Exploring the Influence of Soil Salinity on Microbiota Dynamics in Vitis vinifera cv. “Glera”: Insights into the Rhizosphere, Carposphere, and Yield Outcomes
Abstract
1. Introduction
2. Materials and Methods
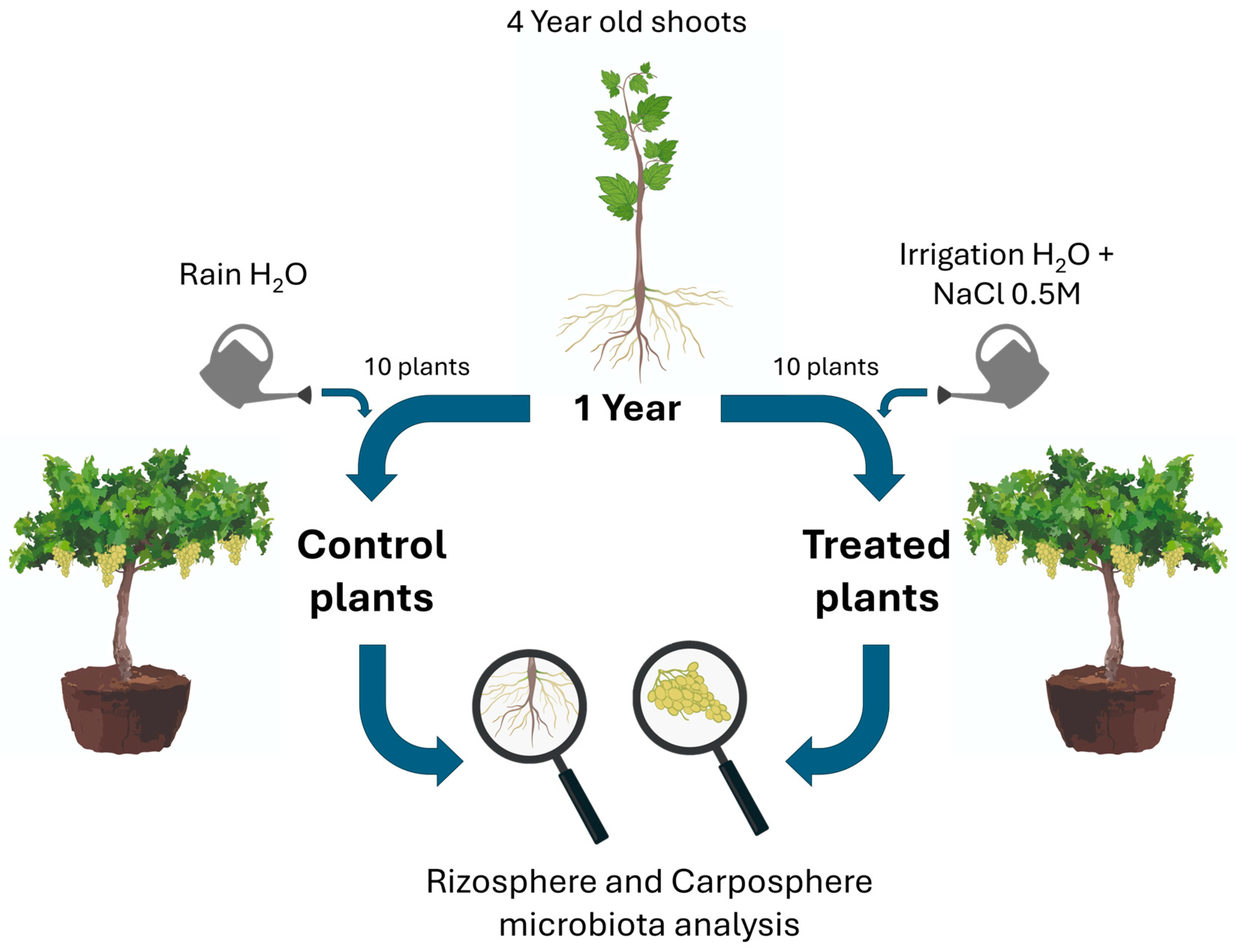
2.1. Experimental Design
2.2. Field Sampling, DNA Extraction, and Amplification
2.3. Bioinformatic and Statistical Analysis
2.4. Yield, Must Chemical Composition, and Radical System Study
3. Results
3.1. Soil and Plant Growth Differences
3.2. Sequencing Results
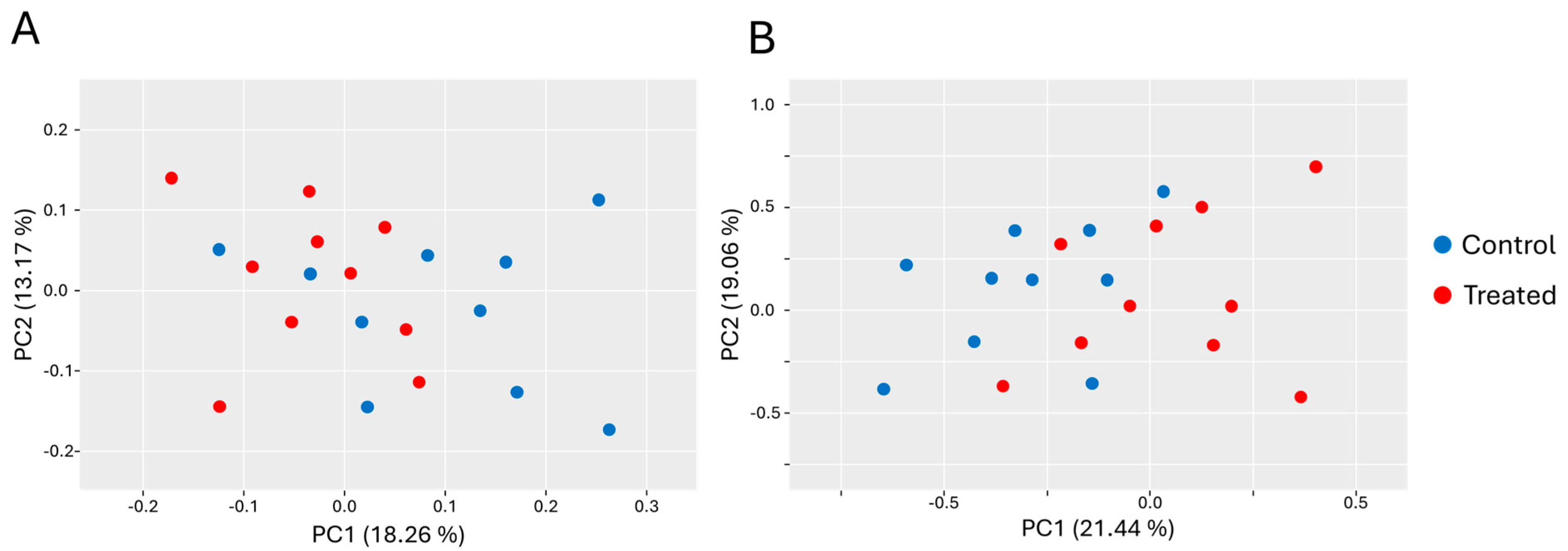
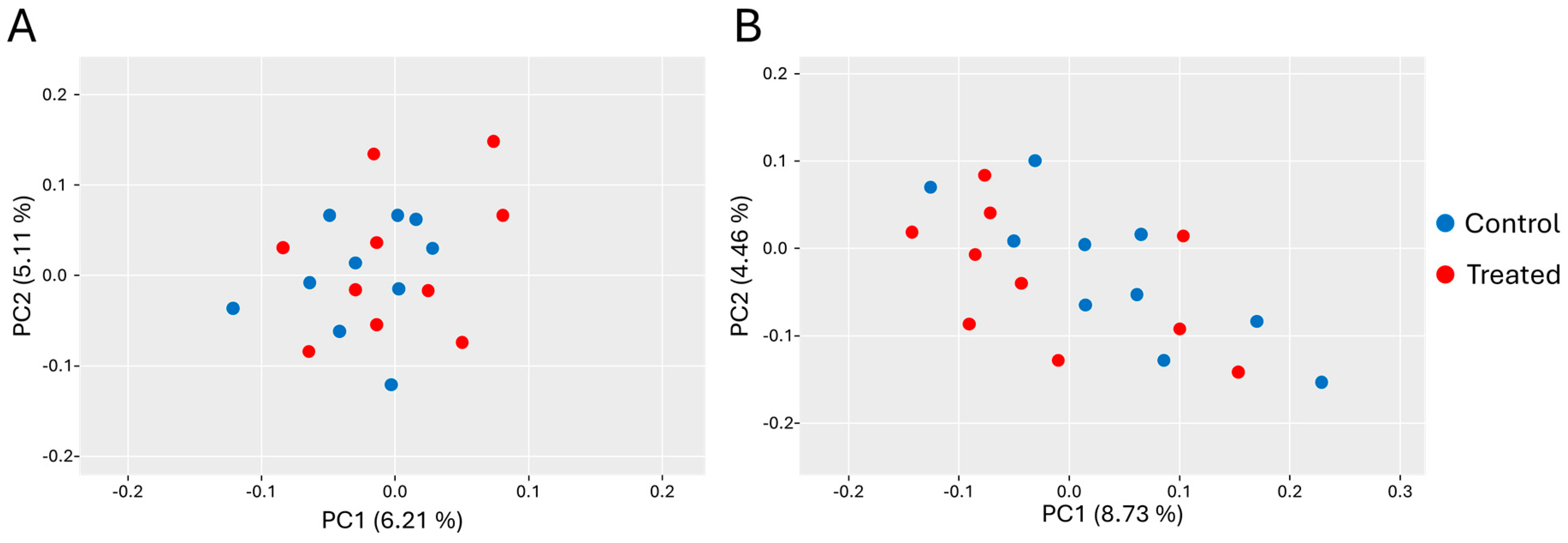
3.3. Alpha and Beta Diversity
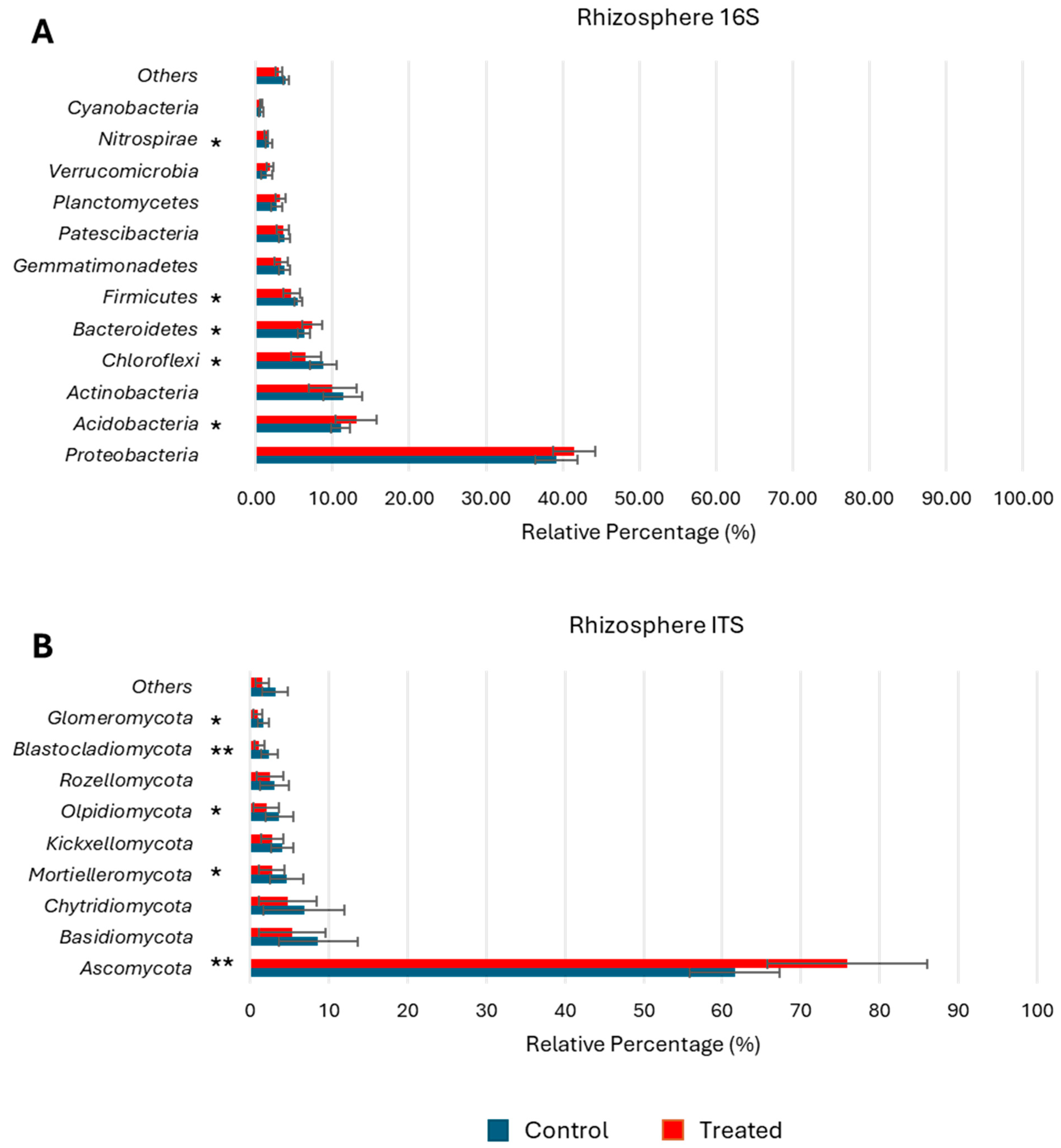
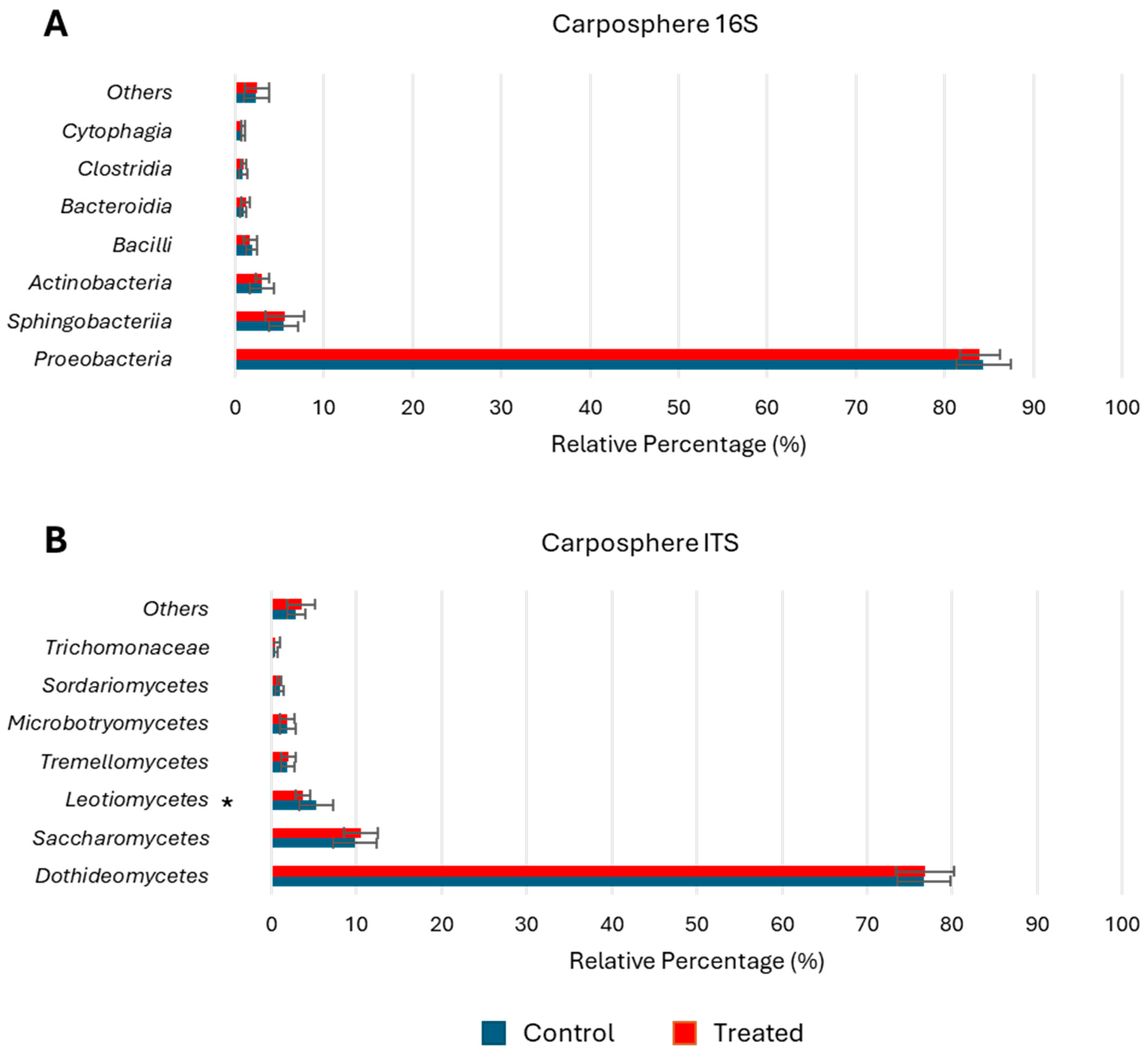
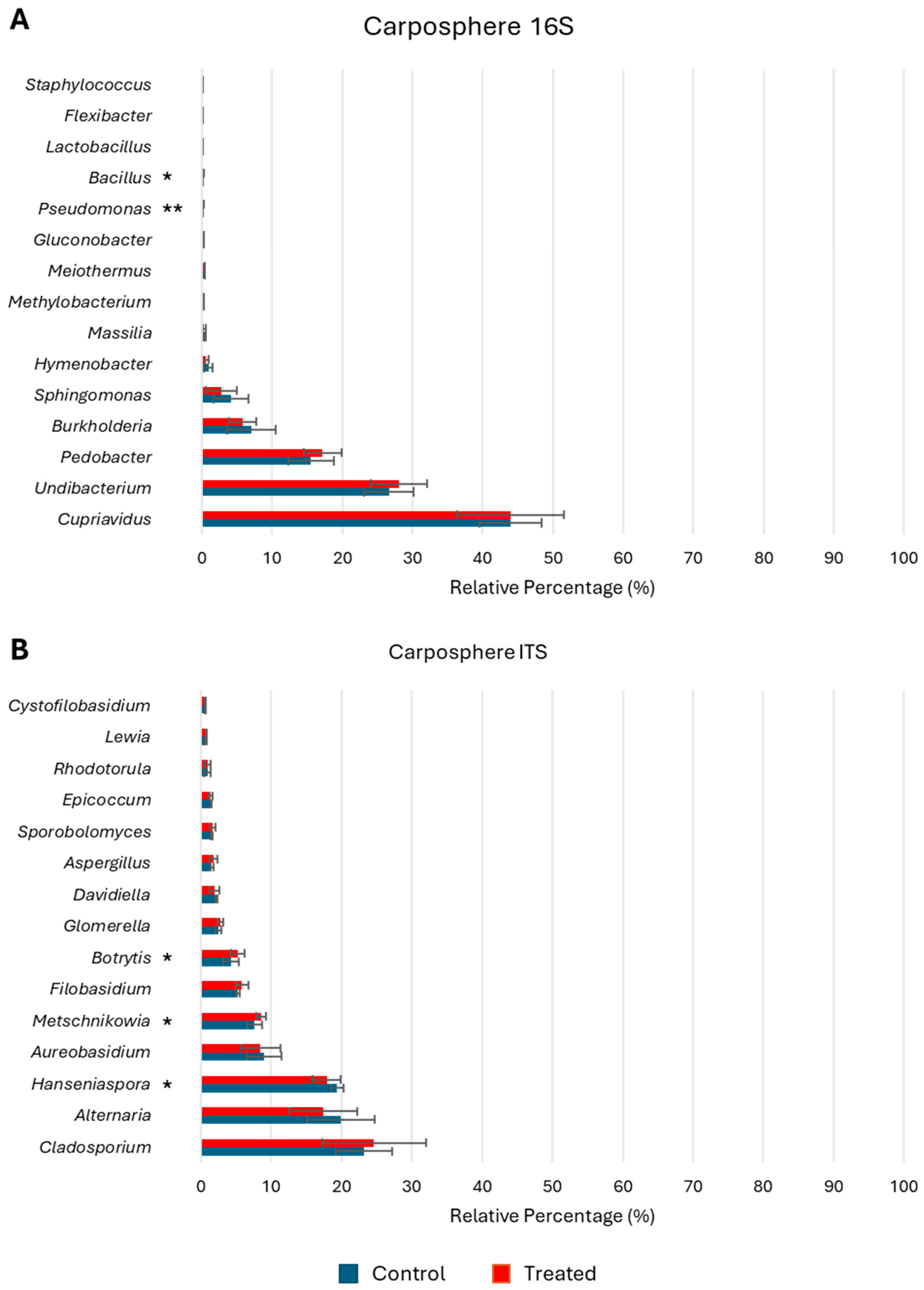
3.4. Microbial Composition
4. Discussion
5. Conclusions and Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Corwin, D.L. Climate change impacts on soil salinity in agricultural areas. Eur. J. Soil Sci. 2021, 72, 842–862. [Google Scholar] [CrossRef]
- Ullah, A.; Bano, A.; Khan, N. Climate Change and Salinity Effects on Crops and Chemical Communication Between Plants and Plant Growth-Promoting Microorganisms Under Stress. Front. Sustain. Food Syst. 2021, 5, 618092. [Google Scholar] [CrossRef]
