Evaluation of Deep Neural Networks for Semantic Segmentation of Prostate in T2W MRI
Abstract
1. Introduction
2. Related Work
3. Material and Methods
3.1. Deep CNN Architectures for Prostate Segmentation
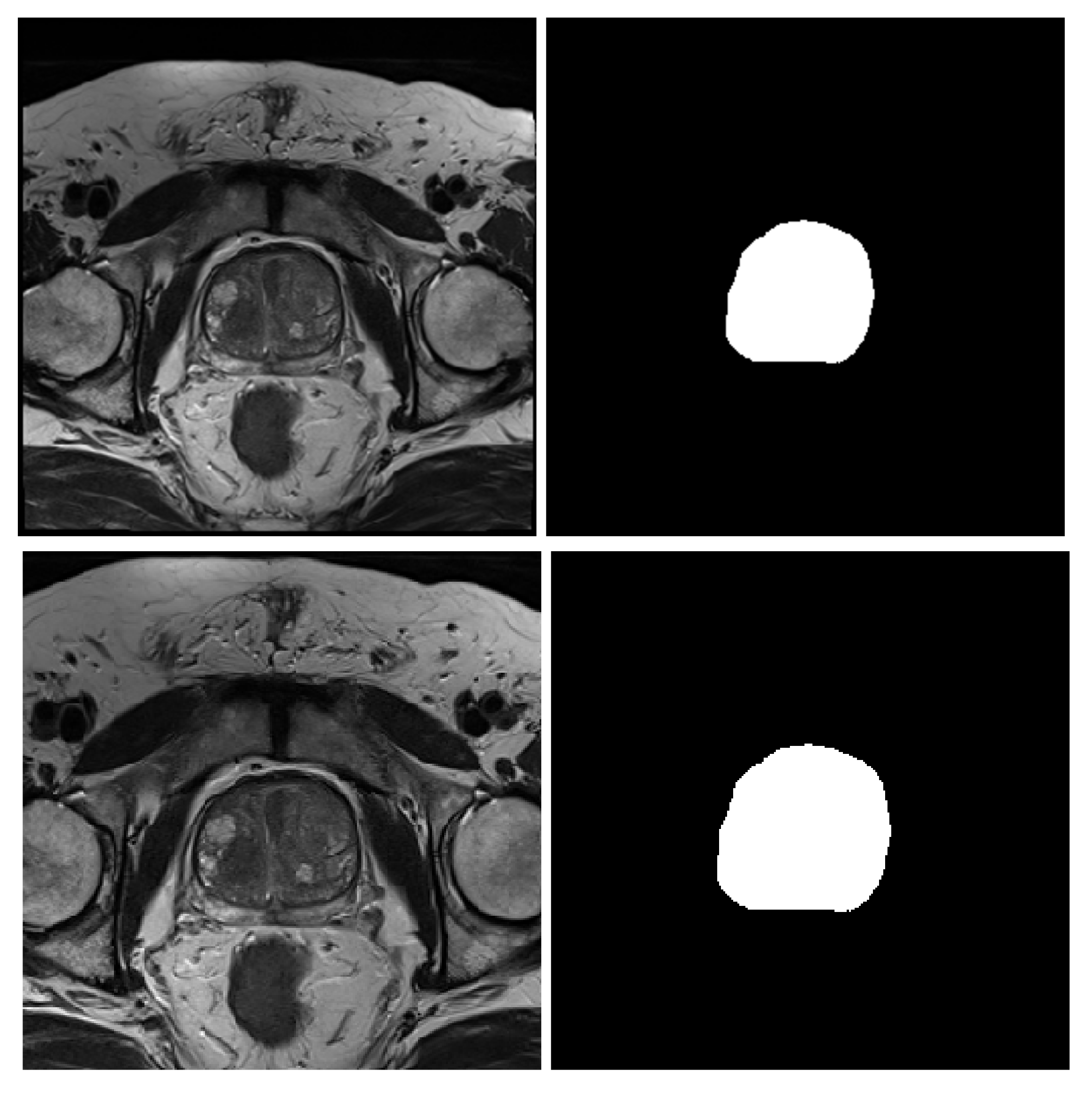
3.2. MRI Dataset
- NCI-ISBI 2013 is a prostate 3T collection of 40 patients having 542 T2-weighted MR slices available at the Cancer Imaging Archive (TCIA) site [52]. The images were collected at Radboud University Medical Center, Netherlands, using Siemens TIM 3 Tesla scanners [53]. This dataset has three different image sizes, , and , having a thickness of 4 mm. The image masks are labeled to 3-class, peripheral zone (PZ), Central Gland (CG), and background.
- UKMMC dataset is a prostate 3T collection consists of 11 patients having 229 T2-weighted MR slices that are collected from UKMMC. The images were acquired using a 3 Tesla Siemens TIM MRI scanner with a surface coil. The image dimensions are and , with a thickness of 3 mm. The image masks are labeled to 2-class, whole prostate gland (WG), and background.
