed in Human Papillary Thyroid Carcinomas and Enhances Thyroid Carcinoma Cell Invasiveness. J. Clin. Endocrinol. Metab. 2005, 90, 5270–5278. [Google Scholar] [PubMed]
- Kiss, T.; Ecsedi, S.; Vizkeleti, L.; Koroknai, V.; Emri, G.; Kovács, N.; Adany, R.; Balazs, M. The role of osteopontin expression in melanoma progression. Tumor Biol. 2015, 36, 7841–7847. [Google Scholar]
- Primasari, M.; Soehartati, A.; Gondhowiardjo, A.; Handjari, D.R.; Matondang, S.; Kantaatmadja, A.B. Osteopontin level correlates negatively with tumor shrinkage in neoadjuvant chemoradiation of locally advanced rectal cancer. Adv. Mod. Oncol. Res. 2015, 1, 56–61. [Google Scholar]
- Cao, Y.; Chen, D.; Chen, W.; Yu, J.; Chen, Z.; Li, G. Aptamer-based homogeneous protein detection using cucurbit[7]uril functionalized electrode. Anal. Chim. Acta 2014, 812, 45–49. [Google Scholar]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Development of an electrochemical RNA-aptasensor to detect human osteopontin. Biosens. Bioelectron. 2015, 71, 332–341. [Google Scholar]
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Electrochemical aptasensor for human osteopontin detection using a DNA aptamer selected by SELEX. Anal. Chim. Acta 2017, 987, 25–37. [Google Scholar]
- Zhou, S.; Gu, C.; Li, Z.; Yang, L.; He, L.; Wang, M.; Huang, X.; Zhou, N.; Zhang, Z. Ti3C2Tx MXene and polyoxometalate nanohybrid embedded with polypyrrole: Ultra-sensitive platform for the detection of osteopontin. Appl. Surf. Sci. 2019, 498, 143889. [Google Scholar]
- Zhou, S.; Hu, M.; Huang, X.; Zhou, N.; Zhang, Z.; Wang, M.; Liu, Y.; He, L. Electrospun zirconium oxide embedded in graphene-like nanofiber for aptamer-based impedimetric bioassay toward osteopontin determination. Microchim. Acta 2020, 187, 219. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xu, Y.; Zhang, L. Serum CA153 as biomarker for cancer and noncancer diseases. Prog. Mol. Biol. Transl. Sci. 2019, 162, 265–276. [Google Scholar] [PubMed]
- Nath, S.; Mukherjee, P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014, 20, 332–342. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. Mucins in cancer: Function, prognosis and therapy. Nat. Rev. Cancer 2009, 9, 874–885. [Google Scholar] [CrossRef] [PubMed]
- Bastu, E.; Mutlu, M.F.; Yasa, C.; Dural, O.; Nehir, A.A.; Celik, C.; Buyru, F.; Yeh, J. Role of Mucin 1 and Glycodelin A in recurrent implantation failure. Fertil. Steril. 2015, 103, 1059–1064. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhang, Q.; Qian, C.; Hao, N.; Xu, L.; Yao, C. Electrochemical aptasensors for mucin1 based on dual signal amplification of poly(o-phenylenediamine) carrier and functionalized carbon nanotubes tracing tag. Biosens. Bioelectron. 2015, 64, 485–492. [Google Scholar] [CrossRef]
- Liu, X.; Qin, Y.; Deng, C.; Xiang, J.; Li, Y. A simple and sensitive impedimetric aptasensors for the detection of tumor markers based on gold nanoparticles signal amplification. Talanta 2015, 132, 150–154. [Google Scholar] [CrossRef]
- Wang, H.; Sun, J.; Lu, L.; Yang, X.; Xia, J.; Zhang, F.; Wang, Z. Competitive electrochemical aptasensor based on a cDNA-ferrocene/MXene probe for detection of breast cancer marker Mucin1. Anal. Chim. Acta 2020, 1094, 18–25. [Google Scholar] [CrossRef]
- Gupta, P.; Bharti, A.; Kaur, N.; Singh, S.; Prabhakar, N. An electrochemical aptasensor based on gold nanoparticles and graphene oxide doped poly(3,4-ethylenedioxythiophene) nanocomposite for detection of MUC1. J. Electroanal. Chem. 2018, 813, 102–108. [Google Scholar] [CrossRef]
- Karpik, A.E.; Crulhas, B.P.; Rodrigues, C.B.; Castro, G.R.; Pedrosa, V.A. Aptamer-based biosensor developed to monitor MUC1 released by prostate cancer cells. Electroanalysis 2017, 29, 2246–2253. [Google Scholar] [CrossRef]
- Florea, A.; Ravalli, A.; Cristea, C.; Săndulescu, R.; Marrazza, G. An optimized bioassay for Mucin1 detection in serum samples. Electroanalysis 2015, 27, 1594–1601. [Google Scholar] [CrossRef]
- Wen, W.; Hu, R.; Bao, T.; Zhang, X.; Wang, S. An insertion approach electrochemical aptasensor for mucin 1 detection based on exonuclease-assisted target recycling. Biosens. Bioelectron. 2015, 71, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Farzin, L.; Sadjadi, S.; Shamsipur, M.; Chabok, A.; Sheibani, S. A sandwich-type electrochemical aptasensor for determination of MUC 1 tumor marker based on PSMA-capped PFBT dots platform and high conductive rGO-N’(1),N’(3)-dihydroxymalonimidamide/thionine nanocomposite as a signal tag. J. Electroanal. Chem. 2017, 807, 108–118. [Google Scholar] [CrossRef]
- Song, J.; Zhou, Y.; Chen, B.; Lou, W.; Gu, J. Development of electrochemical aptamer biosensor for tumor marker MUC1 determination. Int. J. Electrochem. Sci. 2017, 12, 5618–5627. [Google Scholar] [CrossRef]
- Wang, M.; Hu, B.; Ji, H.; Song, Y.; Liu, J.; Peng, D.; He, L.; Zhang, Z. Aptasensor based on hierarchical core–shell nanocomposites of zirconium hexacyanoferrate nanoparticles and mesoporous mFe3O4@mC: Electrochemical quantitation of epithelial tumor marker Mucin-1. ACS Omega 2017, 2, 6809–6818. [Google Scholar] [CrossRef]
- Lin, C.; Zheng, H.; Huang, Y.; Chen, Z.; Luo, F.; Wang, J.; Guo, L.; Qiu, B.; Lin, Z.; Yang, H. Homogeneous electrochemical aptasensor for mucin 1 detection based on exonuclease I-assisted target recycling amplification strategy. Biosens. Bioelectron. 2018, 117, 474–479. [Google Scholar] [CrossRef]
- Ma, C.; Liu, H.; Zhang, L.; Li, H.; Yan, M.; Song, X.; Yu, J. Multiplexed aptasensor for simultaneous detection of carcinoembryonic antigen and mucin-1 based on metal ion electrochemical labels and Ru(NH3)63+ electronic wires. Biosens. Bioelectron. 2018, 99, 8–13. [Google Scholar] [CrossRef]
- Zheng, J.; Peng, X.; Wang, Y.; Bao, T.; Wen, W.; Zhang, X.; Wang, S. An exonuclease-assisted triple-amplified electrochemical aptasensor for mucin 1 detection based on strand displacement reaction and enzyme catalytic strategy. Anal. Chim. Acta 2019, 1086, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Aithal, A.; Rauth, S.; Kshirsagar, P.; Shah, A.; Lakshmanan, I.; Junker, W.M.; Jain, M.; Ponnusamy, M.P.; Batra, S.K. MUC16 as a novel target for cancer therapy. Expert Opin. Ther. Targets 2018, 22, 675–686. [Google Scholar] [CrossRef]
- Suh, H.; Valle, S.; Morris, D.L. Targeting MUC16 in cancer therapy. Chemother. Open Access 2017, 6, 235. [Google Scholar] [CrossRef]
- Milman, S.; Whitney, K.D.; Fleischer, N. Metastatic medullary thyroid cancer presenting with elevated levels of CA 19–9 and CA 125. Thyroid 2011, 21, 913–916. [Google Scholar] [CrossRef]
