Abstract
Convolvulus arvensis is an herbaceous dicotyledonous plant in the Convolvulaceae family that is native to Europe and Asia. It is a perennial soboliferous plant and is one of the most harmful weeds. This weed is successful in many types of climates, including temperate, tropical, and Mediterranean climates, but it is most troublesome for agriculture throughout the temperate zone. In this study, several pathogenic isolates were collected from this host. The internal transcribed spacer (ITS) and large subunit (28S) or ribosomal DNA, partial DNA-directed RNA polymerase II subunit (rpb2), and β-tubulin (tub2) genes were amplified and sequenced for all the isolates studied. Further, both a multilocus phylogenetic analysis of DNA sequences and an analysis of morphological features were implemented. Based on the results obtained, all the studied isolates were found to be distinct from any described species in the genus Ascochyta and are, therefore, described here as a new species Ascochyta erotica sp. nov. The pathogenicity of A. erotica sp. nov. was also tested and confirmed on leaf segments of C. arvensis.
1. Introduction
The genus Ascochyta Lib. (Dothideomycetes, Pleosporomycetidae, Pleosporales, Didymellaceae) was established in 1830, with Ascochyta pisi Lib. as the type species [1]. It was initially assumed that the most informative taxonomic character of species in this genus was two-celled conidia in thin-walled pycnidia developing in leaves and branches [2]. In 2004, as a result of a comprehensive analysis of the morphological features of pycnidial fungi [3], the Ascochyta species was found to be associated with Phoma Sacc. sensu lato. Subsequently, it was stated that classification systems based on morphological characters are very artificial and do not reflect evolutionary relationships.
In 2015, Chen and Cai [4] emended the protologue for Ascochyta, which had previously been classified in the Didymellaceae family, that included most Phoma-like species [5]. Thereat, several Didymella Sacc. and Phoma, Septoria Sacc. species were transferred to the genus Ascochyta, and on the contrary, other Ascochyta species were reclassified into Boeremia Aveskamp, Gruyter & Verkley, Calophoma Qian Chen & L. Cai, Didymella, Neoascochyta Qian Chen & L. Cai, Remotididymella Valenz.-Lopez, Crous, Cano, Guarro & Stchigel, Sclerotiophoma L.W. Hou, L. Cai & Crous, Stagonosporopsis Died., Vacuiphoma Valenz.-Lopez, J.F. Cano, Crous, Guarro & Stchigel [4,6,7]. According to the actual taxonomy of the Didymellaceae family, the identification of its members, especially Ascochyta species, should be based on the consolidated species concept (CSC), which relies on the integration of phylogenetic, morphological, and biological characters. Phylogenetic data should be obtained by genealogical concordant phylogenetic species recognition (GCPSR) using a multilocus phylogenetic analysis to identify fungal species [4,6,7,8,9,10].
From the mid-19th to the mid-20th century, over 1100 Ascochyta species were described [11]. Over the past five years, only nine new Ascochyta species have been described according to the CSC. To date, the Mycobank has listed 1518 items (=species epithets) for Ascochyta, and among them, 225 are illegitimate or invalid or an orthographic variant. Of these, the molecular multilocus phylogeny was reconstructed for only 24 Ascochyta species.
Ascochyta species could be associated with plants from more than 150 families [11]. In most cases, the host range of species belonging to this genus is very limited and is found mainly on the Apiaceae, Asteraceae, Brassicaceae, Fabaceae, Lamiaceae, Ranunculaceae, Rosaceae, Poaceae, etc., and several species are associated with one specific host but may also be revealed on other related species of the same genus or family. [11,12].
Field bindweed, Convolvulus arvensis L., is native to Europe and Asia. This is a perennial soboliferous plant and is one of the most harmful weeds. This weed is successful in many types of climates, including temperate, tropical, and Mediterranean climates, but it is most destructive to agriculture throughout the temperate zone [13].
Among the Ascochyta species associated with Convolvulaceae, there are Ascochyta convolvuli Lib. (current name Sphaeria lichenoides var. convolvulicola DC.) (Russia, North Ossetia-Alania, former USSR, Uzbekistan, and France), Ascochyta calystegiae Sacc. (Russia, Voronezh and Leningrad regions, Europe, Bulgaria, Czech Republic, Germany, Estonia, France, and Slovakia) [11,14], and Ascochyta kleinii Bubák, (Armenia, Bulgaria, China, Hungary, and Poland) [11,14]
Across the other Phoma-like fungi from Convolvulus spp., the following have been described: Phoma convolvuli Wehm. in India [15] and Pakistan [14], Phoma capsularum Cooke & Harkn. in Uzbekistan [14], Phoma proboscis Heiny in USA [16], Didymella macrostoma (Mont.) Qian Chen & L. Cai in Turkey [14], Paraphoma melnikii Gomzhina & Gasich in Russia and Ukraine, and Paraphoma convolvuli (Dearn. & House) Gomzhina & Gasich in Kazakhstan [17].
The aim of this study was to identify Ascochyta sp. isolated from C. arvensis with leaf spots collected in the Kyrgyz Republic according to the CSC by their phylogenetic, micromorphological, and cultural features. An additional goal was to validate the pathogenicity of the identified isolates.
2. Materials and Methods
2.1. Plant Material
One specimen of C. arvensis included leaves with necrotic lesions that were collected in the Tchui region (Kyrgyz Republic) in August 2002. For fungal isolation, the pycnidia from these leaves were transferred with a needle in a drop of sterile water into Petri dishes, and after the release of spores from the pycnidia, a potato–sucrose agar (PSA) [18] was added. The Petri dishes were incubated at 24 °C in the dark and were analyzed after 7–10 days of cultivation. Then, five obtained isolates were transferred to fresh PSA. Samples of the infected leaves were deposited in the mycological herbarium (LEP) of the All-Russian Institute of Plant Protection (VIZR). All the isolates used in this study were stored in microtubes on PSA at 4 °C in the mycological collection of pure cultures in the A. A. Jaczewskii Laboratory of Mycology and Phytopathology (MF; VIZR).