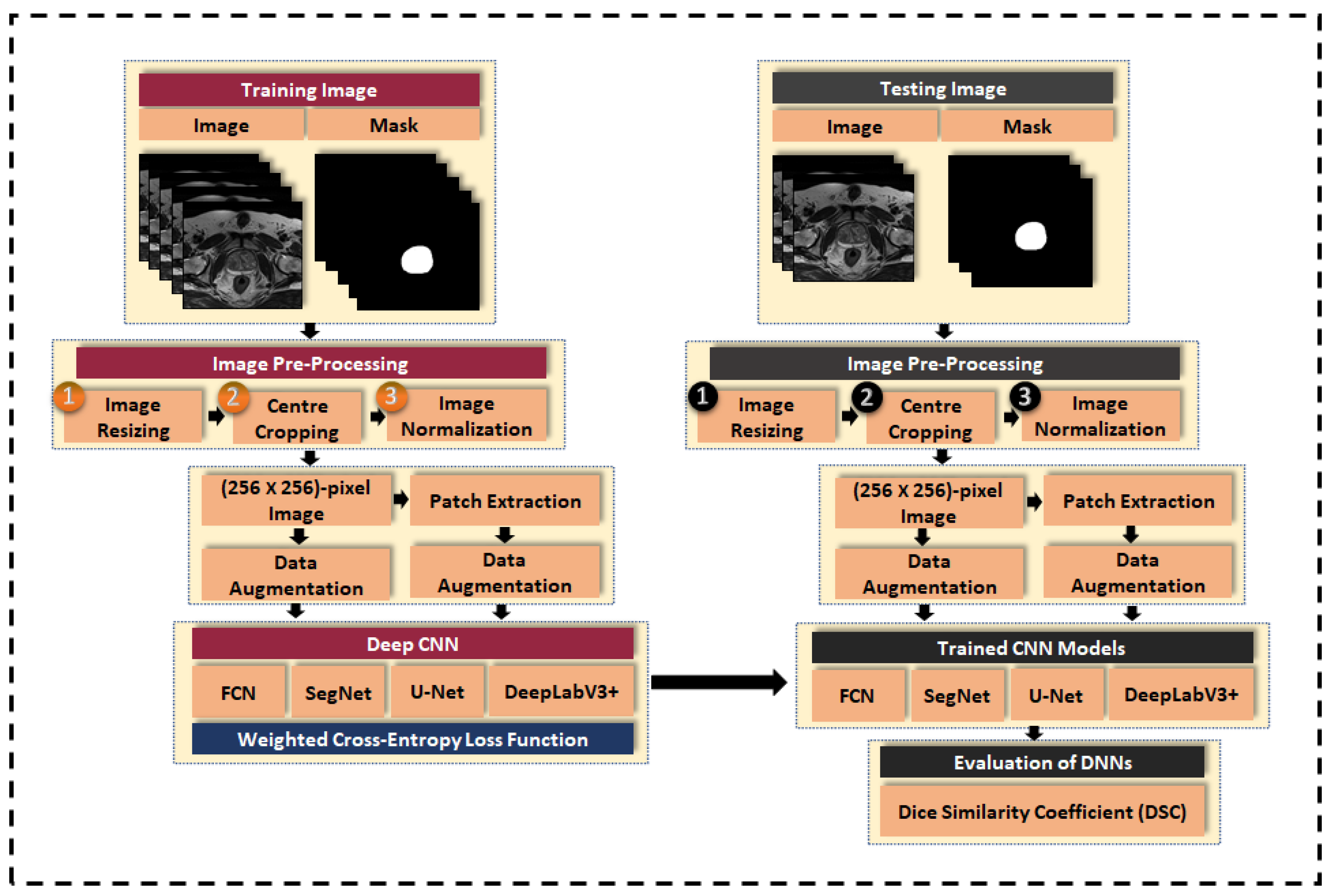
3.3. Image Pre-Processing
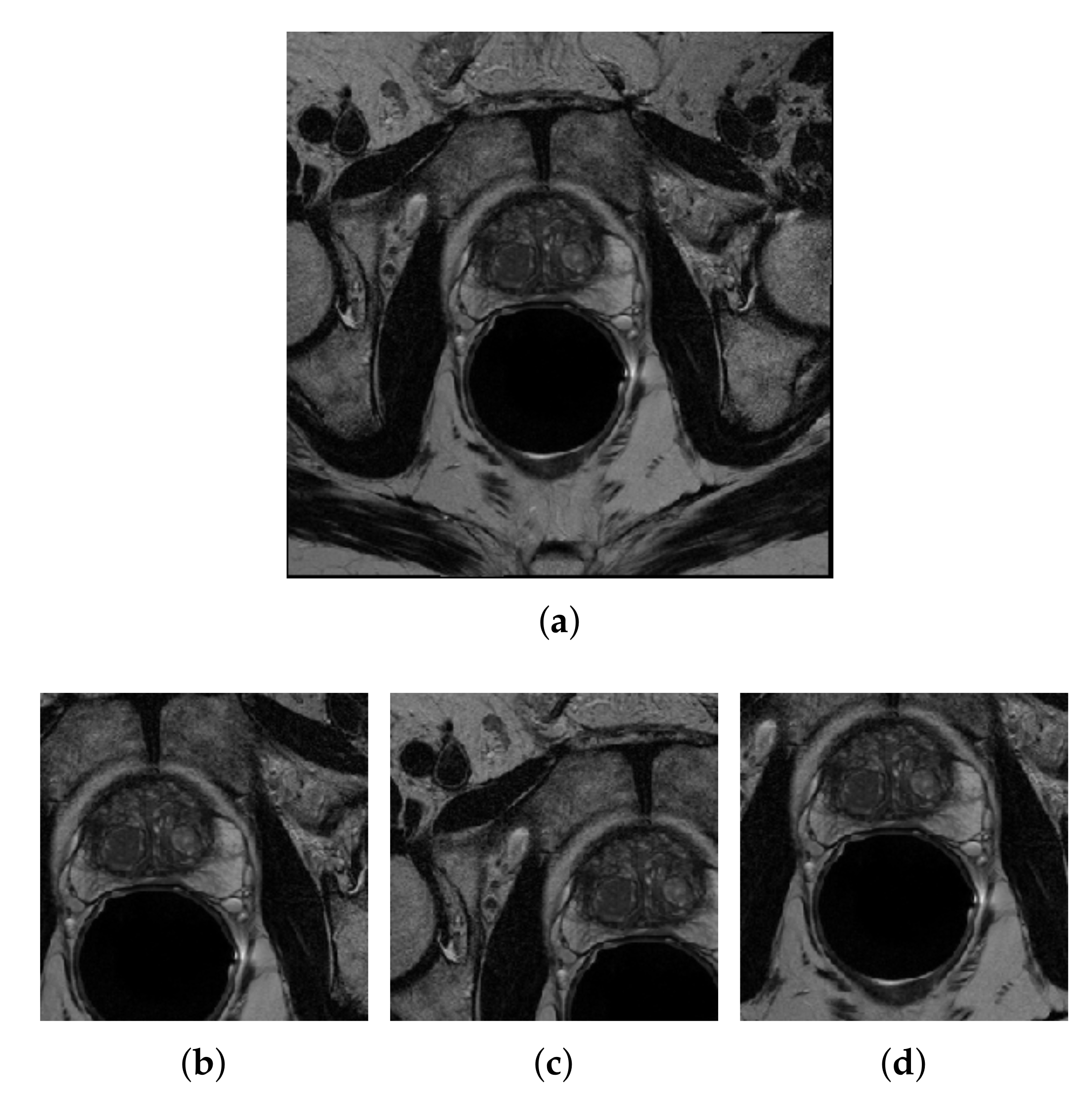
3.3.1. Image Resizing
3.3.2. Center-Cropping
3.3.3. Intensity Normalization
3.4. Data Augmentation
3.5. Patch Extraction
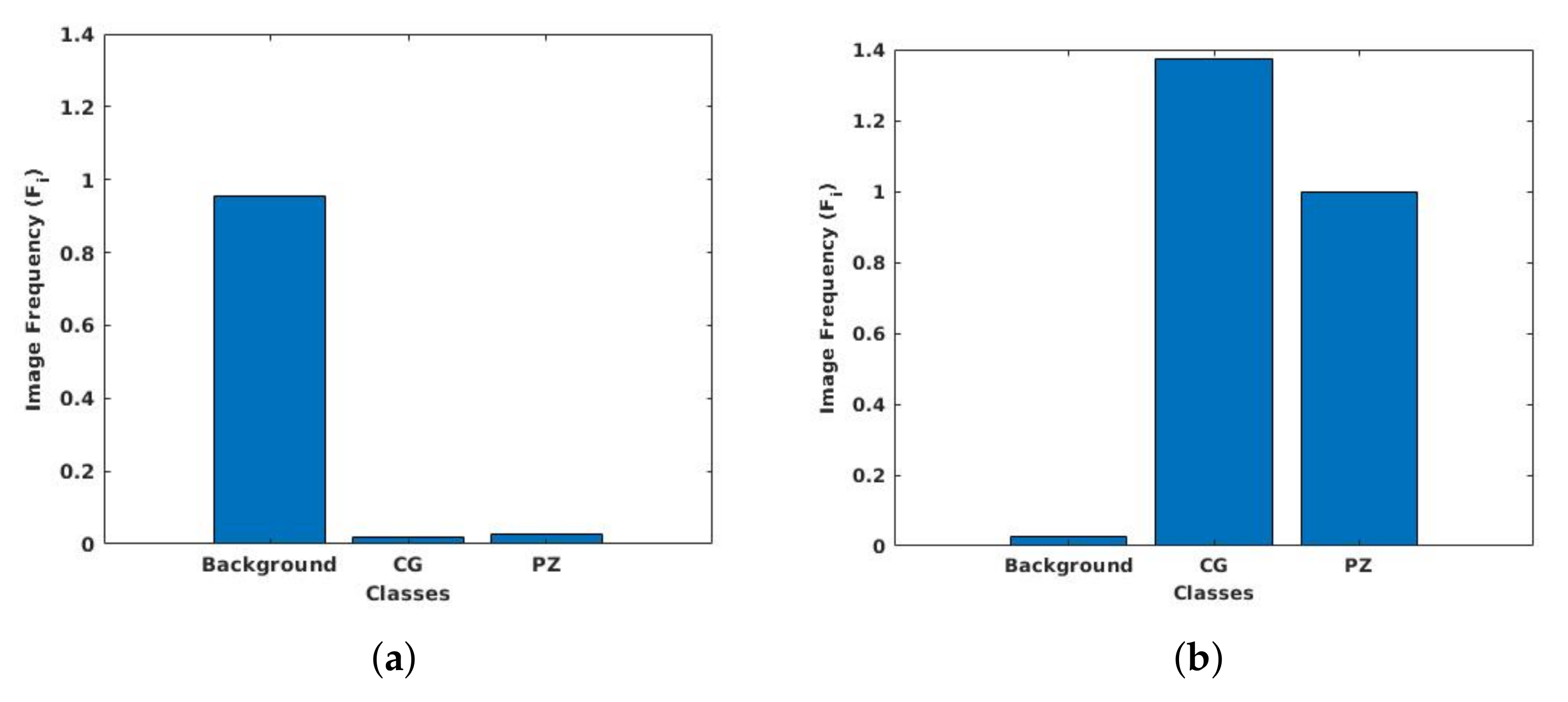
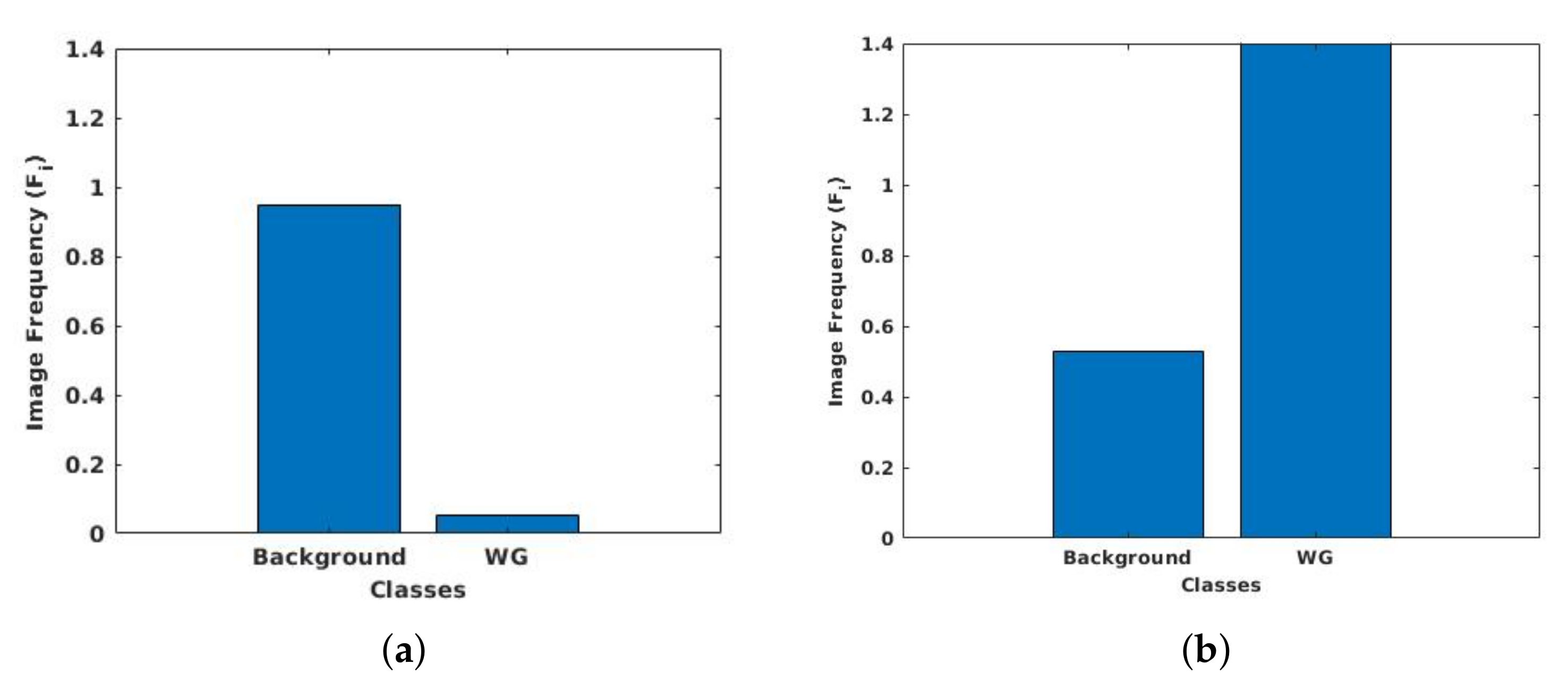
3.6. Class Weight Balancing
3.7. Loss Function
3.8. Evaluation Metric
4. Results and Discussion
4.1. Experimental Set-up
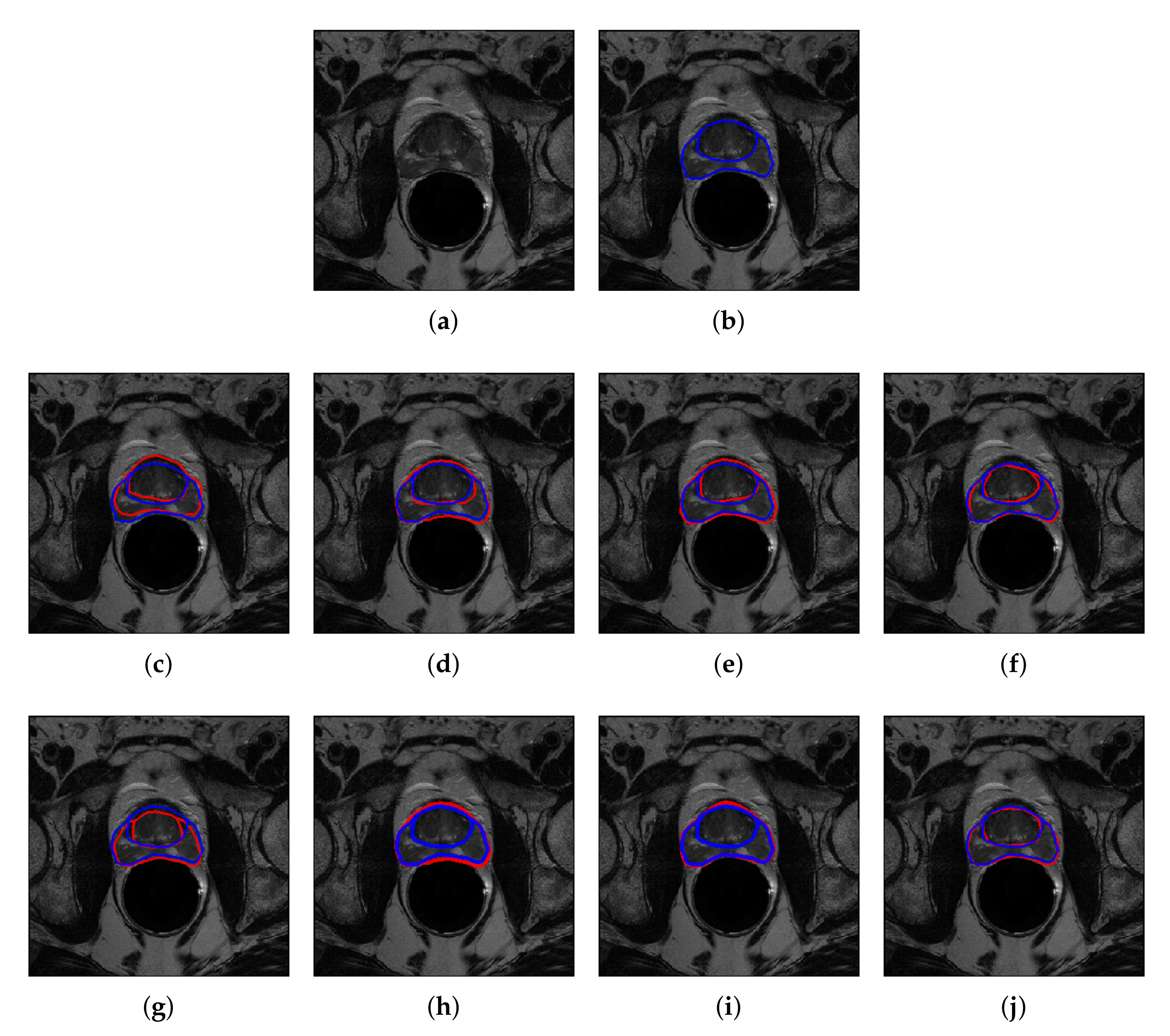
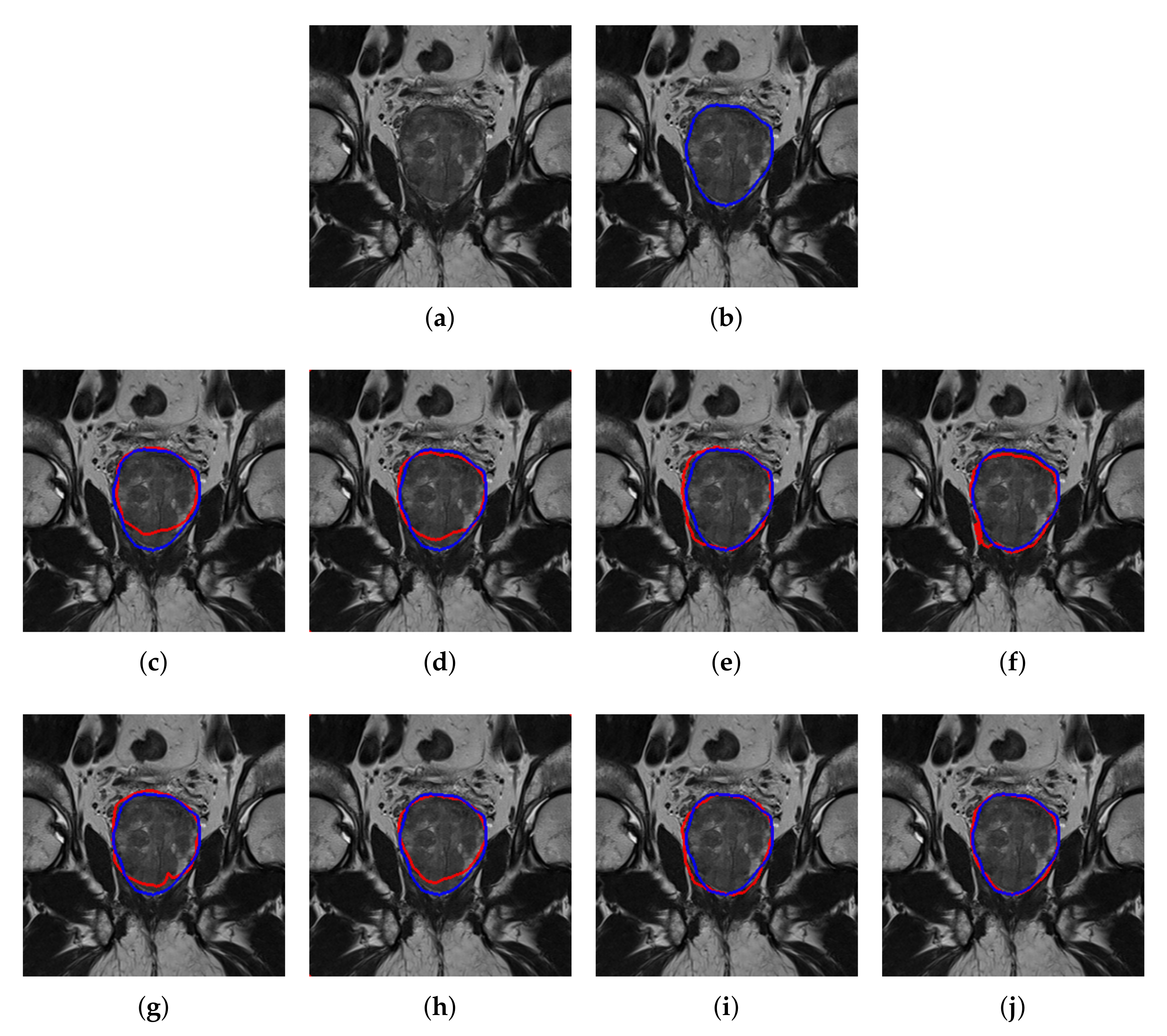
4.2. Quantitative and Qualitative Performance
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953. [Google Scholar] [CrossRef]
- Rawla, P. Epidemiology of Prostate Cancer. World J. Oncol. 2019, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Gandellini, P.; Folini, M.; Zaffaroni, N. Emerging role of microRNAs in prostate cancer: Implications for personalized medicine. Discov. Med. 2010, 9, 212–218. [Google Scholar] [PubMed]
- Loeb, S.; Catalona, W.J. What to do with an abnormal PSA test. The Oncologist 2008, 13, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Ogden, J.M.; O’Keefe, S.J.; Marks, I.S. Development of a method for the purification of human trypsin by single step affinity chromatography suitable for human isotope incorporation studies. Clin. Chim. Acta 1992, 212, 141–147. [Google Scholar] [CrossRef]
- Backer, H. Prostate cancer screening: Exploring the debate. Permanente J. 1999, 3, 330–340. [Google Scholar]
- Turkbey, B.; Choyke, P.L. Multiparametric MRI and prostate cancer diagnosis and risk stratification. Curr. Opin. Urol. 2012, 22, 310. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Barentsz, J.O.; Richenberg, J.; Clements, R.; Choyke, P.; Verma, S.; Villeirs, G.; Rouviere, O.; Logager, V.; Fütterer, J.J. ESUR prostate MR guidelines 2012. Eur. Radiol. 2012, 22, 746–757. [Google Scholar] [CrossRef]
- McNeal, J.E.; Redwine, E.A.; Freiha, F.S.; Stamey, T.A. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am. J. Surg. Pathol. 1988, 12, 897–906. [Google Scholar] [CrossRef]