- Bast Jr, R.C.; Feeney, M.; Lazarus, H.; Nadler, L.M.; Colvin, R.B.; Knapp, R.C. Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Investig. 1981, 68, 1331–1337. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Shen, J.; Wang, J.; Cai, P.; Huang, Y. Clinical analysis of four serum tumor markers in 458 patients with ovarian tumors: Diagnostic value of the combined use of HE4, CA125, CA19–9, and CEA in ovarian tumors. Cancer Manag. Res. 2018, 10, 1313–1318. [Google Scholar] [CrossRef] [PubMed]
- Majd, S.M.; Salimi, A. Ultrasensitive flexible FET-type aptasensor for CA 125 cancer marker detection based on carboxylated multiwalled carbon nanotubes immobilized onto reduced graphene oxide film. Anal. Chim. Acta 2018, 1000, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hu, W.; Wei, J.; Yu, F.; Wu, L.; Wang, C.; Wang, W.; Zuo, S.; Shang, B.; Chen, Q. An electrochemical aptasensing platform for carbohydrate antigen 125 based on the use of flower-like gold nanostructures and target-triggered strand displacement amplification. Microchim. Acta 2019, 186, 388. [Google Scholar] [CrossRef] [PubMed]
- Farzin, L.; Sadjadi, S.; Shamsipur, M.; Sheibani, S.; Mousazadeh, M.H. Employing AgNPs doped amidoxime-modified polyacrylonitrile (PANoxime) nanofibers for target induced strand displacement-based electrochemical aptasensing of CA125 in ovarian cancer patients. Mater. Sci. Eng. C 2019, 97, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Hu, M.; Li, Z.; He, L.; Song, Y.; Jia, Q.; Zhang, Z.; Du, M. Construction of Tb-MOF-on-Fe-MOF conjugate as a novel platform for ultrasensitive detection of carbohydrate antigen 125 and living cancer cells. Biosens. Bioelectron. 2019, 142, 111536. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Liu, B.; Leng, J.; Ma, X.; Peng, H. Electrochemical mixed aptamer-antibody sandwich assay for mucin protein 16 detection through hybridization chain reaction amplification. Anal. Bioanal. Chem. 2020, 412, 7169–7178. [Google Scholar] [CrossRef]
- Sadasivam, M.; Sakthivel, A.; Nagesh, N.; Hansda, S.; Veerapandian, M.; Alwarappan, S.; Manickam, P. Magnetic bead-amplified voltammetric detection for carbohydrate antigen 125 with enzyme labels using aptamer-antigen-antibody sandwiched assay. Sens. Actuators B Chem. 2020, 312, 127985. [Google Scholar] [CrossRef]
- Niu, G.; Chen, X. Vascular endothelial growth factor as an anti-angiogenic target for cancer therapy. Curr. Drug Targets 2010, 11, 1000–1017. [Google Scholar] [CrossRef]
- Goel, H.L.; Mercurio, A.M. VEGF targets the tumour cell. Nat. Rev. Cancer 2013, 13, 871–882. [Google Scholar] [CrossRef] [PubMed]
- Razy, N.H.M.P.; Rahman, W.F.W.A.; Win, T.T. Expression of Vascular Endothelial Growth Factor and Its Receptors in Thyroid Nodular Hyperplasia and Papillary Thyroid Carcinoma: A Tertiary Health Care Centre Based Study. Asian Pac. J. Cancer Prev. 2019, 20, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Ławicki, S.; Będkowska, G.E.; Gacuta-Szumarska, E.; Szmitkowski, M. The plasma concentration of VEGF, HE4 and CA125 as a new biomarkers panel in different stages and sub-types of epithelial ovarian tumors. J. Ovarian Res. 2013, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Shamsipur, M.; Farzin, L.; Tabrizi, M.A.; Molaabasi, F. Highly sensitive label free electrochemical detection of VGEF165 tumor marker based on “signal off” and “signal on” strategies using an anti-VEGF165 aptamer immobilized BSA-gold nanoclusters/ionic liquid/ glassy carbon electrode. Biosens. Bioelectron. 2015, 74, 369–375. [Google Scholar] [CrossRef]
- Dehghani, S.; Nosrati, R.; Yousefi, M.; Nezami, A.; Soltani, F.; Taghdisi, S.M.; Abnous, K.; Alibolandi, M.; Ramezani, M. Aptamer-based biosensors and nanosensors for the detection of vascular endothelial growth factor (VEGF): A review. Biosens. Bioelectron. 2018, 110, 23–37. [Google Scholar] [CrossRef]
- Wei, T.; Dong, T.; Xing, H.; Liu, Y.; Dai, Z. Cucurbituril and azide co-functionalized graphene oxide for ultrasensitive electro-click biosensing. Anal. Chem. 2017, 89, 12237–12243. [Google Scholar] [CrossRef]
- Ye, H.; Qin, B.; Sun, Y.; Li, J. Electrochemical detection of VEGF165 lung cancer marker based on Au-Pd alloy assisted aptasensor. Int. J. Electrochem. Sci. 2017, 12, 818–1828. [Google Scholar]
- Hongxia, C.; Zaijun, L.; Ruiyi, L.; Guangli, W.; Zhiguo, G. Molecular machine and gold/graphene quantum dot hybrid based dual amplification strategy for voltammetric detection of VEGF165. Microchim. Acta 2019, 186, 242. [Google Scholar] [CrossRef]
- Wang, H.; Ma, Y.; Guo, C.; Yang, Y.; Peng, Z.; Liu, Z.; Zhang, Z. Templated seed-mediated derived Au nanoarchitectures embedded with nanochitosan: Sensitive electrochemical aptasensor for vascular endothelial growth factor and living MCF-7 cell detection. Appl. Surf. Sci. 2019, 481, 505–514. [Google Scholar] [CrossRef]
- Ni, S.; Qiao, L.; Shen, Z.; Gao, Y.; Liu, G. Physical absorption vs covalent binding of graphene oxide on glassy carbon electrode towards a robust aptasensor for ratiometric electrochemical detection of vascular endothelial growth factor (VEGF) in serum. Electrochim. Acta 2020, 331, 135321. [Google Scholar] [CrossRef]
- Ni, S.; Shen, Z.; Zhang, P.; Liu, G. Enhanced performance of an electrochemical aptasensor for real-time detection of vascular endothelial growth factor (VEGF) by nanofabrication and ratiometric measurement. Anal. Chim. Acta 2020, 1121, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Tatsumi, A.; Tsukakoshi, K.; Wilson, E.D.; Abe, K.; Sode, K.; Ikebukuro, K. Application of a glucose dehydrogenase-fused with zinc finger protein to label DNA aptamers for the electrochemical detection of VEGF. Sensors 2020, 20, 3878. [Google Scholar] [CrossRef]
- Feng, Y.; Xiao, S.; Xiong, X.; Wang, H.; Kong, F.; Li, Y.; Zhang, Y.; Chen, L. An impedimetric aptasensor based on a novel line-pad-line electrode for the determination of VEGF165. Electroanalysis 2020, 32, 1843–1849. [Google Scholar] [CrossRef]
- Qureshi, A.; Gurbuz, Y.; Niazi, J.H. Capacitive aptamer-antibody based sandwich assay for the detection of VEGF cancer biomarker in serum. Sens. Actuators B Chem. 2015, 209, 645–651. [Google Scholar] [CrossRef]
- Ravalli, A.; Rivas, L.; De la Escosura-Muñiz, A.; Pons, J.; Merkoçi, A.; Marrazza, G. A DNA aptasensor for electrochemical detection of vascular endothelial growth factor. J. Nanosci. Nanotechnol. 2015, 15, 3411–3416. [Google Scholar] [CrossRef]