2.2. DNA Isolation, PCR, and Sequencing
Mycelium was collected from the cultures on PSA and grinded with 0.3 mm glass sand in an MM400 mixer mill (Retsch, Haan, Germany). Genomic DNA was extracted according to the standard CTAB/chloroform protocol [19].
The internal transcribed spacer (ITS) and large subunit (28S) or ribosomal DNA, partial DNA-directed RNA polymerase II subunit (rpb2), and β-tubulin (tub2) genes were amplified and sequenced for all the isolates studied. The primers ITS1F [20] and ITS4 [21] and LR0R [22]/LR5 [21] were used to amplify the ITS and 28S regions, respectively. The primers fRPB2-5f2 [23] and fRPB2-7cR [24] and βtub2Fw/βtub4Rd [25] were used to amplify the partial rpb2 and tub2 genes, respectively.
The amplification reactions had a total volume of 25 μL, including dNTPs (200 μM), forward and reverse primers (0.5 μM each), Taq DNA polymerase (5 U/μL), 10× PCR buffer with Mg2+, and NH4+ and 1–10 ng of total genomic DNA.
The PCR conditions were as follows: 95 °C for 5 min, followed by 35 cycles of 92 °C for 50 s, 55 °C for 40 s (ITS1F/ITS4 and LR0R/LR5), 52 °C for 40 s (βtub2Fw/βtub4Rd), 72 °C for 75 s, and a final elongation for 5 min at 72 °C.
The partial rpb2 gene was amplified according to the touchdown program. All the steps were the same as described above, but the annealing temperature consequently declined from 5 cycles of 60 °C for 40 s and 5 cycles of 58 °C for 40 s to 30 cycles of 54 °C for 40 s.
PCR amplification products were checked using electrophoresis in 1% agarose gel stained with ethidium bromide. Amplicons were purified according to a standard method [26]. Then, the DNA of the amplicons was sequenced from both strands by Sanger’s method [27] on an ABI Prism 3500 analyzer (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA) with a Bigdye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Thermo Fisher Scientific) according to the manufacturer’s instructions.
The nucleotide sequences of the ITS, 28S, rpb2, and tub2 genes obtained were deposited in the GenBank database with corresponding accession numbers (Table 1).

Table 1.
GenBank accession numbers of studied and reference isolates.
2.3. Phylogenetic Analyses
To implement GCPSR and explore the possibility of constructing a set of four loci, ITS, 28S, rpb2, and tub2 were examined in a multilocus phylogenetic analysis, and single-locus trees were generated and compared to detect conflict. The multilocus phylogenetic analysis included sequences of all the studied isolates and 24 Ascochyta strains representing all the 24 species currently accepted in this genus. The tree was rooted at the ex-type Phomatodes nebulosa (Persoon) Q. Chen & L. Cai CBS 117.93.
Sequences were assembled using Vector NTI advance v. 11.0 (Invitrogen, Thermo Fisher Scientific) and aligned with ClustalX 1.8 [28]. The alignments were optimized with Molecular Evolutionary Genetics Analysis 10 (MEGA X) [29] and concatenated using Sequence Matrix [30].
The phylogenetic analysis of the concatenated alignment included maximum likelihood (ML), maximum parsimony (MP), and Bayesian inference. The maximum likelihood analyses were conducted using RaxML in IQtree [31], and the MP analyses were conducted using MEGA X. The Bayesian inference was conducted using MrBayes v. 3.2.1. in ARMADILLO v. 1.1 [32].
The maximum likelihood analyses were performed on a neighbor-joining starting tree that was automatically generated by the software. The nearest-neighbor interchange was used as the heuristic method for the tree inference, and 1000 bootstrap replicates were performed. IQtree was also used to determine the best nucleotide substitution model according to the Bayesian information criterion (BIC) that was used for building the maximum likelihood trees. Bootstrap values with 1000 replicates were calculated for the tree branches.
The maximum parsimony analyses were performed using the heuristic search option with 100 random taxon additions and the subtree pruning regrafting method as the branch-swapping algorithm. All the characters were unordered and of equal weight, and gaps were treated as missing values. The input parameter “maxtrees” was set to 100, and branches of zero length were collapsed. Clade stability was assessed using a bootstrap analysis with 1000 replicates. Afterwards, parameters including the consistency index, retention index, tree length, rescaled consistency index, and homoplasy index were calculated.
The Bayesian analyses were performed using the Markov chain Monte Carlo (MCMC) sampling method. The general time-reversible model of evolution, including the estimation of invariable sites and assuming a gamma distribution with six rate categories, was used for the Bayesian inference analyses. Four MCMC chains were run simultaneously, starting from random trees for 1000 generations and sampling every hundredth generation for a total of 2 000,000 trees. The final matrices used for the phylogenetic analyses and phylogenetic trees were deposited in TreeBASE (www.treebase.org; accession number 31140, accessed on 19 April 2024).
2.4. Morphology
Pure cultures of the studied isolates were incubated on PSA, oatmeal agar (OA), and malt extract agar (MEA) [3]. The petri dishes were placed for 7 d in darkness and then for 7 d under a 12 h near-ultraviolet light/12 h dark cycle to stimulate sporulation. Colony diameter was measured after 7 d, and colony morphology was examined after 14 d [3]. Morphological features of spore-bearing structures were described from the colonies on OA [3]. Observations and measurements of 100 conidia, 50 pycnidia, and 30 conidiogenous cells were conducted with an Olympus SZX16 stereomicroscope (Olympus, Tokyo, Japan) and an Olympus BX53 microscope. Images were captured with a PROKYON camera (Jenoptik, Jena, Germany) with Nomarski differential interference contrast.