- Muller, B.G.; Shih, J.H.; Sankineni, S.; Marko, J.; Rais-Bahrami, S.; George, A.K.; de la Rosette, J.J.; Merino, M.J.; Wood, B.J.; Pinto, P.; et al. Prostate cancer: Interobserver agreement and accuracy with the revised prostate imaging reporting and data system at multiparametric MR imaging. Radiology 2015, 277, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Xia, Y.; Song, Y.; Cai, W.; Fulham, M.; Feng, D.D. Atlas registration and ensemble deep convolutional neural network-based prostate segmentation using magnetic resonance imaging. Neurocomputing 2018, 275, 1358–1369. [Google Scholar] [CrossRef]
- Fasihi, M.S.; Mikhael, W.B. Overview of current biomedical image segmentation methods. In Proceedings of the 2016 International Conference on Computational Science and Computational Intelligence (CSCI), Las Vegas, NV, USA, 15–17 December 2016; pp. 803–808. [Google Scholar]
- Vincent, G.; Guillard, G.; Bowes, M. Fully automatic segmentation of the prostate using active appearance models. MICCAI Grand Chall. Prostate MR Image Segmentation 2012, 2012, 2. [Google Scholar]
- Kirschner, M.; Jung, F.; Wesarg, S. Automatic prostate segmentation in MR images with a probabilistic active shape model. In Proceedings of the International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Nice, France, 1 October 2012; pp. 1–8. [Google Scholar]
- Cheng, R.; Roth, H.R.; Lu, L.; Wang, S.; Turkbey, B.; Gandler, W.; McCreedy, E.S.; Agarwal, H.K.; Choyke, P.; Summers, R.M.; et al. Active appearance model and deep learning for more accurate prostate segmentation on MRI. Proc. SPIE 2016, 9784. [Google Scholar] [CrossRef]
- Martin, S.; Troccaz, J.; Daanen, V. Automated segmentation of the prostate in 3D MR images using a probabilistic atlas and a spatially constrained deformable model. Med. Phys. 2010, 37, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Baig, S.; Wong, A.; Haider, M.A.; Khalvati, F. Segmentation of prostate in diffusion MR images via clustering. In Proceedings of the International Conference on Image Analysis and Recognition (ICIAR), Springer, At Montreal, QC, Canada, 2 June 2017; pp. 471–478. [Google Scholar]