- Shani, U.; Ben-Gal, A. Long-term response of grapevines to salinity: Osmotic effects and ion toxicity. Am. J. Enol. Vitic. 2005, 56, 148–154. [Google Scholar] [CrossRef]
- Al-Taey, D.K.A.; Al-Ameer, A. A Effect of Salinity on the Growth and Yield of Grapes: A review. IOP Conf. Ser. Earth Environ. Sci. 2023, 1262, 042038. [Google Scholar] [CrossRef]
- Elnashar, W.; Elyamany, A. Managing Risks of Climate Change on Irrigation Water in Arid Regions. Water Resour. Manag. 2023, 37, 2429–2446. [Google Scholar] [CrossRef]
- Lee, J.L.; Huang, W.C. Impact of climate change on the irrigation water requirement in Northern Taiwan. Water 2014, 6, 3339–3361. [Google Scholar] [CrossRef]
- Incoom, A.B.M.; Adjei, K.A.; Odai, S.N.; Akpoti, K.; Siabi, E.K. Impacts of climate change on crop and irrigation water requirement in the Savannah regions of Ghana. J. Water Clim. Chang. 2022, 13, 3338–3356. [Google Scholar] [CrossRef]
- Austin, J.; Zhang, L.; Jones, R.N.; Durack, P.; Dawes, W.; Hairsine, P. Climate change impact on water and salt balances: An assessment of the impact of climate change on catchment salt and water balances in the Murray-Darling Basin, Australia. Clim. Chang. 2010, 100, 607–631. [Google Scholar] [CrossRef]
- Colombani, N.; Osti, A.; Volta, G.; Mastrocicco, M. Impact of Climate Change on Salinization of Coastal Water Resources. Water Resour. Manag. 2016, 30, 2483–2496. [Google Scholar] [CrossRef]