- Tabrizi, M.A.; Shamsipur, M.; Farzin, L. A high sensitive electrochemical aptasensor for the determination of VEGF165 in serum of lung cancer patient. Biosens. Bioelectron. 2015, 74, 764–769. [Google Scholar] [CrossRef]
- Feng, L.; Lyu, Z.; Offenhäusser, A.; Mayer, D. Electrochemically triggered aptamer immobilization via click reaction for vascular endothelial growth factor detection. Eng. Life Sci. 2016, 16, 550–559. [Google Scholar] [CrossRef]
- Fu, X.-M.; Liu, Z.-J.; Cai, S.-X.; Zhao, Y.-P.; Wu, D.-Z.; Li, C.-Y.; Chen, J.-H. Electrochemical aptasensor for the detection of vascular endothelial growth factor (VEGF) based on DNA-templated Ag/Pt bimetallic nanoclusters. Chin. Chem. Lett. 2016, 27, 920–926. [Google Scholar] [CrossRef]
- Crulhas, B.P.; Karpik, A.E.; Delella, F.K.; Castro, G.R.; Pedrosa, V.A. Electrochemical aptamer-based biosensor developed to monitor PSA and VEGF released by prostate cancer cells. Anal. Bioanal. Chem. 2017, 409, 6771–6780. [Google Scholar] [CrossRef]
- Tabrizi, M.A.; Shamsipur, M.; Saber, R.; Sarkar, S. Simultaneous determination of CYC and VEGF165 tumor markers based on immobilization of flavin adenine dinucleotide and thionine as probes on reduced graphene oxide-poly(amidoamine)/gold nanocomposite modified dual working screen-printed electrode. Sens. Actuators B Chem. 2017, 240, 1174–1181. [Google Scholar] [CrossRef]
- Rawla, P. Epidemiology of prostate cancer. World J. Oncol. 2019, 10, 63–89. [Google Scholar] [CrossRef]
- Diamandis, E.P. Prostate-specific antigen: A cancer fighter and a valuable messenger? Clin. Chem. 2000, 46, 896–900. [Google Scholar] [CrossRef]
- Ankerst, D.P.; Thompson, I.M. Sensitivity and specificity of prostate-specific antigen for prostate cancer detection with high rates of biopsy verification. Arch. Ital. Urol. Androl. 2006, 78, 125–129. [Google Scholar]
- Healy, D.A.; Hayes, C.J.; Leonard, P.; McKenna, L.; O’Kennedy, R. Biosensor developments: Application to prostate-specific antigen detection. Trends Biotechnol 2007, 25, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Moradi, A.; Srinivasan, S.; Clements, J.; Batra, J. Beyond the biomarker role: Prostate-specific antigen (PSA) in the prostate cancer microenvironment. Cancer Metastasis Rev. 2019, 38, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J. Biomarkers for prostate cancer: Prostate-specific antigen and beyond. Clin. Chem. Lab. Med. 2020, 58, 326–339. [Google Scholar] [CrossRef]
- Jolly, P.; Formisano, N.; Estrela, P. DNA aptamer-based detection of prostate cancer. Chem. Pap. 2015, 69, 77–89. [Google Scholar] [CrossRef]
- Damborska, D.; Bertok, T.; Dosekova, E.; Holazova, A.; Lorencova, L.; Kasak, P.; Tkac, J. Nanomaterial-based biosensors for detection of prostate specific antigen. Microchim. Acta 2017, 184, 3049–3067. [Google Scholar] [CrossRef] [PubMed]
- Najeeb, M.A.; Ahmad, Z.; Shakoor, R.A.; Mohamed, A.M.A.; Kahraman, R. A novel classification of prostate specific antigen (PSA) biosensors based on transducing elements. Talanta 2017, 168, 52–61. [Google Scholar] [CrossRef]
- Chang, Y.; Wang, M.; Wang, L.; Xia, N. Recent Progress in electrochemical biosensors for detection of prostate-specific antigen. Int. J. Electrochem. Sci. 2018, 13, 4071–4084. [Google Scholar] [CrossRef]
- Xu, L.; Wen, Y.; Pandit, S.; Mokkapati, V.R.S.S.; Mijakovic, I.; Li, Y.; Ding, M.; Ren, S.; Li, W.; Liu, G. Graphene-based biosensors for the detection of prostate cancer protein biomarkers: A review. BMC Chem. 2019, 13, 112. [Google Scholar] [CrossRef]
- Negahdary, M.; Sattarahmady, N.; Heli, H. Advances in prostate specific antigen biosensors-impact of nanotechnology. Clin. Chim. Acta 2020, 504, 43–55. [Google Scholar] [CrossRef] [PubMed]
- Perry, G.; Cortezon-Tamarit, F.; Pascu, S.I. Detection and monitoring prostate specific antigen using nanotechnology approaches to biosensing. Front. Chem. Sci. Eng. 2020, 14, 4–18. [Google Scholar] [CrossRef]
- Traynor, S.M.; Pandey, R.; Maclachlan, R.; Hosseini, A.; Didar, T.F.; Li, F.; Soleymani, L. Review—Recent advances in electrochemical detection of Prostate Specific Antigen (PSA) in clinically-relevant samples. J. Electrochem. Soc. 2020, 167, 037551. [Google Scholar] [CrossRef]
- Zhang, G.; Liu, Z.; Fan, L.; Guo, Y. Electrochemical prostate specific antigen aptasensor based on hemin functionalized graphene-conjugated palladium nanocomposites. Microchim. Acta 2018, 185, 159. [Google Scholar] [CrossRef]
- Jalalvand, A.R. Fabrication of a novel and ultrasensitive label-free electrochemical aptasensor for detection of biomarker prostate specific antigen. Int. J. Biol. Macromol. 2019, 126, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Hassani, S.; Maghsoudi, A.S.; Akmal, M.R.; Rahmani, S.; Sarihi, P.; Ganjali, M.R.; Norouzi, P.; Abdollahi, M. A sensitive aptamer-based biosensor for electrochemical quantification of PSA as a specific diagnostic marker of prostate cancer. J. Pharm. Pharm. Sci. 2020, 23, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Soleimani, S.; Arkan, E.; Jalalvand, A.R.; Goicoechea, H.C. Fabrication of a novel electrochemical aptasensor assisted by a novel computerized monitoring system for real-time determination of the prostate specific antigen: A computerized experimental method brought elegancy. Microchem. J. 2020, 157, 104898. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, H.; Shi, L.; Zheng, W.; Jing, X. Electroactive Cu2O nanoparticles and Ag nanoparticles driven ratiometric electrochemical aptasensor for prostate specific antigen detection. Sens. Actuators B Chem. 2020, 315, 128155. [Google Scholar] [CrossRef]
- Raica, M.; Cimpean, A.M. Platelet-Derived Growth Factor (PDGF)/PDGF receptors (PDGFR) axis as target for antitumor and antiangiogenic therapy. Pharmaceuticals 2010, 3, 572–599. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Yamakuchi, M.; Shimizu, T.; Kadono, J.; Furoi, A.; Gejima, K.; Komokata, T.; Koriyama, C.; Hashiguchi, T.; Imoto, Y. Predictive value of diminished serum PDGF-BB after curative resection of hepatocellular cancer. J. Oncol. 2019, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Razmi, N.; Baradaran, B.; Hejazi, M.; Hasanzadeh, M.; Mosafer, J.; Mokhtarzadeh, A.; De la Guardia, M. Recent advances on aptamer-based biosensors to detection of platelet derived growth factor. Biosens. Bioelectron. 2018, 113, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.X.; Huang, K.J.; Liu, Y. Novel electrochemical dual-aptamer-based sandwich biosensor using molybdenum disulfide/carbon aerogel composites and Au nanoparticles for signal amplification. Biosens. Bioelectron. 2015, 71, 171–178. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, W.; Cho, S.; Jun, J.; Cho, K.H.; Jang, J. Multidimensional hybrid conductive nanoplate-based aptasensor for platelet-derived growth factor detection. J. Mater. Chem. B 2016, 4, 4447–4454. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, C.; Zhang, S.; He, L.; Wang, M.; Peng, D.; Tian, J.; Fang, S. Carbon based nanocomposites with aptamer-templated silver nanoclusters for the highly sensitive and selective detection of platelet-derived growth factor. Biosens. Bioelectron. 2017, 89, 735–742. [Google Scholar] [CrossRef]