2.5. Optimal Temperature for Growth
The optimal temperatures for the growth of all the isolates were determined on potato dextrose agar (PDA), [18], OA, and MEA incubated at 20, 25, and at 30 °C in darkness in Petri dishes with a diameter of 80 mm. Agar plugs that were 5 mm in diameter were taken from 7-day-old PSA cultures and inverted in the center of each plate. All treatments were replicated 4 times. Measurements of radial growth were recorded after 4, 7, and 14 days. Images were captured at 10 d of growth.
2.6. Pathogenicity
For the pathogenicity tests, leaf segments of C. arvensis were inoculated with all the studied isolates. Mycelial suspension was used as the inoculum. For inoculation, isolates were grown in liquid soybean nutrient medium with the following composition: KH2PO4 (0.2%), (NH4)2SO4 (0.1%), MgSO4 (0.1%), glucose (2%), and soy flour (1%). A total of 50 mL of medium in 250 mL flasks was inoculated with three 5 mm mycelial discs cut from 2-week-old colonies grown on PSA and kept on an orbital shaker (200 rpm) for four days at 24 °C. The mycelium was separated from the liquid culture by filtering and then dried with filter paper and ground in sterile water to prepare a mycelial suspension of 100 mg/mL. Leaf segments of C. arvensis collected from a natural habitat (1.5 × 1.5 cm) were placed in Petri dishes. To create a moisture chamber effect, filter paper premoistened with sterile water was added to each Petri dish. A drop of mycelial suspension (10 µL) was applied to the center of the previously wounded or intact leaf segments on the abaxial or adaxial surface of the leaf segments. In the control, the wounded or non-wounded leaf segments were treated with 10 µL of sterile distilled water. After inoculation, closed Petri dishes with leaf segments were kept at room temperature and evaluated for the presence of symptoms at 3, 4, and 7 d post treatment (dpt). Subsequent reisolation of fungus from the inoculated leaf segments and identification were carried out to fulfill the Koch postulates.
3. Results
3.1. Phylogenetic Analyses
The multilocus phylogenetic analysis based on ITS, 28S, rpb2, and tub2 sequences inferred intraspecific relationships within all 24 Ascochyta species currently accepted in this genus. The parsimony informative characters and the nucleotide substitution models used for each analysis are summarized in Table 2.

Table 2.
Alignment properties and nucleotide substitution models used for phylogenetic analyses.
The analysis of the single-gene phylogenies revealed no conflicts. The individual gene trees (Supplementary Figures S1–S4), as well the combined tree (Figure 1), demonstrated that the studied isolates clustered in a single distinct, monophyletic, well-supported phylogenetic clade with high bootstrap support that did not include any ex-type or representative Ascochyta strains and are considered to represent the new Ascochyta species. All five isolates were identical in the ITS, 28S, rpb2, and tub2 sequences.
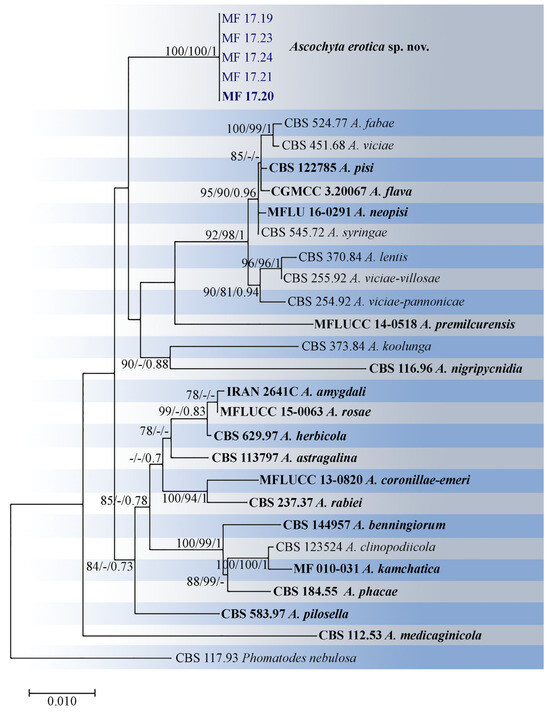
Figure 1.
Phylogenetic tree of Ascochyta species inferred from a maximum likelihood analysis based on a concatenated alignment of ITS, 28S, rpb2, and tub2. The maximum likelihood bootstrap support values (MLBS ≥ 70%), maximum parsimony bootstrap support values (MPBS ≥ 70%), and Bayesian posterior probabilities (BPP ≥ 0.70) are given at the nodes (MLBS/MPBS/BPP). Tree is rooted by Phomatodes nebulosa (CBS 117.93). The studied isolates are in blue, and the ex−type and representative strains are in bold.
3.2. Optimal Temperature for Growth
The colonies with the largest diameter on all three media for most isolates were obtained at 25 °C (Table 3, Figure 2). Cultivation at 20 °C on MEA led to a decrease in colony diameter after 7 d of 5–9%, on PDA, the decrease was 3–16%, and on OA (except MF 17.21), the decrease was 0.2–10.5% compared with cultivation at 25 °C. The growth of MF 17.21 on OA at 25 °C was slower (10%) than when grown at 20 °C. Also, the growth of all isolates was recorded at 30 °C, and the colony diameter after 7 d on MEA was 40–50% less, on PDA it was 36–52% less, and on OA it was 23–46% less than when grown at 25 °C. Among the tested media, the colonies with the largest diameter were developed on PDA, and the smallest were developed on MEA.

Table 3.
Colony diameters of Phoma proboscis and Ascochyta erotica isolates grown at different temperature on various media.