- Nielsen, D.L.; Brock, M.A. Modified water regime and salinity as a consequence of climate change: Prospects for wetlands of Southern Australia. Clim. Chang. 2009, 95, 523–533. [Google Scholar] [CrossRef]
- Zarraonaindia, I.; Owens, S.M.; Weisenhorn, P.; West, K.; Hampton-Marcell, J.; Lax, S.; Bokulich, N.A.; Mills, D.A.; Martin, G.; Taghavi, S.; et al. The soil microbiome influences grapevine-associated microbiota. mBio 2015, 6, e02527-14. [Google Scholar] [CrossRef] [PubMed]
- Griggs, R.G.; Steenwerth, K.L.; Mills, D.A.; Cantu, D.; Bokulich, N.A. Sources and Assembly of Microbial Communities in Vineyards as a Functional Component of Winegrowing. Front. Microbiol. 2021, 12, 673810. [Google Scholar] [CrossRef] [PubMed]
- Xia, F.; Hao, H.; Qi, Y.; Bai, H.; Li, H.; Shi, Z.; Shi, L. Effect of Salt Stress on Microbiome Structure and Diversity in Chamomile (Matricaria chamomilla L.) Rhizosphere Soil. Agronomy 2023, 13, 1444. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mirza, B.S.; Mehnaz, S.; Mirza, M.S.; Mclean, J.; Malik, K.A. Impact of soil salinity on the microbial structure of halophyte rhizosphere microbiome. World J. Microbiol. Biotechnol. 2018, 34, 136. [Google Scholar] [CrossRef]