- Guo, Y.; Gao, Y.; Shen, D. Deformable MR prostate segmentation via deep feature learning and sparse patch matching. IEEE Trans. Med. Imaging 2015, 35, 1077–1089. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Van Der Heide, U.A.; Lips, I.M.; Van Vulpen, M.; Staring, M.; Pluim, J.P. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Med. Phys. 2008, 35, 1407–1417. [Google Scholar] [CrossRef]
- Langerak, T.R.; van der Heide, U.A.; Kotte, A.N.; Viergever, M.A.; Van Vulpen, M.; Pluim, J.P. Label fusion in atlas-based segmentation using a selective and iterative method for performance level estimation (SIMPLE). IEEE Trans. Med. Imaging 2010, 29, 2000–2008. [Google Scholar] [CrossRef]
- Dowling, J.A.; Fripp, J.; Chandra, S.; Pluim, J.P.W.; Lambert, J.; Parker, J.; Denham, J.; Greer, P.B.; Salvado, O. Fast automatic multi-atlas segmentation of the prostate from 3D MR images. In Proceedings of the International Workshop on Prostate Cancer Imaging, Toronto, ON, Canada, 22 September 2011; pp. 10–21. [Google Scholar]
- Litjens, G.; Debats, O.; van de Ven, W.; Karssemeijer, N.; Huisman, H. A pattern recognition approach to zonal segmentation of the prostate on MRI. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Nice, France, 1–5 October 2012; pp. 413–420. [Google Scholar]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Tian, Z.; Liu, L.; Zhang, Z.; Fei, B. PSNet: Prostate segmentation on MRI based on a convolutional neural network. J. Med. Imaging 2018, 5, 021208. [Google Scholar] [CrossRef]
- Everingham, M.; Van Gool, L.; Williams, C.K.; Winn, J.; Zisserman, A. The pascal visual object classes (VOC) challenge. Int. J. Comput. Vis. 2010, 88, 303–338. [Google Scholar] [CrossRef]
- Yosinski, J.; Clune, J.; Bengio, Y.; Lipson, H. How transferable are features in deep neural networks? Adv. Neural Inf. Process. Syst. 2014, 3320–3328. [Google Scholar]
- Zeiler, M.D.; Fergus, R. Visualizing and understanding convolutional networks. In Proceedings of the European Conference on Computer Vision, Zurich, Switzerland, 6–12 September 2014; pp. 818–833. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Milletari, F.; Navab, N.; Ahmadi, S.A. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar]