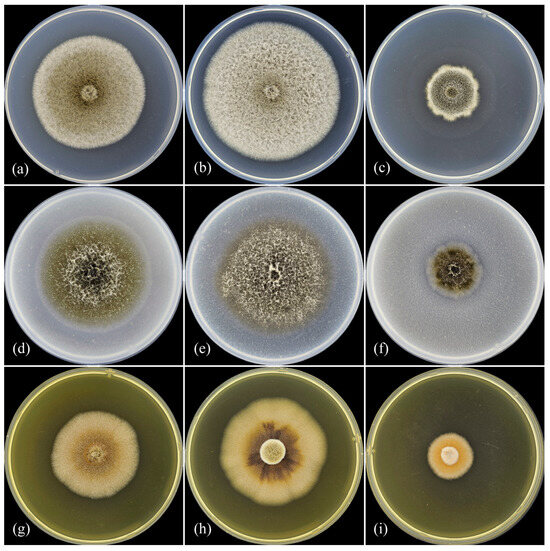
Figure 2.
Ascochyta erotica sp. nov. (ex-holotype culture MF-17.20). Colonies grown at different temperatures on various media after 10 days of growth. (a) PDA, 20 °C; (b) PDA, 25 °C; (c) PDA, 30 °C; (d) OA, 20 °C; (e) OA, 25 °C; (f) OA, 30 °C; (g) MEA, 20 °C; (h) MEA, 25 °C; (i) MEA, 30 °C.
3.3. Taxonomy
Based on the multilocus analysis, the five Ascochyta isolates MF-17.19, MF-17.20, MA-17.21, MF-17.23, and MF-17.24 from C. arvensis were found to be distinct from any described species in the genus Ascochyta and are, therefore, described here as a new species.

Figure 3.
Ascochyta erotica sp. nov. from the holotype material LEP 124536. (a) Necrotic lesions on Convolvulus arvensis leaf; (b) pycnidia on necrotic lesion; (c) conidia. Scale bars: (b) 1 mm; (c) 20 µm. Arrows indicate pycnidia.
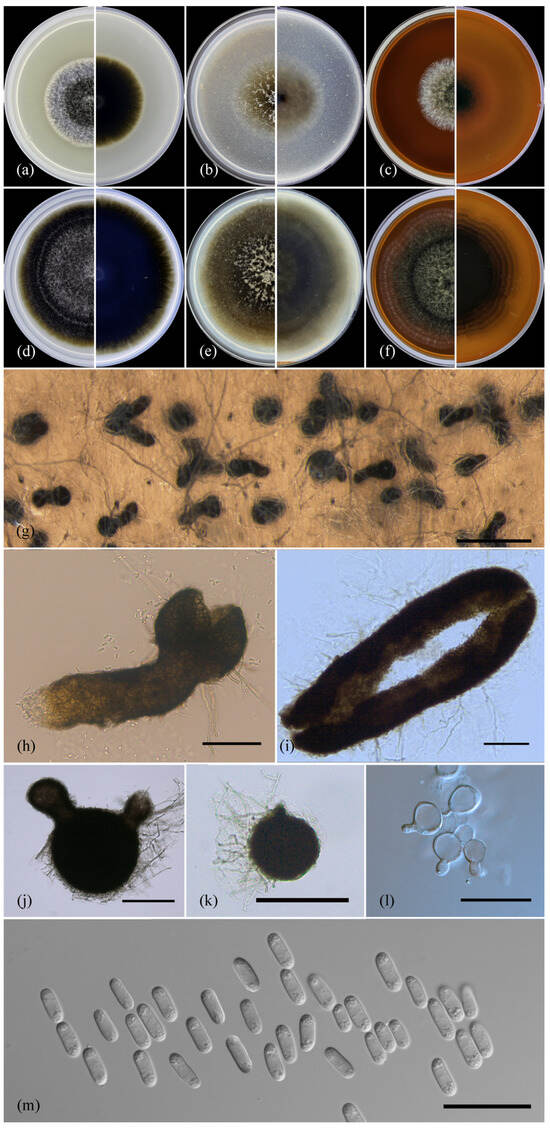
Figure 4.
Ascochyta erotica sp. nov. (ex-holotype culture MF-17.20). (a) Culture on PSA after 7 d of growth (front and reverse); (b) culture on OA after 7 d of growth (front and reverse); (c) culture on MEA after 7 d of growth (front and reverse); (d) culture on PSA after 14 d of growth (front and reverse); (e) culture on OA after 14 d of growth (front and reverse); (f) culture on MEA after 14 d of growth (front and reverse); (g–k) pycnidia from OA after 14 d of growth; (l) conidiogenous cells; (m) conidia. Scale bars: (g), 500 µm; (h,i), 100 µm; (j,k), 200 µm; (l,m), 20 µm.
MycoBank: 852086.
Typification: Kyrgyz Republic, Tchui region, from necrotic Convolvulus arvensis leaves, 15 Aug 2002, collected by Bilder I.V. (holotype LEP 124536). Ex-holotype living culture MF-17.20, isolated by Gasich E.L. GenBank: ITS = MH651559, 28S = MH651600, rpb2 = PP197205, tub2 = MH725219.
Etymology: The specific epithet refers to the extraordinary and unusual form of the pycnidia, resembling human genitals.
Description from the Holotype: The holotype represents both infected plant material and the metabolic inactive culture grown for 14 d on OA. Large lesions with a concentric zonation were detected on the C. arvensis leaves. The pycnidia from the plant material were (sub)globose or pyriform and narrow with long necks that measured 46.15–103.69 (70.61 ± 9.24) × 26.58–42.77 (32.49 ± 1.7) μm developed on the lesions and contained ellipsoidal, smooth, thin-walled, hyaline, aseptate conidia that measured 3.53–6.53 (5.08 ± 0.14) × 1.9–2.76 (2.37 ± 0.04) μm. A description of the spore-bearing structures from the metabolic inactive culture grown for 14d on OA is given below.