- Miransari, M. Arbuscular Mycorrhizal Fungi and Soil Salinity. Mycorrhizal Mediation of Soil: Fertility, Structure, and Carbon Storage; Elsevier Inc.: Amsterdam, The Netherlands, 2017. [Google Scholar] [CrossRef]
- Wang, B.; Wang, X.; Wang, Z.; Zhu, K.; Wu, W. Comparative metagenomic analysis reveals rhizosphere microbial community composition and functions help protect grapevines against salt stress. Front. Microbiol. 2023, 14, 1102547. [Google Scholar] [CrossRef]
- Bettenfeld, P.; Cadena i Canals, J.; Jacquens, L.; Fernandez, O.; Fontaine, F.; van Schaik, E.; Courty, P.-E.; Trouvelot, S. The microbiota of the grapevine holobiont: A key component of plant health. J. Adv. Res. 2022, 40, 1–15. [Google Scholar] [CrossRef]
- Garde-Cerdán, T.; Ancín-Azpilicueta, C. Contribution of wild yeasts to the formation of volatile compounds in inoculated wine fermentations. Eur. Food Res. Technol. 2006, 222, 15–25. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the Vineyard to the Winery: How Microbial Ecology Drives Regional Distinctiveness of Wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef] [PubMed]
- Mian, G.; Cipriani, G.; Firrao, G.; Martini, M.; Ermacora, P. Genetic diversity of Actinidia spp. shapes the oomycete pattern associated with Kiwifruit Vine Decline Syndrome (KVDS). Sci. Rep. 2023, 13, 16449. [Google Scholar] [CrossRef]