- Yu, L.; Yang, X.; Chen, H.; Qin, J.; Heng, P.A. Volumetric convnets with mixed residual connections for automated prostate segmentation from 3D MRI images. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Rundo, L.; Han, C.; Nagano, Y.; Zhang, J.; Hataya, R.; Militello, C.; Tangherloni, A.; Nobile, M.S.; Ferretti, C.; Besozzi, D.; et al. USE-Net: Incorporating Squeeze-and-Excitation blocks into U-Net for prostate zonal segmentation of multi-institutional MRI datasets. Neurocomputing 2019, 365, 31–43. [Google Scholar] [CrossRef]
- Zhang, W.; Li, R.; Deng, H.; Wang, L.; Lin, W.; Ji, S.; Shen, D. Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 2015, 108, 214–224. [Google Scholar] [CrossRef] [PubMed]
- Tajbakhsh, N.; Shin, J.Y.; Gurudu, S.R.; Hurst, R.T.; Kendall, C.B.; Gotway, M.B.; Liang, J. Convolutional neural networks for medical image analysis: Full training or fine tuning? IEEE Trans. Med. Imaging 2016, 35, 1299–1312. [Google Scholar] [CrossRef]
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251. [Google Scholar] [CrossRef]
- Moeskops, P.; Viergever, M.A.; Mendrik, A.M.; de Vries, L.S.; Benders, M.J.; Išgum, I. Automatic segmentation of MR brain images with a convolutional neural network. IEEE Trans. Med. Imaging 2016, 35, 1252–1261. [Google Scholar] [CrossRef]
- Kooi, T.; Litjens, G.; Van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R.; den Heeten, A.; Karssemeijer, N. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef]
- Milletari, F.; Ahmadi, S.A.; Kroll, C.; Plate, A.; Rozanski, V.; Maiostre, J.; Levin, J.; Dietrich, O.; Ertl-Wagner, B.; Bötzel, K.; et al. Hough-CNN: Deep learning for segmentation of deep brain regions in MRI and ultrasound. Comput. Vis. Image Underst. 2017, 164, 92–102. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Q.; Geng, J.; Ding, M.; Li, J. Efficient Patch-Wise Semantic Segmentation for Large-Scale Remote Sensing Images. Sensors 2018, 18, 3232. [Google Scholar] [CrossRef]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 801–818. [Google Scholar]