Culture characteristics: The colonies grown on PSA were 41–56 (av. 46.7 ± 3) mm in diameter after 7 d and 82–89 (av. 81 ± 3) mm after 14 d, and they had a regular margin. The entire colony was deep brown, yellow olive near the periphery, reverse concolorous, aerial mycelia were abundant and pearlescent light grey, had a velvety-felty near the center, and were sparse near the margin. Pycnidia were absent. The colonies grown on OA were 43–47 (av. 44.5 ± 1) mm in diameter after 7 d and 76–85 (av. 78.6 ± 2) mm after 14 d. They were mottled due to numerous submerged pycnidia, had a regular margin, were dark olive brown in the center, yellow olive brown near the periphery, reverse concolorous, tufted aerial mycelia were not abundant, and were pebble grey. Pycnidia were abundant and superficial, semi-immersed and immersed in the agar media, solitary and in conglomerates, and formed in concentric rings. The optimal temperature for growth on OA was 25 °C. The colonies grown on MEA were 34–43 (av. 37.9 ± 2) mm in diameter after 7 d and 63–73 (av. 69 ± 2) mm after 14 d, had an irregular margin, were slightly lobate, greenish dark brown, had a periphery covered with a scanty light grey aerial mycelia in concentric rings, were reverse concolorous, the aerial mycelia were green grey and had velvety-felty near the center, and were sparse near the margin. The pycnidia were immersed in the agar media formed wavy concentric rings from the edge of the aerial mycelia to the colony margin. An NaOH spot test on the MEA was negative.
The conidiomata were pycnidial, mostly solitary, sometimes aggregated or confluent, superficial, and semi-immersed and submerged in the agar media. Almost half of the pycnidia were (sub)globose or pyriform with single or plural (1–3) short papillate ostioles measuring 104.25–337.24 (212.2 ± 33.69) × 117.81–397.39 (220.16 ± 38.13) μm. The other half of the pycnidia were narrow with single or plural long necks, often branched, and the pycnidial body was immersed and ascended from the bottom of the Petri dish, breaking through the surface of the agar media, and measured 195.32–932.93 (382.91 ± 72.84) × 93.09–228.77 (149.93 ± 13.63) μm. In the older cultures, the number of such long pycnidia with necks was significantly predominant. The pycnidial wall was pseudoparenchymatous and composed of isodiametric cells, and the outer layers had a brown pigment. The conidiogenous cells were phialidic, formed from the inner cells of the pycnidial wall, hyaline, smooth, flask-shaped, and measured 6.88–34 (12.41 ± 1.59) × 4.21–8.9 (6.27 ± 0.34) μm. The conidia were ellipsoidal to oblong and cylindrical, smooth, thin-walled, hyaline, aseptate, had small guttules located in groups in the polar ends, and measured 5.98–11.06 (8.51 ± 0.11) × 2.43–4.26 (3.35 ± 0.04) μm.
No chlamydospores were observed. Ascomata were absent.
Ascochyta erotica pathogenic to the hosts in the Convolvulaceae: C. arvensis (present study).
Notes: Among the Phoma-like fungi, A. erotica might morphologically resemble Phoma proboscis Heiny [16]. Below, we list the similarities and differences between A. erotica and P. proboscis. Among the similarities are the long necks of the pycnidia and the host plant. The differences are numerous and significant. Firstly, A. erotica differs from P. proboscis in its cultural features and optimal temperature for growth. Notably, the optimum growth rate for P. proboscis is 20 °C, and at 25 °C, the fungus did not actually grow; after 14 days, the colony grew only 2 mm, whereas the optimum temperature for growth of A. erotica was 25 °C. Comparing the optimum temperature for all the studied media, the A. erotica colonies grew faster than the P. proboscis ones.
The average diameter of the A. erotica colonies after 7 d on MEA at 20 °C was 32.2 mm as opposed to the diameter of 24 mm for the P. proboscis colonies; At 25 °C the diameter was 34.6 mm as opposed to 7.5 mm; and at 30 °C, the diameter was 19.2 mm as opposed to 5 mm. On PDA, at 20 °C, the diameter was 41.3 mm as opposed to 29.0 mm; at 25 °C the diameter was 45.9 mm as opposed to 7 mm; and at 30 °C, the diameter was 24.2 mm as opposed to 5 mm. On OA, at 20 °C, the diameter was 39.2 mm as opposed to 34.0 mm; at 25 °C, the diameter was 40.2 mm as opposed to 7 mm; and at 30 °C, the diameter was 24.6 mm as opposed to 5 mm, respectively.
Also, the colonies P. proboscis grown on PDA were yellow-brown with a white margin, the colonies of A. erotica grown on PDA were brown-olive with a whitish margin, and the reverse was dark brown-olive, and the colonies grown on PSA were deep brown and yellow-olive near the periphery. On OA, the P. proboscis strain produced a dense mat of pycnidia, whereas the pycnidia of A. erotica grown on OA were superficial, semi-immersed and immersed in the agar media, and formed in concentric rings. The pycnidia of P. proboscis developed elongate necks within 7 days on all the media, especially on PDA. Ascochyta erotica produced pycnidia only on OA and MEA, and pycnidia with necks only formed on OA, even during exposure to UV light. No chlamydospores occurred in the A. erotica cultures; by comparison, in the P. proboscis cultures, unicellular chlamydospores are abundant. Additionally, there were differences in the conidial morphology. The P. proboscis conidia are salmon-colored in mass, and no conidial exudate was observed in the A. erotica spore-bearing structures. The conidia of P. proboscis are cylindrical or narrowly ellipsoidal, often slightly larger at one end, foot-shaped, slightly curved, and several conidia are occasionally one-septate, while the A. erotica conidia were similar in form and size, ellipsoidal to oblong and cylindrical, and only aseptate. The A. erotica conidia, on average, were smaller and narrower, measuring 5.98–11.06 (8.51 ± 0.11) × 2.43–4.26 (3.35 ± 0.04) μm, than the conidia of P. proboscis, measuring 5.5–15.0 (17) × 2.3–5.0 μm and averaging 10.5 × 3.5 μm.