- Degefa, T.H.; Kwak, J. Label-free aptasensor for platelet-derived growth factor (PDGF) protein. Anal. Chim. Acta 2008, 613, 163–168. [Google Scholar] [CrossRef]
- Lai, R.Y.; Plaxco, K.W.; Heeger, A.J. Aptamer-based electrochemical detection of picomolar platelet-derived growth factor directly in blood serum. Anal. Chem. 2007, 79, 229–233. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Chen, Y.-X.; Wu, X.; Huang, K.-J. Electrochemical biosensor based on Se-doped MWCNTs-graphene and Y shaped DNA-aided target-triggered amplification strategy. Colloids Surf. B 2018, 172, 407–413. [Google Scholar] [CrossRef]
- Kim, W.; Lee, J.S.; Jang, J. Aptamer-functionalized three-dimensional carbon nanowebs for ultrasensitive and free-standing PDGF biosensor. ACS Appl. Mater. Interfaces 2020, 12, 20882–20890. [Google Scholar] [CrossRef]
- Hu, X.; Zhang, S.B.; Lu, J.L.; Zheng, L.Y.; Geng, H.E.; Qiu, Y.Q.; Huang, Y.Y.; Qiu, L.P.; Chen, Y.D. Structure-switching hairpin probe based electrochemical aptasensor for highly sensitive detection of protein. In 5th International Conference on Advanced Design and Manufacturing Engineering; Jiang, Z., Ed.; Atlantis Press: Zhengzhou, China, 2015. [Google Scholar]
- Huang, K.-J.; Liu, Y.-J.; Zhai, Q.-F. Ultrasensitive biosensing platform based on layered vanadium disulfide-graphene composites coupling with tetrahedron-structured DNA probes and exonuclease III assisted signal amplification. J. Mater. Chem. B 2015, 3, 8180–8187. [Google Scholar] [CrossRef]
- Liu, X.; Shuai, H.-L.; Huang, K.-J. A label-free electrochemical aptasensor based on leaf-like vanadium disulfide-Au nanoparticles for the sensitive and selective detection of platelet-derived growth factor BB. Anal. Methods 2015, 7, 8277–8284. [Google Scholar] [CrossRef]
- Zhang, S.; Hu, X.; Yang, X.; Sun, Q.; Xu, X.; Liu, X.; Shen, G.; Lu, J.; Shen, G.; Yu, R. Background eliminated signal-on electrochemical aptasensing platform for highly sensitive detection of protein. Biosens. Bioelectron. 2015, 66, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Dong, X.; Liu, S.; Penng, D.; He, L.; Wang, M.; Fu, G.; Feng, X.; Zhang, Z. A label-free multi-functionalized electrochemical aptasensor based on a Fe3O4@3D-rGO@plasma-polymerized (4-vinyl pyridine) nanocomposite for the sensitive detection of proteins in whole blood. Electrochim. Acta 2016, 212, 1–9. [Google Scholar] [CrossRef]
- He, L.; Zhang, S.; Ji, H.; Wang, M.; Peng, D.; Yan, F.; Fang, S.; Zhang, H.; Jia, C.; Zhang, Z. Protein-templated cobaltous phosphate nanocomposites for the highly sensitive and selective detection of platelet-derived growth factor-BB. Biosens. Bioelectron. 2016, 79, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.-J.; Shuai, H.-L.; Zhang, J.-Z. Ultrasensitive sensing platform for platelet-derived growth factor BB detection based on layered molybdenum selenide–graphene composites and Exonuclease III assisted signal amplification. Biosens. Bioelectron. 2016, 77, 69–75. [Google Scholar] [CrossRef]
- Lin, M.-Y.; Hsu, W.-Y.; Yang, Y.-S.; Huang, J.-W.; Chung, Y.-L.; Chen, H. Immobilized rolling circle amplification on extended-gate field-effect transistors with integrated readout circuits for early detection of platelet-derived growth factor. Anal. Bioanal. Chem. 2016, 408, 4785–4797. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Tian, D.; Zhang, L.; Guo, Q.; Cui, Y.; Yang, M. Dual signal amplification strategy for amperometric aptasensing using hydroxyapatite nanoparticles. Application to the sensitive detection of the cancer biomarker platelet-derived. Microchim. Acta 2017, 184, 4375–4381. [Google Scholar] [CrossRef]
- Xie, B.; Zhou, N.; Ding, R.; Zhao, Y.; Zhang, B.; Li, T.; Yang, M. Dual signal amplification strategy for electrochemical detection of platelet-derived growth factor BB. Anal. Methods 2017, 9, 6569–6573. [Google Scholar] [CrossRef]
- Yu, Y.; Su, G.; Zhu, H.; Zhu, Q.; Chen, Y.; Xu, B.; Li, Y.; Zhang, W. Proximity hybridization-mediated isothermal exponential amplification for ultrasensitive electrochemical protein detection. Int. J. Nanomed. 2017, 12, 5903–5914. [Google Scholar] [CrossRef][Green Version]
- Zhao, C.-L.; Hua, M.; Yang, C.-Y.; Yang, Y.-H. A novel aptasensor based on 3D inorganic hybrid composite as immobilized substrate for sensitive detection of platelet-derived growth factor. Chin. Chem. Lett. 2017, 28, 1417–1423. [Google Scholar] [CrossRef]
- Hasanzadeh, M.; Razmi, N.; Mokhtarzadeh, A.; Shadjou, N.; Mahboob, S. Aptamer based assay of plated-derived grow factor in unprocessed human plasma sample and MCF-7 breast cancer cell lysates using gold nanoparticle supported α-cyclodextrin. Int. J. Biol. Macromol. 2018, 108, 69–80. [Google Scholar] [CrossRef] [PubMed]
- Naz, Z.; Usman, S.; Saleem, K.; Ahmed, S.; Bashir, H.; Bilal, M.; Sumrin, A. Alpha-fetoprotein: A fabulous biomarker in hepatocellular, gastric and rectal cancer diagnosis. Biomed. Res. 2018, 29, 2478–2483. [Google Scholar] [CrossRef]
- Liu, Y.; Deng, Y.; Li, T.; Chen, Z.; Chen, H.; Li, S.; Liu, H. Aptamer-Based Electrochemical Biosensor for Mercury Ions Detection Using AuNPs-Modified Glass Carbon Electrode. J. Biomed. Nanotechnol. 2018, 14, 2156–2161. [Google Scholar] [CrossRef] [PubMed]
- Sauzay, C.; Petit, A.; Bourgeois, A.; Barbare, J.; Chauffert, B.; Galmiche, A.; Houessinon, A. Alpha-foetoprotein (AFP): A multi-purpose marker in hepatocellular carcinoma. Clin. Chim. Acta 2016, 463, 39–44. [Google Scholar] [CrossRef]
- Mohammadinejad, A.; Oskuee, R.K.; Eivazzadeh-Keihan, R.; Rezayi, M.; Baradaran, B.; Maleki, A.; Hashemzaei, M.; Mokhtarzadeh, A.; De la Guardia, M. Development of biosensors for detection of alpha-fetoprotein: As a major biomarker for hepatocellular carcinoma. Tractrends Anal. Chem. 2020, 130, 115961. [Google Scholar] [CrossRef]