- Chun, C.; Ryu, S.K. Road Surface Damage Detection Using Fully Convolutional Neural Networks and Semi-Supervised Learning. Sensors 2019, 19, 5501. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.M.; Kim, J.M. Vision-Based Autonomous Crack Detection of Concrete Structures Using a Fully Convolutional Encoder Decoder Network. Sensors 2019, 19, 4251. [Google Scholar] [CrossRef] [PubMed]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 2012, 1097–1105. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Zhao, X.; Yuan, Y.; Song, M.; Ding, Y.; Lin, F.; Liang, D.; Zhang, D. Use of Unmanned Aerial Vehicle Imagery and Deep Learning UNet to Extract Rice Lodging. Sensors 2019, 19, 3859. [Google Scholar] [CrossRef]
- Zhu, Y.; Luo, K.; Ma, C.; Liu, Q.; Jin, B. Superpixel Segmentation Based Synthetic Classifications with Clear Boundary Information for a Legged Robot. Sensors 2018, 18, 2808. [Google Scholar] [CrossRef]
- Yao, X.; Yang, H.; Wu, Y.; Wu, P.; Wang, B.; Zhou, X.; Wang, S. Land Use Classification of the Deep Convolutional Neural Network Method Reducing the Loss of Spatial Features. Sensors 2019, 19, 2792. [Google Scholar] [CrossRef]
- Lobo Torres, D.; Queiroz Feitosa, R.; Nigri Happ, P.; Elena Cu La Rosa, L.; Marcato Junior, J.; Martins, J.; Ol Bressan, P.; Gonalves, W.N.; Liesenberg, V. Applying Fully Convolutional Architectures for Semantic Segmentation of a Single Tree Species in Urban Environment on High Resolution UAV Optical Imagery. Sensors 2020, 20, 563. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Prior, F.; Smith, K.; Sharma, A.; Kirby, J.; Tarbox, L.; Clark, K.; Bennett, W.; Nolan, T.; Freymann, J. The public cancer radiology imaging collections of The Cancer Imaging Archive. Sci. Data 2017, 4, 170124. [Google Scholar] [CrossRef]
- Bloch, N.; Madabhushi, A.; Huisman, H.; Freymann, J.; Kirby, J.; Grauer, M.; Enquobahrie, A.; Jaffe, C.; Clarke, L.; Farahani, K. NCI-ISBI 2013 challenge: Automated segmentation of prostate structures. Cancer Imaging Arch. 2015, 370. [Google Scholar]
- Bovik, A.C. The Essential Guide to Image Processing; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Feng, X.; Qing, K.; Tustison, N.J.; Meyer, C.H.; Chen, Q. Deep convolutional neural network for segmentation of thoracic organs-at-risk using cropped 3D images. Med. Phys. 2019, 46, 2169–2180. [Google Scholar] [CrossRef] [PubMed]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. arXiv 2014, arXiv:1409.4842. [Google Scholar]
