Abstract
In this article, we analyzed the origin of wild polyploid oats (Avena L., Poaceae) using the region 18S rDNA (partially)–ITS1–5.8S rDNA obtained via NGS. There are six tetraploid (2n = 28) and four hexaploid (2n = 42) wild species differing by specific genome combinations: A. barbata, A. vaviloviana (AB), A. agadiriana (AB or BB), A. magna, A. murphyi, A. insularis (AC or CD), A. ludoviciana, A. sterilis, A. fatua, and A. occidentalis (ACD). We compared the pool of marker sequences of polyploid oats with those of their putative diploid ancestors: A. atlantica (As-genome), A. hirtula (As), A. canariensis (Ac), A. ventricosa (Cv), and A. clauda (paleopolyploid with Cp and A-related rDNA). We found 15 major ribotypes (more than 1000 reads per rDNA pool) in polyploid oats. Comparing them, we found that the AB-tetraploid oats possibly inherited their A-genome ribotypes from A. atlantica (As1-ribotype), whereas their B-genome ribotype is specific and can be a derivative of the A-genome family. Our data do not support the hypothesis of the CD-genome set in A. magna, A. murphyi, and A. insularis: they have an AC-genome ribotype constitution instead. The C-genome-related sequences could have been obtained from A. ventricosa. Hexaploids show a different ribotype pattern than tetraploids; the main ribotypes of A. fatua, A. ludoviciana, and A. sterilis probably belong to the D-group and are also shared with one of the major ribotypes of A. clauda.
1. Introduction
The genus Avena L. (Poaceae) is one of the most economically important cereals. It comprises about 30 species that form a polyploid row (from diploids, 2n = 14, to hexaploids, 2n = 42). All wild oats historically belong to the ancient Mediterranean region and, as many researchers suppose, originated from multiple hybridizations. Recent cytogenetic and molecular phylogenetic studies have described three genome types that characterize the oat species: A, B, C, and D. The Avena ancestor probably have had a diploid chromosome set with “symmetrical” chromosomes, similar in this respect to the karyotypes of diploid oats with the A genome and that of Arrhenatherum P. Beauv. [1], as well as to the chromosome sets (CmCm) of A. macrostachya Balansa & Durieu. Further, there was a divergence of oat phylogenetic lines with the genomes A and C, accompanied by the accumulation of differences in dispersed repeats (apparently determining the results of GISH hybridization) and the accumulation of transitions and transversions specific to each branch. According to cytogenetic data [2] the A-genome oats have a symmetrical karyotype, whereas in Avena species, with the C-genome, chromosome rearrangements occurred, resulting in the appearance of multiple small heterochromatine blocks and desymmetrization of chromosomes. From a cytological point of view, the A-genome oats can have a more primitive karyotype [2] than the C-genome species, which tend to have more primitive morphological features [3]. This pattern indicates evolutionary heterogeneity: taxa evolve rather slowly, retaining archaic morphological features, but cannot be called truly primitive [4].
Considering the evolution of tetraploid oat species, we can trace two main independent lines: species with AB genomes and species with AC genomes. The genomic structure of the AB-genome oats probably derives from the As-genome variant [2,5,6,7] though some other researchers consider the Ac-genome oat to be the progenitor of the AB-genome tetraploids [3,8]. The B-genome in oats is known only in tetraploids and does not occur in diploids. This genome variant is thought to be a derivative of the A-genome [9,10,11]. Probably, the AB-genome species originated from the As-genome species via autopolyploidization [6,12], and thus can be considered the species with the AA’ genome. The AC-genome oats show a close relationship with the hexaploid ones [13]. In accordance with cytogenetic data, some researchers considered A. insularis Ladiz. as the tetraploid progenitor of hexaploid Avena species [3,13,14,15], while others regarded A. maroccana Gand. (=A. magna Murphy & Terrell) and A. murphyi Ladiz. as possible ancestors of hexaploids [7]. The C-genome in the AC-genome tetraploids has probably been inherited from A. ventricosa Balansa [11] or from A. clauda Durieu [16]. In addition to some challenges about relationships between tetraploids and hexaploids, there are also ambiguities concerning their very genomic constitution. Some previous studies determined the tetraploids with the C-genome as AC [17]. However, later works based on Acc-nuclear and chloroplast gene sequences suggested that these tetraploids are in fact the bearers of the CD-genome [18,19]. The D-genome was thought to be related to the A-variant [20,21].
Evolution of hexaploid Avena species are the most complicated; the A-genome of hexaploid oats is different from that of diploid species [22]. Cytogenetic research also showed that the putative A-genome donor to hexaploid oats could have been tetraploid A. magna [23]. Recent phylogenetic studies estimate some Al/Ad-genome species as the A-genome ancestor of hexaploids, whereas the C and D-genomes were obtained by them from some CD-tetraploids [19]. The D-genome diploid progenitor may be extinct according to the genomic studies [19]. In addition, A. sterilis L. is thought to be the first in the hexaploid oat line and the progenitor of all their diversity [3,11,24]. Avena occidentalis Durieu, a relic of the ancient African flora, is probably a relative of A. vaviloviana (Malzew) Mordv. (the AB-genome tetraploid) and thus can represent the second ancestral line of hexaploid oats [11,25].