- Li, G.; Li, S.; Wang, Z.; Xue, Y.; Dong, C.; Zeng, J.; Huang, Y.; Liang, J.; Zhou, Z. Label-free electrochemical aptasensor for detection G. of alpha-fetoprotein based on AFP-aptamer and thionin/reduced graphene oxide/gold nanoparticles. Anal. Biochem. 2018, 547, 37–44. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, F.; Wang, Z.; Liang, Q. A graphene oxide-based label-free electrochemical aptasensor for the detection of alpha-fetoprotein. Biosens. Bioelectron. 2018, 112, 186–192. [Google Scholar] [CrossRef]
- Zhang, B.; Ding, H.; Chen, Q.; Wang, T.; Zhang, K. Prussian blue nanoparticle-labeled aptasensing platform on graphene oxide for voltammetric detection of α-fetoprotein in hepatocellular carcinoma with target recycling. Analyst 2019, 144, 4858–4864. [Google Scholar] [CrossRef] [PubMed]
- Heiat, M.; Negahdary, M. Sensitive diagnosis of alpha-fetoprotein by a label free nanoaptasensor designed by modified Au electrode with spindle-shaped gold nanostructure. Microchem. J. 2019, 148, 456–466. [Google Scholar] [CrossRef]
- Huang, X.; Cui, B.; Ma, Y.; Yan, X.; Xia, L.; Zhou, N.; Wang, M.; He, L.; Zhang, Z. Three-dimensional nitrogen-doped mesoporous carbon nanomaterials derived from plant biomass: Cost-effective construction of label-free electrochemical aptasensor for sensitively detecting alpha-fetoprotein. Anal. Chim. Acta 2019, 1078, 125–134. [Google Scholar] [CrossRef]
- Li, J.; Wei, H.; Peng, Y.; Geng, L.; Zhu, L.; Cao, X.-Y.; Liu, C.-S.; Pang, H. A multifunctional self-healing G-PyB/KCl hydrogel: Smart conductive, rapid room-temperature phase-selective gelation, and ultrasensitive detection of alpha-fetoprotein. Chem. Commun. 2019, 55, 7922. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Peng, Y.; Li, J.; Liu, C.-S.; Pang, H. Controllable synthesis of copper ion guided MIL-96 octadecahedron: Highly sensitive aptasensor toward alpha-fetoprotein. Appl. Mater. Today 2020, 20, 100745. [Google Scholar] [CrossRef]
- Abi, A.; Ferapontova, E.E. Electroanalysis of single nucleotide polymorphism by hairpin DNA architectures. Anal. Bioanal. Chem. 2013, 405, 3693–3703. [Google Scholar] [CrossRef]
- Gao, T.; Zhi, J.; Mu, C.; Gu, S.; Xiao, J.; Yang, J.; Wang, Z.; Xiang, Y. One-step detection for two serological biomarker species to improve the diagnostic accuracy of hepatocellular carcinoma. Talanta 2018, 178, 89–93. [Google Scholar] [CrossRef]
- Yang, X.; Zhao, C.; Zhang, C.; Wen, K.; Zhu, Y. Bi-directionally amplified ratiometric electrochemical aptasensor for the ultrasensitive detection of alpha-fetoprotein. Sens. Actuators B Chem. 2020, 323, 128666. [Google Scholar] [CrossRef]
- Han, B.; Dong, L.; Li, L.; Sha, L.; Cao, Y.; Zhao, J. Mild reduction-promoted sandwich aptasensing for simple and versatile detection of protein biomarkers. Sens. Actuators B Chem. 2020, 325, 128762. [Google Scholar] [CrossRef]
- Cui, M.; Wang, Y.; Jiao, M.; Jayachandran, S.; Wu, Y.; Fan, X.; Luo, X. Mixed self-assembled aptamer and newly designed zwitterionic peptide as antifouling biosensing interface for electrochemical detection of alpha-fetoprotein. ACS Sens. 2017, 2, 490–494. [Google Scholar] [CrossRef]
- Dasgupta, A.; Wahed, A. Clinical Chemistry, Immunology and Laboratory Quality Control; Elsevier Inc.: Amsterdam, The Netherlands, 2014; pp. 229–247. [Google Scholar]
- Asad-Ur-Rahman, F.; Saif, M.W. Elevated level of serum carcinoembryonic antigen (CEA) and search for a malignancy: A case report. Cureus 2016, 8, e648. [Google Scholar] [CrossRef] [PubMed]
- Fakih, M.G.; Padmanabhan, A. CEA Monitoring in colorectal cancer. What you should know. Oncology 2006, 20, 579–587. [Google Scholar] [PubMed]
- Liu, Z.; Wang, Y.; Guo, Y.; Dong, C. Label-free electrochemical aptasensor for carcinoembryonic antigen based on ternary nanocomposite of gold nanoparticles, hemin and graphene. Electroanalysis 2016, 28, 1023–1028. [Google Scholar] [CrossRef]
- Wen, W.; Huang, J.-Y.; Bao, T.; Zhou, J.; Xia, H.-X.; Zhang, X.-H.; Wang, S.-F.; Zhao, Y.-D. Increased electrocatalyzed performance through hairpin oligonucleotide aptamer-functionalized gold nanorods labels and graphene-streptavidin nanomatrix: Highly selective and sensitive electrochemical biosensor of carcinoembryonic antigen. Biosens. Bioelectron. 2016, 83, 142–148. [Google Scholar] [CrossRef] [PubMed]
- He, B. Differential pulse voltammetric assay for the carcinoembryonic antigen using a glassy carbon electrode modified with layered molybdenum selenide, graphene, and gold nanoparticles. Microchim. Acta 2017, 184, 229–235. [Google Scholar] [CrossRef]
- Si, Z.; Xie, B.; Chen, Z.; Tang, C.; Li, T.; Yang, M. Electrochemical aptasensor for the cancer biomarker CEA based on aptamer induced current due to formation of molybdophosphate. Microchim. Acta 2017, 184, 3215–3221. [Google Scholar] [CrossRef]
- Zhou, X.; Guo, S.; Gao, J.; Zhao, J.; Xue, S.; Xu, W. Glucose oxidase-initiated cascade catalysis for sensitive impedimetric aptasensor based on metal-organic frameworks functionalized with Pt nanoparticles and hemin/G-quadruplex as mimicking peroxidases. Biosens. Bioelectron. 2017, 98, 83–90. [Google Scholar] [CrossRef]
- Huang, J.-Y.; Zhao, L.; Lei, W.; Wen, W.; Wang, Y.-J.; Bao, T.; Xiong, H.-Y.; Zhang, X.-H.; Wang, S.-F. A high-sensitivity electrochemical aptasensor of carcinoembryonic antigen based on graphene quantum dots-ionic liquid-nafion nanomatrix and DNAzyme-assisted signal amplification strategy. Biosens. Bioelectron. 2018, 99, 28–33. [Google Scholar] [CrossRef]
- Mazloum-Ardakani, M.; Tavakolian-Ardakani, Z.; Sahraei, N.; Moshtaghioun, S.M. Fabrication of an ultrasensitive and selective electrochemical aptasensor to detect carcinoembryonic antigen by using a new nanocomposite. Biosens. Bioelectron. 2019, 129, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Hong, F.; Wang, Q.; Wang, W.; Chen, X.; Cao, Y.; Dong, Y.; Gan, N.; Wu, D.; Hu, F. Background signal-free and highly sensitive electrochemical aptasensor for rapid detecting tumor markers with Pb-MOF functionalized dendritic DNA probes. J. Electroanal. Chem. 2020, 861, 113956. [Google Scholar] [CrossRef]
- Xu, L.; Zou, L.; Guo, J.; Cao, Y.; Feng, C.; Ye, B. simple “signal-off” electrochemical aptasensor based on aptamer-Cu3(PO4)2 hybrid nanoflowers/graphene oxide for carcinoembryonic antigen detection. ChemElectroChem 2020, 7, 1660–1665. [Google Scholar] [CrossRef]