- Colautti, A.; Golinelli, F.; Iacumin, L.; Tomasi, D.; Cantone, P.; Mian, G. Triacontanol (long-chain alcohol) positively enhances the microbial ecology of berry peel in Vitis vinifera cv. ‘Glera’ yet promotes the must total soluble sugars content. OENO One 2023, 57, 477–488. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2′s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, 590–596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef]
- Kandlikar, G.S.; Gold, Z.J.; Cowen, M.C.; Meyer, R.S.; Freise, A.C.; Kraft, N.J.B.; Moberg-Parker, J.; Sprague, J.; Kushner, D.J.; Curd, E.E. Ranacapa: An R package and shiny web app to explore environmental DNA data with exploratory statistics and interactive visualizations [version 1; referees: 1 approved, 2 approved with reservations]. F1000Research 2018, 7, 1734. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: A bioconductor package for handling and analysis of high-throughput phylogenetic sequence data. Pacific. Symp. Biocomput. 2012, 2011, 235–246. [Google Scholar] [CrossRef]
- Dixon, P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003, 14, 927–930. [Google Scholar] [CrossRef]
- Colautti, A.; Civilini, M.; Contin, M.; Celotti, E.; Iacumin, L. Organic vs. conventional: Impact of cultivation treatments on the soil microbiota in the vineyard. Front. Microbiol. 2023, 14, 1242267. [Google Scholar] [CrossRef]