- Zhou, X.Y.; Yang, G.Z. Normalization in training U-Net for 2-D biomedical semantic segmentation. IEEE Robot. Autom. Lett. 2019, 4, 1792–1799. [Google Scholar] [CrossRef]
- Zhu, Q.; Du, B.; Turkbey, B.; Choyke, P.L.; Yan, P. Deeply-supervised CNN for prostate segmentation. In Proceedings of the 2017 International Joint Conference on Neural Networks (IJCNN), San Francisco, CA, USA, 4–9 February 2017; pp. 178–184. [Google Scholar]
- Sekou, T.B.; Hidane, M.; Olivier, J.; Cardot, H. From patch to image segmentation using fully convolutional networks-application to retinal images. arXiv 2019, arXiv:1904.03892. [Google Scholar]
- Dhivya, J.J.; Ramaswami, M. A Perusal Analysis on Hybrid Spectrum Handoff Schemes in Cognitive Radio Networks. In Proceedings of the International Conference on Intelligent Systems Design and Applications, Vellore, India, 6–8 December 2018; pp. 312–321. [Google Scholar]
- Christ, P.F.; Elshaer, M.E.A.; Ettlinger, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; Rempfler, M.; Armbruster, M.; Hofmann, F.; D Anastasi, M.; et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; pp. 415–423. [Google Scholar]
- Litjens, G.; Toth, R.; van de Ven, W.; Hoeks, C.; Kerkstra, S.; van Ginneken, B.; Vincent, G.; Guillard, G.; Birbeck, N.; Zhang, J.; et al. Evaluation of prostate segmentation algorithms for MRI: The PROMISE12 challenge. Med. Image Anal. 2014, 18, 359–373. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Gandhi, T.; Panigrahi, B.K.; Bhatia, M.; Anand, S. Expert model for detection of epileptic activity in EEG signature. Expert Syst. Appl. 2010, 37, 3513–3520. [Google Scholar] [CrossRef]
- Kohavi, R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In Proceedings of the 14th International Joint Conference on Artificial Intelligence (IJCAI), San Francisco, CA, USA; 1995; pp. 1137–1145. [Google Scholar]
| Without Patch | Patch-Wise | |||
|---|---|---|---|---|
| PZ | CG | PZ | CG | |
| FCN | 69.2±5.8 | 85.1±4.8 | 72.7±5.1 | 88.6±2.9 |
| SegNet | 75.3±4.8 | 88.9±3.8 | 76.0±3.9 | 90.8±1.2 |
| U-Net | 76.3±3.5 | 89.8±2.6 | 76.8±3.3 | 91.6±1.4 |