- Yen, Y.-K.; Chao, C.-H.; Yeh, Y.-S. A graphene-PEDOT: PSS modified paper-based aptasensor for electrochemical impedance spectroscopy detection of tumor marker. Sensors 2020, 20, 1372. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Su, F.; Song, Y.; Hu, B.; Wang, M.; He, L.; Peng, D.; Zhang, Z. Aptamer-templated silver nanoclusters embedded in zirconium metal-organic framework for bifunctional electrochemical and SPR aptasensors toward carcinoembryonic antigen. ACS Appl. Mater. Interfaces 2017, 9, 41188–41199. [Google Scholar] [CrossRef]
- Jimenez-Falcao, S.; Parra-Nieto, J.; Pérez-Cuadrado, H.; Martínez-Máñez, R.; Martínez-Ruiz, P.; Villalonga, R. Avidin-gated mesoporous silica nanoparticles for signal amplification in electrochemical biosensor. Electrochem. Commun. 2019, 108, 106556. [Google Scholar] [CrossRef]
- Zheng, J.; Wang, J.; Song, D.; Xu, J.; Zhang, M. Electrochemical aptasensor of carcinoembryonic antigen based on concanavalin A-functionalized magnetic copper silicate carbon microtubes and gold-nanocluster-assisted signal amplification. ACS Appl. Nano Mater. 2020, 3, 3449–3458. [Google Scholar] [CrossRef]
- Xu, Z.; Han, R.; Liu, N.; Gao, F.; Luo, X. Electrochemical biosensors for the detection of carcinoembryonic antigen with low fouling and high sensitivity based on copolymerized polydopamine and zwitterionic polymer. Sens. Actuators B Chem. 2020, 319, 128253. [Google Scholar] [CrossRef]
- Zhang, R.; Liu, L.; Mao, D.; Luo, D.; Cao, F.; Chen, Q.; Chen, J. Construction of electrochemical aptasensor of carcinoembryonic antigen based on toehold-aided DNA recycling signal amplification. Bioelectrochemistry 2020, 133, 107492. [Google Scholar] [CrossRef]
- Cheng, H.; Xu, L.; Zhang, H.; Yu, A.; Lai, G. Enzymatically catalytic signal tracing by a glucose oxidase and ferrocene dually functionalized nanoporous gold nanoprobe for ultrasensitive electrochemical measurement of a tumor biomarker. Analyst 2016, 141, 4381–4387. [Google Scholar] [CrossRef]
- Park, J.W.; Na, W.; Jang, J. One-pot synthesis of multidimensional conducting polymer nanotubes for superior performance field-effect transistor-type carcinoembryonic antigen biosensors. RSC Adv. 2016, 6, 14335–14343. [Google Scholar] [CrossRef]
- Wang, P.; Wan, Y.; Deng, S.; Yang, S.; Su, Y.; Fan, C.; Aldalbahi, A.; Zuo, X. Aptamer-initiated on-particle template-independent enzymatic polymerization (aptamer-OTEP) for electrochemical analysis of tumor biomarkers. Biosens. Bioelectron. 2016, 86, 536–541. [Google Scholar] [CrossRef]
- Zhou, X.; Xue, S.; Jing, P.; Xu, W. A sensitive impedimetric platform biosensing protein: Insoluble precipitates based on the biocatalysis of manganese (III) meso-tetrakis (4-N-methylpyridiniumyl)-porphyrinin in HCR-assisted dsDNA. Biosens. Bioelectron. 2016, 86, 656–663. [Google Scholar] [CrossRef]
- Taghdisi, S.M.; Danesh, N.M.; Ramezani, M.; Emrani, A.S.; Abnous, K. A novel electrochemical aptasensor for carcinoembryonic antigen detection based on target-induced bridge assembly. Electroanalysis 2018, 30, 1734–1739. [Google Scholar] [CrossRef]
- Wang, Q.-L.; Cui, H.-F.; Song, X.; Fan, S.-F.; Chen, L.-L.; Li, M.-M.; Li, Z.-Y. A label-free and lectin-based sandwich aptasensor for detection of carcinoembryonic antigen. Sens. Actuators B Chem. 2018, 260, 48–54. [Google Scholar] [CrossRef]
- He, L.; Li, Z.; Guo, C.; Hu, B.; Wang, M.; Zhang, Z.; Du, M. Bifunctional bioplatform based on NiCo Prussian blue analogue: Label-free impedimetric aptasensor for the early detection of carcino-embryonic, antigen and living cancer cells. Sens. Actuators B Chem. 2019, 298, 126852. [Google Scholar] [CrossRef]
- Liu, Z.; Lei, S.; Zou, L.; Li, G.; Xu, L.; Ye, B. A label-free and double recognition-amplification novel strategy for sensitive and accurate carcinoembryonic antigen assay. Biosens. Bioelectron. 2019, 131, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, G.; Villalonga, A.; Eguílaz, M.; Vegas, B.; Parrado, C.; Rivas, G.; Díez, P.; Villalonga, R. Amperometric aptasensor for carcinoembryonic antigen based on the use of bifunctionalized Janus nanoparticles as biorecognition signaling element. Anal. Chim. Acta 2019, 1061, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Liu, Z.; Lei, S.; Huang, D.; Zou, L.; Ye, B. A sandwich-type electrochemical aptasensor for the carcinoembryonic antigen via biocatalytic precipitation amplification and by using gold nanoparticle composites. Microchim. Acta 2019, 186, 473. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Liu, X.; Qin, Y.; Deng, C.; Xiang, J. A simple regenerable electrochemical aptasensor for the parallel and continuous detection of biomarkers. RSC Adv. 2016, 6, 58469–58476. [Google Scholar] [CrossRef]
- Xiang, J.; Pi, X.; Chen, X.; Xiang, L.; Yang, M.; Ren, H.; Shen, X.; Qi, N.; Deng, C. Integrated signal probe based aptasensor for dual-analyte detection. Biosens. Bioelectron. 2017, 96, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Yang, Y.; Sun, Y.; Cui, L.; Zheng, F.; Zhang, J.; Song, Q.; Xu, C. Shell-encoded Au nanoparticles with tunable electroactivity for specific dual disease biomarkers detection. Biosens. Bioelectron. 2018, 99, 193–200. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, J.; Liu, J.; Sun, S.; Xiong, Y.; Ma, Y.; Yan, S.; Yang, Y.; Yin, H.; Cai, X. Label-free microfluidic paper-based electrochemical aptasensor for ultrasensitive and simultaneous multiplexed detection of cancer biomarkers. Biosens. Bioelectron. 2019, 136, 84–90. [Google Scholar] [CrossRef]
- Wang, W.; Wang, Q.; Xie, H.; Wu, D.; Gan, N. A universal assay strategy for sensitive and simultaneous quantitation of multiplex tumor markers based on the stirring rod-immobilized DNA-LaMnO3 perovskite-metal ions encoded probes. Talanta 2021, 222, 121456. [Google Scholar] [CrossRef]
- Fan, J.; Tang, Y.; Yang, W.; Yu, Y. Disposable multiplexed electrochemical sensors based on electro-triggered selective immobilization of probes for simultaneous detection of DNA and proteins. J. Mater. Chem. B 2020, 8, 7501–7510. [Google Scholar] [CrossRef]
- Marko, N.F.; Weil, R.J.; Toms, S.A. Nanotechnology in proteomics. Expert Rev. Proteom. 2007, 4, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Ehdaie, B. Application of nanotechnology in cancer research: Review of progress in the National Cancer Institute’s Alliance for nanotechnology. Int. J. Biol. Sci. 2007, 3, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.pointofcare.abbott/int/en/offerings/istat (accessed on 2 December 2020).