- Falginella, L.; Gaiotti, F.; Belfiore, N.; Mian, G.; Lovat, L.; Tomasi, D. Effect of early cane pruning on yield components, grape composition, carbohydrates storage and phenology in Vitis vinifera L. cv. Merlot. OENO One 2022, 56, 19–28. [Google Scholar] [CrossRef]
- Mian, G.; Celotti, E.; Falginella, L.; de Oliveria Cantão, F.R.; Belfiore, N. Effect of manure application timing on roots, canopy and must quality in Vitis vinifera “Merlot”: A case study in Italy, North-East. VITIS J. Grapevine Res. 2022, 61, 87–92. [Google Scholar] [CrossRef]
- Mian, G.; Colautti, A.; Belfiore, N.; Marcuzzo, P.; Tomasi, D.; Bell, L.; Celotti, E. Fertigation affects photosynthesis, modulation of secondary metabolism and sensory profiles of Vitis vinifera cv. “Schioppettino” withered grapes and wines. Sci. Hortic. 2024, 328, 112954. [Google Scholar] [CrossRef]
- Mian, G.; Cipriani, G.; Saro, S.; Martini, M.; Ermacora, P. Potential of Different Actinidia Genotypes as Resistant Rootstocks for Preventing Kiwifruit Vine Decline Syndrome. Horticulturae 2022, 8, 627. [Google Scholar] [CrossRef]
- ARPAV. La Carta della Reazione(pH) e della Salinità della Regione Veneto. 2020. Available online: https://www.arpa.veneto.it/temi-ambientali/suolo/file-e-allegati/documenti (accessed on 21 December 2023).