In addition to having morphological differences, these fungi also differ in habitat. Ascochyta erotica has been revealed only in the Kyrgyz Republic, whereas P. proboscis has been revealed only from the central part of the USA.
Phoma proboscis was described for the sole strain ATCC 74032. Since its description in 1990, there have not been any reports about findings of this fungus and the vitality of the ex-type strain. There are still no sequence data on the ex-type P. proboscis strain in GenBank, and thus there is no way to compare A. erotica and P. proboscis in terms of their molecular phylogenetic features. Resolved morphological differences however, might be sufficient to describe studied isolates as new species
3.4. Pathogenicity
The pathogenicity of Ascochyta erotica sp. nov. was tested using C. arvensis leaf segments (Table 4, Figure 5). The first significant symptoms were observed at 3 dpt (with the exception of MF-17.24) on the abaxial sides of the leaf segments, both wounded and intact. The adaxial sides were less susceptible to infection, and symptoms were recorded at 3 dpt only in the wounded leaf segments. At 7 dpt, a significant increase in the size of the developed necrosis was detected. The necrosis that developed was also observed when the inoculum was applied to the intact adaxial sides of the leaves. To confirm Koch’s postulates, fungal cultures were reisolated in all cases from the necrosis formed on leaf segments. These isolates had morphological characteristics identical to those of all the isolates tested.

Table 4.
Pathogenicity of Ascochyta erotica sp. nov. isolates on leaf segments of Convolvulus arvensis.
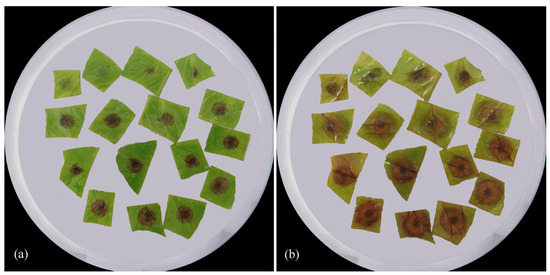
Figure 5.
Pathogenicity of the Ascochyta erotica ex-type isolate MF-17.20 on leaf segments of Convolvulus arvensis. First line in each set (Petri dish) contains leaf segments inoculated with intact adaxial part; second line is adaxial part wounded with needle; third line is intact abaxial part; fourth line is abaxial part wounded with needle. Inoculation with mycelial suspension of isolate: (a) 3 days post treatment; (b) 7 days post treatment.
4. Discussion
Leaf spot of C. arvensis is a widespread disease. Field bindweed leaves exhibiting this symptom can be collected from almost their entire habitat. As a result of an extensive phytosanitary examination of industrial fields and natural areas in different regions of Russia as well several countries of the former USSR, conducted from 1990 to date, more than 200 specimens of infected C. arvensis leaves were collected. From these leaves, fungal isolates were obtained, maintained, and studied. In total, this collection listed more 200 fungal isolates. Among them, 34% were Sphaeria lichenoides var. convolvulicola DC. (syn. Septoria convolvuli Desm.) isolates, 22% were Paraphoma convolvuli (syn. Stagonospora convolvuli Dearn. & House), 19% were Septoria calystegiae Westend. (syn. Stagonospora calystegiae (Westend.) Bubák), 13% were Phoma s.l., and 8% were Paraphoma spp. Ascochyta species accounted for less than 4% of all the isolates collected. Among them, Ascochyta erotica comprised only five isolates that were collected only in one location in the Kyrgyz Republic. Apparently, this fungus is local and requires specific optimal growth conditions. And, this is apparently not common, e.g., P. proboscis was not known to occur outside the central part of the USA (Colorado, Kansas, Nebraska) [33] and is specialized to Convolvulaceae plants. There have been no reports of P. proboscis findings since 1994.
In Melnik’s Ascochyta identification manual [11], three Ascochyta species are associated with Convolvulaceae plants: A. calystegiae, A. convolvuli (=Sphaeria lichenoides var. convolvulicola), and A. kleinii. The pycnidia of A. calystegiae are lenticular and thin-walled, and the conidia measure 6–13 × 2.5–4 µm. The pycnidia of A. convolvuli are spherical-flattened and thick-walled, and the conidia measure 10–16 × 3.5–4.5 µm. The pycnidia of A. kleinii are spherical-flattened and thin-walled, and the conidia measure 13–18 × 2.5–3 µm. The size of the conidia of A. erotica are comparable to the size of those of A. calystegiae and Phoma capsularum (8–10 × 2.5–3.5 µm) [34]; however, in terms of the pycnidia shape, these three species significantly differ from each other. The shape of the pycnidia of A. erotica somewhat resemble those of Ectophoma iranica M. Mehrabi, Larki & Farokhineja and Ectophoma multirostrata (P.N. Mathur, S.K. Menon & Thirum.) Valenz.-Lopez, Cano, Crous, Guarro & Stchigel. Both E. iranica and E. multirostrata conidia are significantly smaller (4.5–6.5 × 2–2.5 µm. and 1.9–4.4 × 1.3–2.6 µm., respectively) [3,35,36].