- Zhang, Y.; Figueroa-Miranda, G.; Wu, C.; Willbold, D.; Offenhäusser, A.; Mayer, D. Electrochemical dual-aptamer biosensors based on nanostructured multielectrode arrays for the detection of neuronal biomarkers. Nanoscale 2020, 12, 16501–16513. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, R.; Patel, V.; Chikkaveeraiah, B.V.; Munge, B.S.; Cheong, S.C.; Zain, R.B.; Abraham, M.T.; Dey, D.K.; Gutkind, J.S.; Rusling, J.F. Ultrasensitive detection of cancer biomarkers in the clinic by use of a nanostructured microfluidic array. Anal. Chem. 2012, 84, 6249–6255. [Google Scholar] [CrossRef]
- Ilyas, A.; Asghar, W.; Allen, P.B.; Duhon, H.; Ellington, A.D.; Iqbal, S.M. Electrical detection of cancer biomarker using aptamers with nanogap break-junctions. Nanotechnology 2012, 23, 275502. [Google Scholar] [CrossRef]
- Liu, D.; Wang, J.; Wu, L.; Huang, Y.; Zhang, Y.; Zhu, M.; Wang, Y.; Zhu, Z.; Yang, C. Trends in miniaturized biosensors for point-of-care testing. Tractrends Anal. Chem. 2020, 122, 115701. [Google Scholar] [CrossRef]
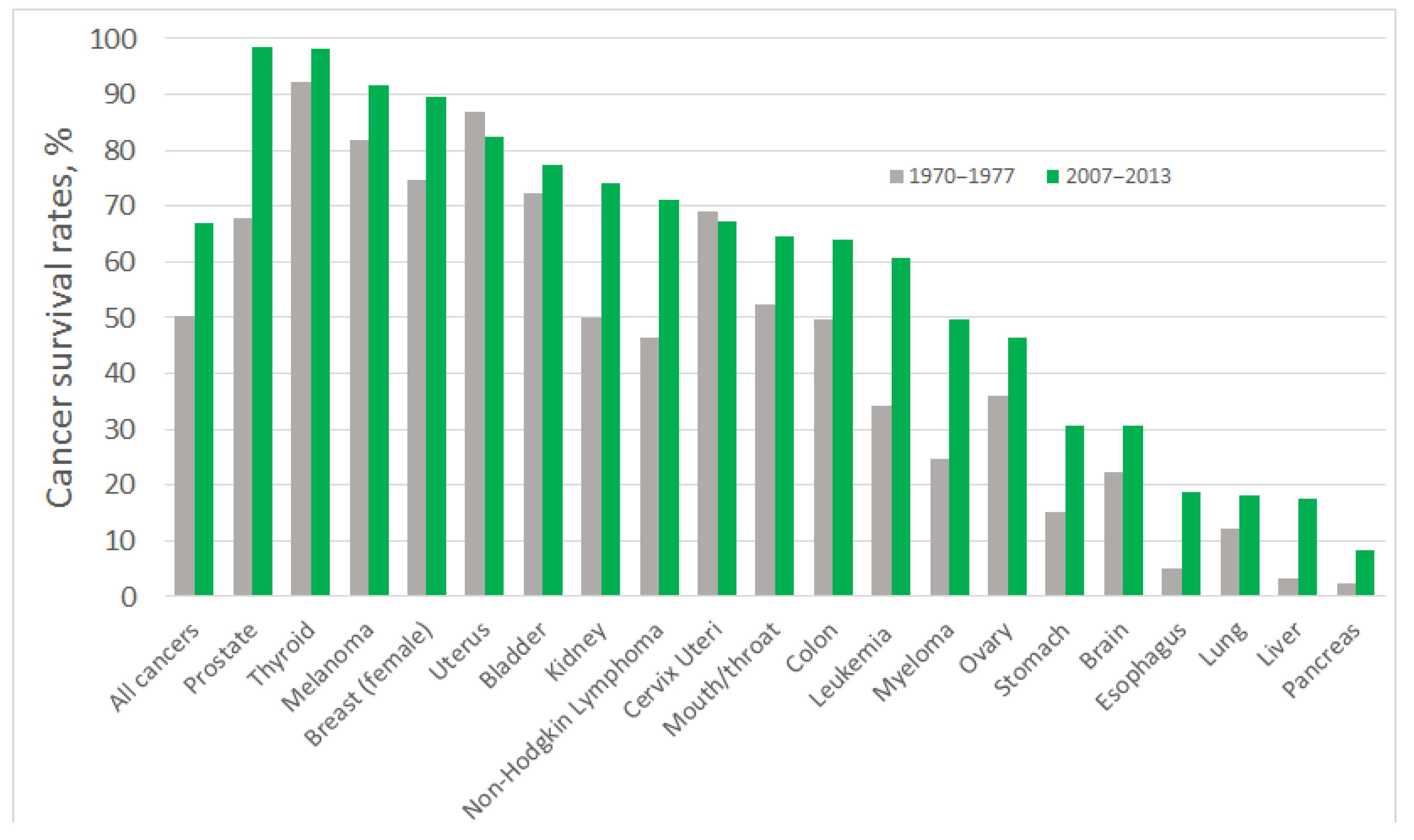


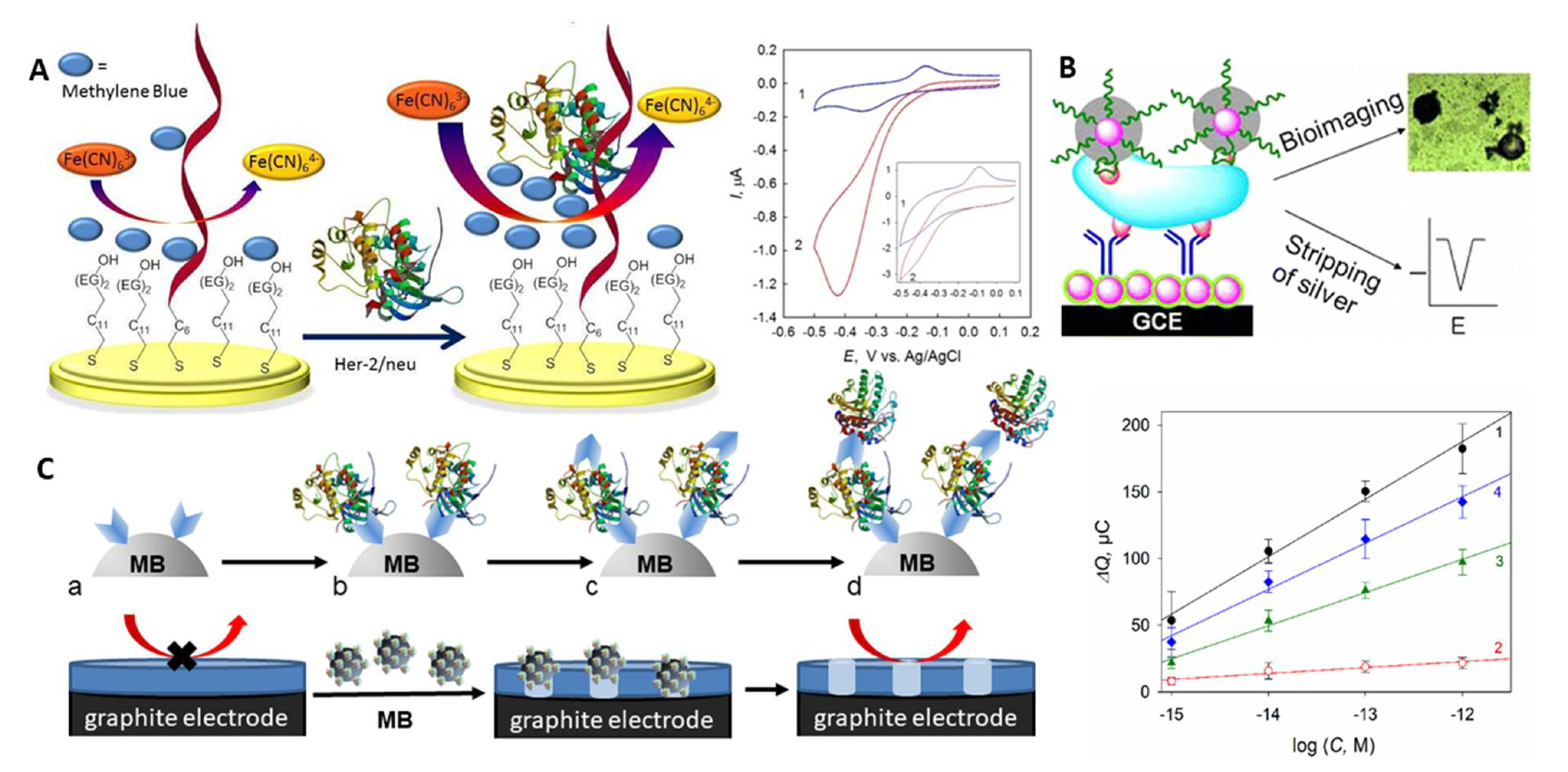

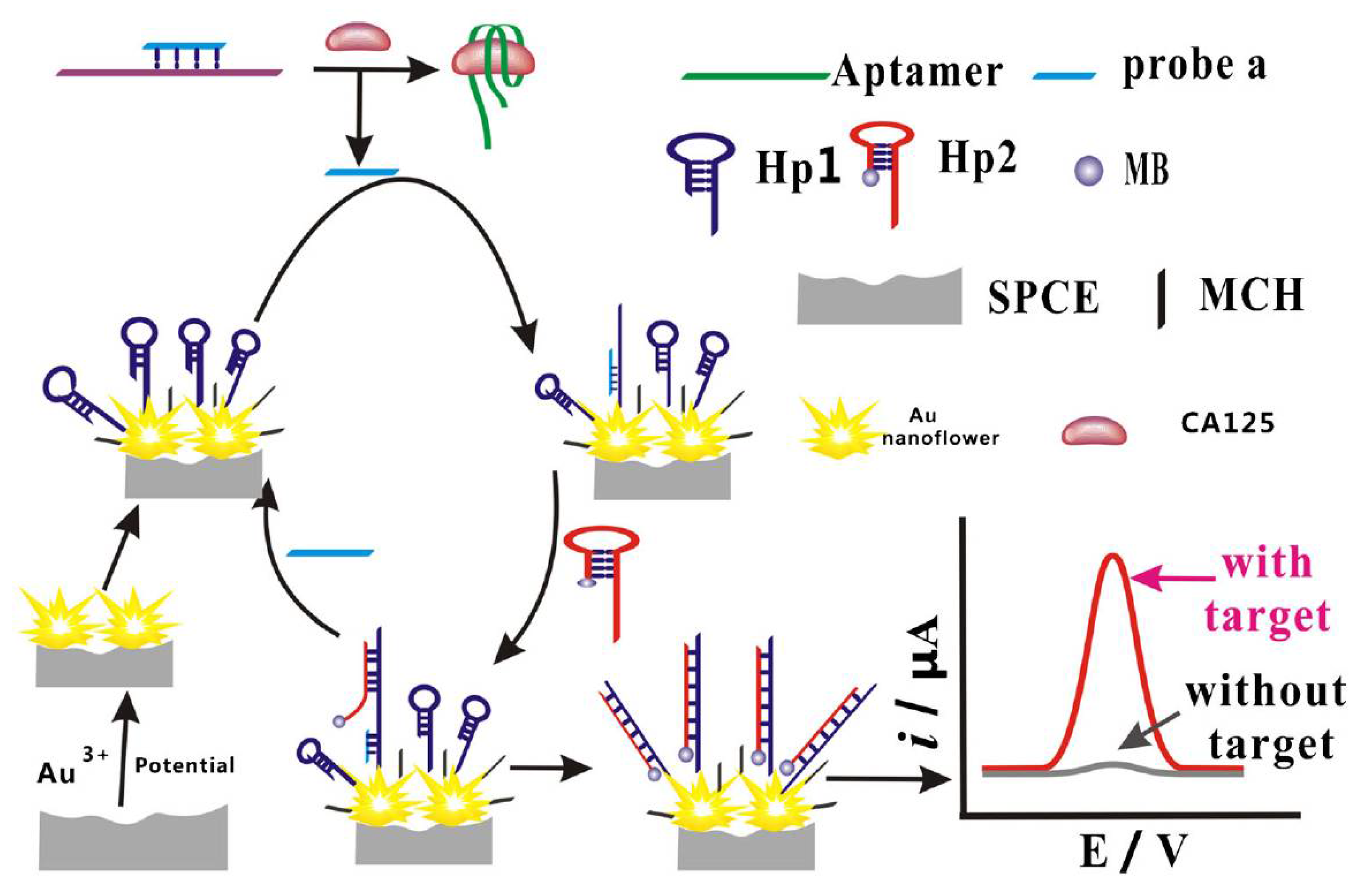
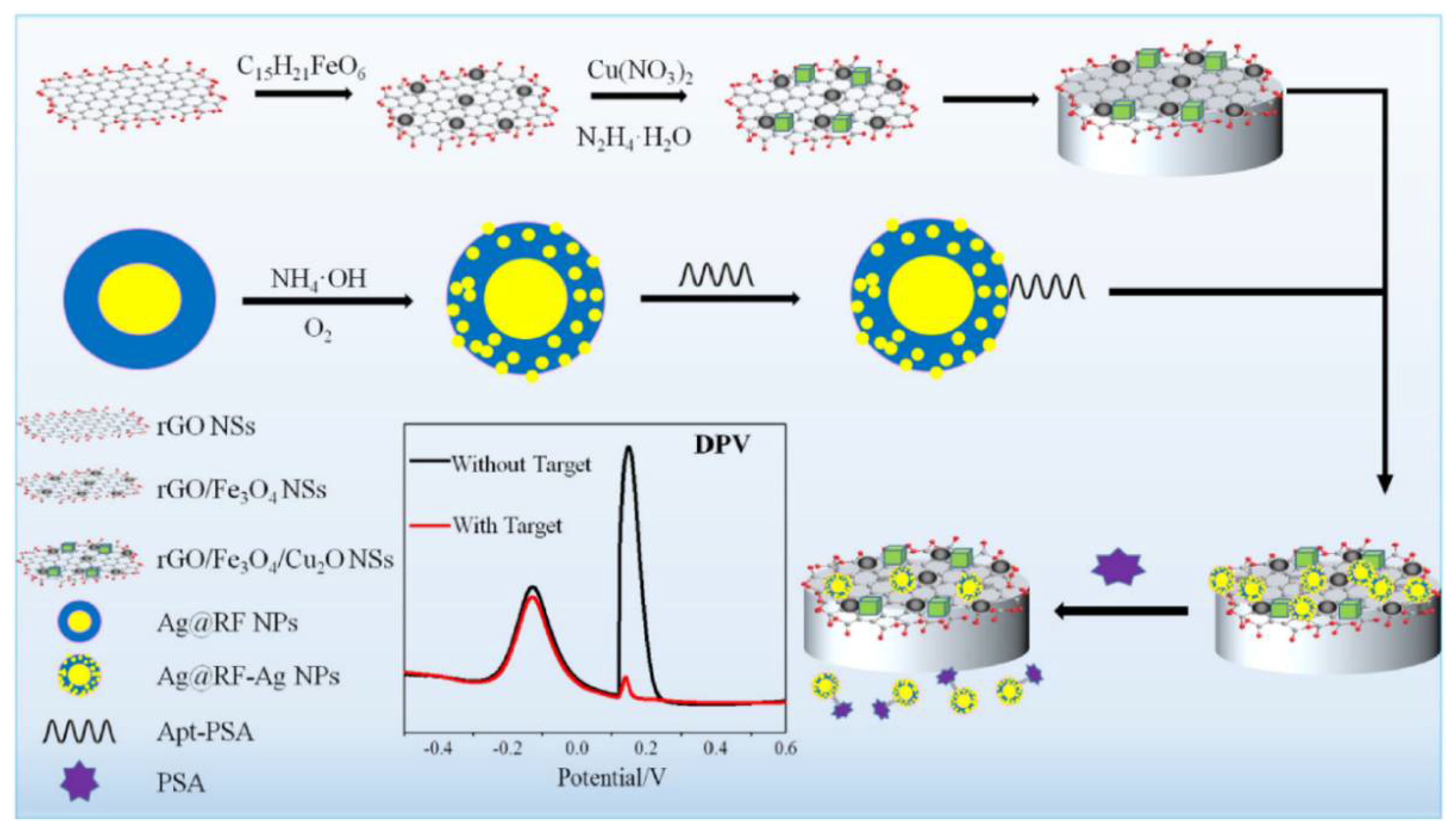
| Immobilisation Strategy | Type of Interaction | Advantages | Disadvantages |
|---|---|---|---|