- Tomasi, D.; Marcuzzo, P.; Nardi, T.; Lonardi, A.; Lovat, L.; Flamini, R.; Mian, G. Influence of Soil Chemical Features on Aromatic Profile of, V. vinifera cv. Corvina Grapes and Wines: A Study-Case in Valpolicella Area (Italy) in a Calcareous and Non-Calcareous Soil. Agriculture 2022, 12, 1980. [Google Scholar] [CrossRef]
- Mian, G.; Nassivera, F.; Sillani, S.; Iseppi, L. Grapevine Resistant Cultivars: A Story Review and the Importance on the Related Wine Consumption Inclination. Sustainability 2022, 15, 390. [Google Scholar] [CrossRef]
- Pereyra, G.; Pellegrino, A.; Ferrer, M.; Gaudin, R. How soil and climate variability within a vineyard can affect the heterogeneity of grapevine vigour and production. OENO One 2023, 57, 297–313. [Google Scholar] [CrossRef]
- Chowdhury, N.; Marschner, P.; Burns, R. Response of microbial activity and community structure to decreasing soil osmotic and matric potential. Plant Soil. 2011, 344, 241–254. [Google Scholar] [CrossRef]
- Elmajdoub, B.; Marschner, P. Responses of soil microbial activity and biomass to salinity after repeated additions of plant residues. Pedosphere 2015, 25, 177–185. [Google Scholar] [CrossRef]
- Polonenko, D.R.; Mayfield, C.I.; Dumbroff, E.B. Microbial responses to salt-induced osmotic stress. Plant Soil 1986, 92, 417–425. [Google Scholar] [CrossRef]
- Rath, K.M.; Rousk, J. Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: A review. Soil Biol. Biochem. 2015, 81, 108–123. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, C.; Yang, J.; Yao, R.; Wang, X.; Xie, W.; Ge, A.-H. Salinity-dependent potential soil fungal decomposers under straw amendment. Sci. Total Environ. 2023, 891, 164569. [Google Scholar] [CrossRef] [PubMed]
- Coller, E.; Cestaro, A.; Zanzotti, R.; Bertoldi, D.; Pindo, M.; Larger, S.; Albanese, D.; Mescalchin, E.; Donati, C. Microbiome of vineyard soils is shaped by geography and management. Microbiome 2019, 7, 140. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Druzhinina, I.S.; Pan, X.; Yuan, Z. Microbially Mediated Plant Salt Tolerance and Microbiome-based Solutions for Saline Agriculture. Biotechnol. Adv. 2016, 34, 1245–1259. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Sun, J. Soil Salinity Drives the Distribution Patterns and Ecological Functions of Fungi in Saline-Alkali Land in the Yellow River Delta, China. Front. Microbiol. 2020, 11, 594284. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, D.J.; Martiny, J.B.H. Patterns of fungal diversity and composition along a salinity gradient. ISME J. 2011, 5, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Krishnamoorthy, R.; Kim, K.; Kim, C.; Sa, T. Changes of arbuscular mycorrhizal traits and community structure with respect to soil salinity in a coastal reclamation land. Soil Biol. Biochem. 2014, 72, 1–10. [Google Scholar] [CrossRef]
- Kim, K.; Samaddar, S.; Chatterjee, P.; Krishnamoorthy, R.; Jeon, S.; Sa, T. Structural and functional responses of microbial community with respect to salinity levels in a coastal reclamation land. Appl. Soil Ecol. 2019, 137, 96–105. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Miao, F.; Li, Z.; Tang, W.; Sun, J. Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl. Soil Ecol. 2020, 155, 103671. [Google Scholar] [CrossRef]
- Valentin, D.N.; Voyron, S.; Soteras, F.; Iriarte, H.J.; Giovannini, A.; Lumini, E.; Lugo, M.A. Modeling geographic distribution of arbuscular mycorrhizal fungi from molecular evidence in soils of Argentinean Puna using a maximum entropy approach. PeerJ 2023, 11, e14651. [Google Scholar] [CrossRef]
- Guo, H.N.; Huang, Z.J.; Li, M.Q.; Min, W. Response of soil fungal community structure and diversity to saline water irrigation in alluvial grey desert soils. Appl. Ecol. Environ. Res. 2020, 18, 4969–4985. [Google Scholar] [CrossRef]
- Zhang, L.; Ge, A.H.; Tóth, T.; Yang, F.; Wang, Z.; An, F. Enrichment of keystone fungal taxa after flue gas desulphurization gypsum application drives reclamation of the saline-sodic soil. Land Degrad. Dev. 2023, 34, 2276–2287. [Google Scholar] [CrossRef]
- Zhao, S.; Liu, J.J.; Banerjee, S.; Zhou, N.; Zhao, Z.Y.; Zhang, K.; Hu, M.; Tian, C. Biogeographical distribution of bacterial communities in saline agricultural soil. Geoderma 2020, 361, 114095. [Google Scholar] [CrossRef]
- Banerjee, S.; Kirkby, C.A.; Schmutter, D.; Bissett, A.; Kirkegaard, J.A.; Richardson, A.E. Network analysis reveals functional redundancy and keystone taxa amongst bacterial and fungal communities during organic matter decomposition in an arable soil. Soil Biol. Biochem. 2016, 97, 188–198. [Google Scholar] [CrossRef]
- Hou, Y.; Zeng, W.; Hou, M.; Wang, Z.; Luo, Y.; Lei, G.; Zhou, B.; Huang, J. Responses of the soil microbial community to salinity stress in maize fields. Biology 2021, 10, 1114. [Google Scholar] [CrossRef] [PubMed]