The fungal biodiversity associated with field bindweed has been intensively studied to search for possible biocontrol agents for this harmful weed. Mycoherbicidal potential has been revealed in several fungi, such as Diaporthe convolvuli (Ormeno-Nuñez, Reeleder & A.K. Watson) R.R. Gomes, Glienke & Crous [37,38,39,40], Phoma proboscis [41,42], and Paraphoma convolvuli [43,44,45]. In the pathogenicity tests, all the A. erotica isolates were pathogenic to C. arvensis; thus, A. erotica may be of interest for studying its mycoherbicidal activity as an agent of biocontrol against this weed.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/d16040246/s1, Figure S1: Phylogenetic tree of Ascochyta species inferred from a maximum likelihood analysis based on an alignment of ITS. The maximum likelihood bootstrap support values (MLBS ≥ 70%), maximum parsimony bootstrap support values (MPBS ≥ 70%), and Bayesian posterior probabilities (BPP ≥ 0.70) are given at the nodes (MLBS/MPBS/BPP). Tree is rooted by Phomatodes nebulosa (CBS 117.93). The studied isolates are in blue, and the ex-type and representative strains are in bold. Figure S2: Phylogenetic tree of Ascochyta species inferred from a maximum likelihood analysis based on an alignment of 28S. The maximum likelihood bootstrap support values (MLBS ≥ 70%), maximum parsimony bootstrap support values (MPBS ≥ 70%), and Bayesian posterior probabilities (BPP ≥ 0.70) are given at the nodes (MLBS/MPBS/BPP). Tree is rooted by Phomatodes nebulosa (CBS 117.93). The studied isolates are in blue, and the ex-type and representative strains are in bold. Figure S3: Phylogenetic tree of Ascochyta species inferred from a maximum likelihood analysis based on an alignment of rpb2. The maximum likelihood bootstrap support values (MLBS ≥ 70%), maximum parsimony bootstrap support values (MPBS ≥ 70%), and Bayesian posterior probabilities (BPP ≥ 0.70) are given at the nodes (MLBS/MPBS/BPP). Tree is rooted by Phomatodes nebulosa (CBS 117.93). The studied isolates are in blue, and the ex-type and representative strains are in bold. Figure S4: Phylogenetic tree of Ascochyta species inferred from a maximum likelihood analysis based on an alignment of tub2. The maximum likelihood bootstrap support values (MLBS ≥ 70%), maximum parsimony bootstrap support values (MPBS ≥ 70%), and Bayesian posterior probabilities (BPP ≥ 0.70) are given at the nodes (MLBS/MPBS/BPP). Tree is rooted by Phomatodes nebulosa (CBS 117.93). The studied isolates are in blue, and the ex-type and representative strains are in bold.
Author Contributions
Both authors contributed to the conception and design of this study. The molecular work and phylogenetic analyses were performed by M.G. The analyses of the morphological features and optimal temperature for growth were conducted by M.G. and E.G. The preparation of illustrations was performed by M.G. The first draft of the manuscript was written by M.G. and E.G. commented on previous versions of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Russian Science Foundation (RSF), grant number 23-74-01035.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The original data presented in the study are openly available in [TreeBASE] at [www.treebase.org; accession number 31140], in [MycoBank] at [https://www.mycobank.org; MB#852086].
Acknowledgments
We are grateful to Irina V. Bilder for the sample collection and to Lyudmila B. Khlopunova for the laboratory assistance and maintaining the culture collection.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Libert, M.-A. Plantae Cryptogamicae, quas in Arduenna Collegit. Fasc. 1:no. 1-no. 1830. 100p. Available online: https://books.google.com.au/books?id=GTZinQEACAAJ (accessed on 14 March 2023).
- Saccardo, P.A. Sylloge Fungorum Omnium Hucusque Cognitorum; 3. Pavia; Sumptibus Auctoris: Patavii, Italy, 1884; 860p. [Google Scholar]
- Boerema, G.H.; de Gruyter, J.; Noordeloos, M.E. Phoma identification manual. In Differentiation of Specific and Infra-Specific Taxa in Culture; CABI Publishing: Wallingford, UK, 2004; 470p. [Google Scholar]
- Chen, Q.; Jiang, J.R.; Zhang, G.Z.; Cai, L.; Crous, P.W. Resolving the Phoma enigma. Stud. Mycol. 2015, 82, 137–217. [Google Scholar] [CrossRef] [PubMed]
- De Gruyter, J.; Aveskamp, M.M.; Woudenberg, J.H.; Verkley, G.J.; Groenewald, J.Z.; Crous, P.W. Molecular phylogeny of Phoma and allied anamorph genera: Towards a reclassification of the Phoma complex. Mycol. Res. 2009, 113, 508–519. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Hou, L.W.; Duan, W.J.; Crous, P.W.; Cai, L. Didymellaceae revisited. Stud. Mycol. 2017, 87, 105–159. [Google Scholar] [CrossRef]
- Hou, L.W.; Groenewald, J.Z.; Pfenning, L.H.; Yarden, O.; Crous, P.W.; Cai, L. The Phoma-like dilemma. Stud. Mycol. 2020, 96, 309–396. [Google Scholar] [CrossRef]
- Crous, P.W.; Hawksworth, D.L.; Wingfield, M.J. Identifying and naming plant-pathogenic fungi: Past, present, and future. Annu. Rev. Phytopathol. 2015, 53, 247–267. [Google Scholar] [CrossRef]
- Ahmadpour, S.A.; Mehrabi-Koushki, M.; Farokhinejad, R.; Asgari, B. New species of the family Didymellaceae in Iran. Mycol. Prog. 2022, 21, 28. [Google Scholar] [CrossRef]
- Chen, T.; Wang, S.; Jiang, X.; Huang, Y.; Mo, M.; Yu, Z. New species of Didymellaceae within aquatic plants from Southwestern China. J. Fungi. 2023, 9, 761. [Google Scholar] [CrossRef] [PubMed]
- Mel’nik, V.A. Opredelitel’ Gribov Roda Ascochyta Lib; Nauka: Leningrad, Russia, 1977; 246p. [Google Scholar]
- Boerema, G.H.; Bollen, G.J. Conidiogenesis and conidial septation as differentiating criteria between Phoma and Ascochyta. Persoonia 1975, 8, 111–444. [Google Scholar]
- Holm, L.G.; Plucknett, D.L.; Pancho, J.V.; Herberger, J.P. The World’s Worst Weeds: Distribution and Biology; HI University Press: Honolulu, HI, USA, 1977; 609p. [Google Scholar]
- Farr, D.F.; Rossman, A.Y. Fungal Databases, Systematic Mycology and Microbiology Laboratory. Agricultural Research Service United States Department of Agriculture. Available online: http://nt.ars-grin.gov/fungaldatabases/ (accessed on 14 March 2023).