| Electrostatic adsorption | negatively charged aptamer on the positively charged surface |
|
|
| Covalent attachment | EDC/NHS coupling between COOH-functionalised electrode surface and the amine-terminated aptamer sequence |
|
|
| Chemisorption | involves chemical bond between the probe and the electrode surface, e.g., gold through the alkanethiol linker | ||
| Affinity interaction | specific interactions such as those between biotin and avidin or streptavidin |
|
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malecka, K.; Mikuła, E.; Ferapontova, E.E. Design Strategies for Electrochemical Aptasensors for Cancer Diagnostic Devices. Sensors 2021, 21, 736. https://doi.org/10.3390/s21030736
Malecka K, Mikuła E, Ferapontova EE. Design Strategies for Electrochemical Aptasensors for Cancer Diagnostic Devices. Sensors. 2021; 21(3):736. https://doi.org/10.3390/s21030736
Chicago/Turabian StyleMalecka, Kamila, Edyta Mikuła, and Elena E. Ferapontova. 2021. "Design Strategies for Electrochemical Aptasensors for Cancer Diagnostic Devices" Sensors 21, no. 3: 736. https://doi.org/10.3390/s21030736
APA StyleMalecka, K., Mikuła, E., & Ferapontova, E. E. (2021). Design Strategies for Electrochemical Aptasensors for Cancer Diagnostic Devices. Sensors, 21(3), 736. https://doi.org/10.3390/s21030736