- Hashmi, I.; Bindschedler, S.; Junier, P. Firmicutes. In Beneficial Microbes in Agro-Ecology; Academic Press: Cambridge, MA, USA, 2020. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mehnaz, S.; Mirza, M.S.; Mirza, B.S.; Malik, K.A. Diversity of Bacillus-like bacterial community in the rhizospheric and non-rhizospheric soil of halophytes (Salsola stocksii and Atriplex amnicola), and characterization of osmoregulatory genes in halophilic Bacilli. Can. J. Microbiol. 2018, 64, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for Plant Growth Promotion and Stress Resilience: What Have We Learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef] [PubMed]
- Martin, V.; Jose Valera, M.; Medina, K.; Boido, E.; Carrau, F. Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines—A review. Fermentation 2018, 4, 76. [Google Scholar] [CrossRef]
- Vicente, J.; Ruiz, J.; Belda, I.; Benito-Vázquez, I.; Marquina, D.; Calderón, F.; Santos, A.; Benito, S. The genus metschnikowia in enology. Microorganisms 2020, 8, 1038. [Google Scholar] [CrossRef]
- Lami, M.J.; Adler, C.; Caram-Di Santo, M.C.; Zenoff, A.M.; de Cristóbal, R.E.; Espinosa-Urgel, M.; Vincent, P.A. Pseudomonas stutzeri MJL19, a rhizosphere-colonizing bacterium that promotes plant growth under saline stress. J. Appl. Microbiol. 2020, 129, 1321–1336. [Google Scholar] [CrossRef]
- Elabed, H.; González-Tortuero, E.; Ibacache-Quiroga, C.; Bakhrouf, A.; Johnston, P.; Gaddour, K.; Blázquez, J.; Rodríguez-Rojas, A. Seawater salt-trapped Pseudomonas aeruginosa survives for years and gets primed for salinity tolerance. BMC Microbiol. 2019, 19, 142. [Google Scholar] [CrossRef] [PubMed]
- Niem, J.M.; Billones-Baaijens, R.; Stodart, B.; Savocchia, S. Diversity Profiling of Grapevine Microbial Endosphere and Antagonistic Potential of Endophytic Pseudomonas Against Grapevine Trunk Diseases. Front. Microbiol. 2020, 11, 477. [Google Scholar] [CrossRef] [PubMed]
- Ibarra-Villarreal, A.L.; Gándara-Ledezma, A.; Godoy-Flores, A.D.; Herrera-Sepúlveda, A.; Díaz-Rodríguez, A.M.; Parra-Cota, F.I.; Santos-Villalobos, S.d.L. Salt-tolerant Bacillus species as a promising strategy to mitigate the salinity stress in wheat (Triticum turgidum subsp. durum). J. Arid Environ. 2021, 186, 104399. [Google Scholar] [CrossRef]
- Sawant, I.S.; Wadkar, P.N.; Rajguru, Y.R.; Mhaske, N.H.; Salunkhe, V.P.; Sawant, S.D.; Upadhyay, A. Biocontrol potential of two novel grapevine associated Bacillus strains for management of anthracnose disease caused by Colletotrichum gloeosporioides. Biocontrol Sci. Technol. 2016, 26, 964–979. [Google Scholar] [CrossRef]
- Bae, S.; Fleet, G.H.; Heard, G.M. Occurrence and significance of Bacillus thuringiensis on wine grapes. Int. J. Food Microbiol. 2004, 94, 301–312. [Google Scholar] [CrossRef]

| Mean | SD | p-Value | |||
|---|---|---|---|---|---|
| Control | Treated | Control | Treated | ||
| Sugar (°Brix) | 23.80 | 21.83 | 1.24 | 1.08 | ** |
| Total acidity (g·L−1) | 5.78 | 6.89 | 0.77 | 0.98 | * |
| pH | 3.57 | 3.28 | 0.23 | 0.14 | * |
| Malic acid (g·L−1) | 2.12 | 2.47 | 0.35 | 0.34 | * |
| Shoots/vine | 29.00 | 25.90 | 3.13 | 3.84 | |
| Leaves number/vine | 610.00 | 588.80 | 27.08 | 30.95 | |
| Total leaf area/vine (m2) | 31.39 | 29.90 | 6.27 | 5.14 | |
| Leaf size (cm2) | 1.92 | 1.76 | 0.42 | 0.32 | |
| Yield/vine (kg) | 3.70 | 3.10 | 0.56 | 0.94 | |
| Clusters/vine | 11.90 | 9.00 | 2.96 | 4.03 | |
| Cluster weight (g) | 308.00 | 341.00 | 11.00 | 20.00 | *** |
| Root density (number of thick roots/10 cm2 profile wall) | 13.20 | 10.09 | 2.00 | 3.00 | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Colautti, A.; Mian, G.; Tomasi, D.; Bell, L.; Marcuzzo, P. Exploring the Influence of Soil Salinity on Microbiota Dynamics in Vitis vinifera cv. “Glera”: Insights into the Rhizosphere, Carposphere, and Yield Outcomes. Diversity 2024, 16, 247. https://doi.org/10.3390/d16040247
Colautti A, Mian G, Tomasi D, Bell L, Marcuzzo P. Exploring the Influence of Soil Salinity on Microbiota Dynamics in Vitis vinifera cv. “Glera”: Insights into the Rhizosphere, Carposphere, and Yield Outcomes. Diversity. 2024; 16(4):247. https://doi.org/10.3390/d16040247
Chicago/Turabian StyleColautti, Andrea, Giovanni Mian, Diego Tomasi, Luke Bell, and Patrick Marcuzzo. 2024. "Exploring the Influence of Soil Salinity on Microbiota Dynamics in Vitis vinifera cv. “Glera”: Insights into the Rhizosphere, Carposphere, and Yield Outcomes" Diversity 16, no. 4: 247. https://doi.org/10.3390/d16040247
APA StyleColautti, A., Mian, G., Tomasi, D., Bell, L., & Marcuzzo, P. (2024). Exploring the Influence of Soil Salinity on Microbiota Dynamics in Vitis vinifera cv. “Glera”: Insights into the Rhizosphere, Carposphere, and Yield Outcomes. Diversity, 16(4), 247. https://doi.org/10.3390/d16040247