- Wehmeyer, L.E. Some fungi imperfecti of the Punjab and Kashmir. Hycologia 1964, 56, 29–52. [Google Scholar] [CrossRef]
- Heiny, D.K. Phoma proboscis sp.nov. pathogenic on Convolvulus arvensis. Mycotaxon 1990, 36, 457–471. [Google Scholar]
- Gomzhina, M.M.; Gasich, E.L.; Khlopunova, L.B.; Gannibal, P.B. Paraphoma species associated with Convolvulaceae. Mycol. Prog. 2020, 19, 185–194. [Google Scholar] [CrossRef]
- Samson, R.A.; Hoekstra, E.S.; Frisvad, J.C.; Filtenborg, O. Introduction to Food- and Airborne Fungi, 6th ed.; Centraal Bureau Voor Schimmel Cultures: Utrecht, The Netherlands, 2002; 389p. [Google Scholar]
- Doyle, J.J.; Doyle, J.L. A rapid total DNA preparation procedure for fresh plant tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for Basidiomycetes application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Rehner, S.A.; Samuels, G.J. Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 1994, 98, 625–634. [Google Scholar] [CrossRef]
- Sung, G.H.; Sung, J.M.; Hywel-Jones, N.L.; Spatafora, J.W. A multi-gene phylogeny of Clavicipitaceae (Ascomycota, Fungi): Identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 2007, 44, 1204–1223. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerse II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Aveskamp, M.M.; Verkley, G.J.; de Gruyter, J.; Murace, M.A.; Perello, A.; Woudenberg, J.H.; Groenewald, J.Z.; Crous, P.W. DNA phylogeny reveals polyphyly of Phoma section Peyronellaea and multiple taxonomic novelties. Mycologia 2009, 101, 363–382. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.S.; Lew, A.M. An inexpensive alternative to glassmilk for DNA purification. Trends Genet. 1995, 11, 8. [Google Scholar] [CrossRef] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, G.; Lohman, D.J.; Meier, R. SequenceMatrix: Concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 2011, 27, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; Von Haeseler, A.; Lanfear, R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 2020, 35, 1530–1534. [Google Scholar] [CrossRef] [PubMed]
- Lord, E.; Leclercq, M.; Boc, A.; Diallo, A.B.; Makarenkov, V. Armadillo 1.1: An original workflow platform for designing and conducting phylogenetic analysis and simulations. PLoS ONE. 2012, 7, e29903. [Google Scholar] [CrossRef]
- Heiny, D.K. Host-specificity of Phoma proboscis in the Convolvulaceae. Phytopathology 1992, 82, 498. [Google Scholar]
- Saccardo, P.A. Sylloge Fungorum Omnium Hucusque Cognitorum; 11(3); R. Friedländer & Sohn: Berlin, Germany, 1895; 753p. [Google Scholar]
- Morgan-Jones, C. Studies in the genus Phoma. XV. Concerning Phoma multirostrata, a leaf spot- inducing and soil-borne species in warm climates. Mycotaxon 1988, 33, 339–351. [Google Scholar]
- Larki, R.; Mehrabi-Koushki, M.; Farokhinejad, R. Ectophoma iranica sp. nov. and new hosts and recods of Allophoma spp. in Iran. J. Phytopathol. 2019, 167, 538–545. [Google Scholar] [CrossRef]
- Ormeno-Nuñez, J.; Reeleder, R.D.; Watson, A.K. A new species of Phomopsis recovered from field bindweed (Convolvulus arvensis). Can. J. Bot. 1988, 66, 2228–2233. [Google Scholar] [CrossRef]
- Ormeno-Nuñez, J.; Reeleder, R.D.; Watson, A.K. A foliar disease of field bindweed (Convolvulus arvensis) caused by Phomopsis convolvulus. Plant Dis. 1988, 72, 338–342. [Google Scholar] [CrossRef]
- Vogelsang, S.; Watson, A.K.; DiTommaso, A. Effect of moisture, inoculum production, and planting substrate on disease reaction of field bindweed (Convolvulus arvensis L.) to the fungal pathogen, Phomopsis convolvulus. Eur. J. Plant Pathol. 1998, 104, 253–262. [Google Scholar] [CrossRef]
- Watson, A.K.; Reeleder, R.D.; Ormeno-Nuñez, J. Fungal Herbicides. U.S. Patent 5212086, 18 May 1993. [Google Scholar]
- Heiny, D.K.; Templeton, G.E. Effects of spore concentration, temperature, and dew period on disease of field bindweed caused by Phoma proboscis. Phytopathol 1991, 81, 905–909. [Google Scholar] [CrossRef]
- Heiny, D.K.; Templeton, G.E. Method and compositions for the biological control of field bindweed. U.S. Patent 5391538, 21 February 1995. [Google Scholar]
- Défago, G.; Ammon, H.U.; Cagan, L.; Draeger, B.; Greaves, M.P.; Guntli, D.; Hoeke, D.; Klimes, L.; Lawrie, J.; Moënne-Loccoz, Y.; et al. Towards the biocontrol of bindweeds with a mycoherbicide. BioControl 2001, 46, 157–173. [Google Scholar] [CrossRef]
- Pfirter, H.A.; Defago, G. The potential of Stagonospora sp. as a mycoherbicide for field bindweed. Biocontrol. Sci. Tech. 1998, 8, 93–101. [Google Scholar] [CrossRef]
- Pfirter, H.A.; Guntli, D.; Ruess, M.; Defago, G. Preservation, mass production and storage of Stagonospora convolvuli, a bioherbicide candidate for field bindweed (Convolvulus arvensis). BioControl 1999, 44, 437–447. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).