| DeepLabV3+ | 76.4±3.3 | 90.7±1.2 | 78.9±1.9 | 92.8±0.7 |
| L. Rundo [32] | - | - | 76.0±4.1 | 91.5±3.2 |
| Without Patch | Patch-Wise | |
|---|---|---|
| FCN | 76.7±6.1 | 86.6±4.8 |
| SegNet | 79.8±5.7 | 84.3±4.2 |
| U-Net | 85.0±3.6 | 88.4±3.7 |
| DeepLabV3+ | 83.2±4.7 | 91.9±2.0 |
| NCI-ISBI 2013 | UKMMC | |
|---|---|---|
| DeepLabV3+ vs. U-Net | 0.025 | 0.041 |
| DeepLabv3+ vs. SegNet | 0.022 | 0.046 |
| DeepLabV3+ vs. FCN | 0.016 | 0.035 |
| Work | Images | Dataset | Network | DSC (WG) or (PZ, CG) |
|---|---|---|---|---|
| Klein et al. 2008 [20] | 50 volumes | 3T MRI | Atlas matching using localized information | 84.4 |
| Guo et al. 2015 [19] | 66 images | University of Chicago Hospital | Deformable model using SSAE | 87.1 |
| Litjen et al. 2012 [23] | 48 images | 3T MRI | Multi atlas-based segmentation | 74, 89 |
| Milletari et al. 2016 [30] | 80 volumes | PROMISE12 | V-Net | 86.9 |
| Yu et al. 2015 [31] | 80 volumes | PROMISE12 | ConvNet with residual connections | 89.4 |
| Rundo et al. 2019 [32] | 40 images | NCI-ISBI 2013 | U-Net with SE block | 76.0±4.1, 91.5±3.2 |
| Our Work | 40 images | NCI-ISBI 2013 | Patch-wise DeepLabV3+ | 78.9±1.9, 92.8±0.7 |
| Our Work | 11 images | UKMMC | Patch-wise DeepLabV3+ | 91.9±2.0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, Z.; Yahya, N.; Alsaih, K.; Ali, S.S.A.; Meriaudeau, F. Evaluation of Deep Neural Networks for Semantic Segmentation of Prostate in T2W MRI. Sensors 2020, 20, 3183. https://doi.org/10.3390/s20113183
Khan Z, Yahya N, Alsaih K, Ali SSA, Meriaudeau F. Evaluation of Deep Neural Networks for Semantic Segmentation of Prostate in T2W MRI. Sensors. 2020; 20(11):3183. https://doi.org/10.3390/s20113183
Chicago/Turabian StyleKhan, Zia, Norashikin Yahya, Khaled Alsaih, Syed Saad Azhar Ali, and Fabrice Meriaudeau. 2020. "Evaluation of Deep Neural Networks for Semantic Segmentation of Prostate in T2W MRI" Sensors 20, no. 11: 3183. https://doi.org/10.3390/s20113183
APA StyleKhan, Z., Yahya, N., Alsaih, K., Ali, S. S. A., & Meriaudeau, F. (2020). Evaluation of Deep Neural Networks for Semantic Segmentation of Prostate in T2W MRI. Sensors, 20(11), 3183. https://doi.org/10.3390/s20113183