As we see, there are some questions about the origin of hexaploid oats and the evolutionary connections between diploids and tetraploids. Besides, we earlier described possible introgression cases within the diploid Avena [26,27]. These introgression events occurred not only within the A- and C-genome species but also between them [26,27]. Avena clauda (previously determined as the Cp-genome species) has three major ribotypes related to the A-genome, especially one that is shared with A. longiglumis Durieu (the ribotype of the minor fraction) [27]. Thus, drastic differences between the A- and C-genome compositions did not prevent the introgression between the genome bearers. Also, we cannot exclude the possible evolution of the A-genome from the C-genome ancestor close to modern A. clauda.
Locus-specified next-generation sequencing can reveal hidden hybridization when morphological features are constant and do not identify thehybrid origin [28]. For our aims, we took marker sequences including 18S–ITS1–5.8S rDNA. This region can be a suitable marker in cases of multiple hybridization between related species and is sometimes more effective than ITS2 because of its conservative structure and higher substitution rate [29,30,31]. Our goal in this research is to find possible ancestral species of polyploid oats among diploid ones and trace evolutionary pathways that could lead to the origin of hexaploids.
2. Results
The aligned sequences were sorted into ribotypes with a certain amount of reads. Major ribotypes were the ribotypes that were presented in the rDNA pool more than 1000 times. Minor ribotypes with a threshold of 10 reads per whole pool were usually variants of the major ribotypes in the case of the A-genome. The C-genome-related sequences were also in the minor fraction (from 10 to 40 reads). Table 1 demonstrates the studied species and their ribotype constitution.

Table 1.
Summary of the oat species (polyploids and possible parental taxa) used in the present study and their genome types and ribotypes.
Table 2 summarizes the major ribotypes of polyploid oats and their possible diploid ancestors: A. atlantica B.R. Baum and Fedak, A. canariensis B.R. Baum, Rajhathy, and D.R. Sampson, A. hirtula Lag. (A-genome oats), A. ventricosa, and A. clauda (C-genome species, paleopolyploid). We took as the consensus sequence that of A. atlantica (As-genome), a possible ancestor of the A-genome component in the polyploid species.

Table 2.
Primary structure of the major ribotypes (more than 1000 reads per rDNA pool) obtained by NGS. Numbers show the position in the alignment of the major ribotypes. D is a deletion.
The majority of the differences between ribotypes are located within the ITS1 region; the differences are represented by SNPs (single nucleotide polymorphisms) and deletions.
We analyzed three datasets obtained via NGS. The first dataset was aimed at studying the phylogeny of tetraploid oat species. It contains 660 sequences of tetraploids and their possible diploid, A-genome donors, A. atlantica and A. canariensis (Supplementary Table S1). The second dataset comprises sequences of hexaploid ACD-oats, their putative tetraploid progenitors of the AC-genome, and diploid species A. atlantica, A. canariensis, and A. ventricosa. It consists of 792 sequences of 18S rDNA–ITS1–5.8S rDNA (Supplementary Table S2). The third dataset contains the marker sequences of hexaploid oats, related tetraploids, and sequences of the diploid A. hirtula and paleopolyploid A. clauda (917 accessions, Supplementary Table S3).
Totally, we found 23 major ribotypes in the studied Avena species. Networks built by the statistical parsimony algorithm are presented in Figure 1, Figure 3, and Figure 5. Avena canariensis, a possible progenitor of the polyploid species [3,8,11,14] has two related major ribotypes that did not participate in the formation of AB and AC-polyploids (Figure 1a,b).
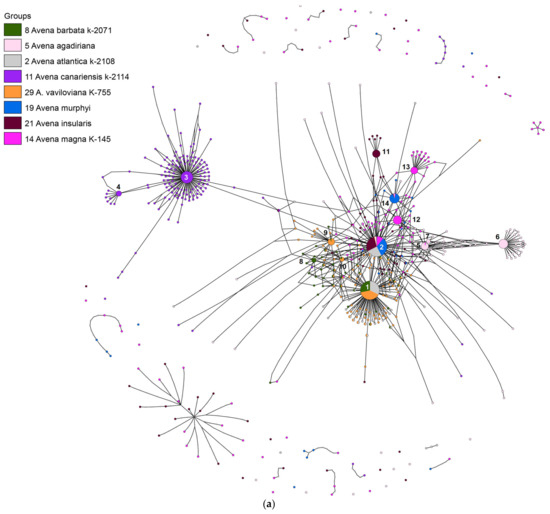
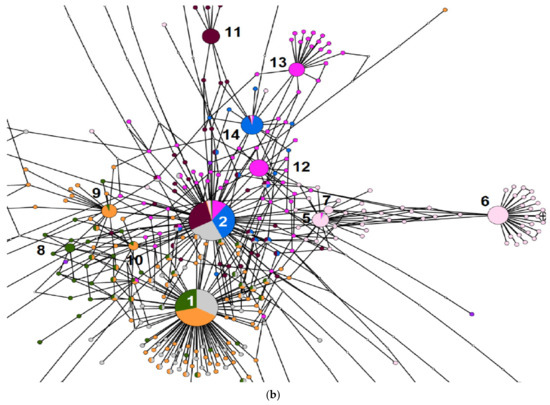
Figure 1.
(a) Ribotype network of wild tetraploid Avena species and their putative parental taxa. The radius of the circles on the ribotype network is proportional to the percent number of reads for each ribotype, as shown in Table 1. Major ribotypes are larger than others (more than 1000 reads per rDNA pool) and marked with numbers. The smallest circles correspond to ITS1 variants that have been read fewer than 1000 times. (b) Ribotype network of wild tetraploid Avena species and their putative parental taxa. More detailed picture of the major ribotypes.
Avena atlantica, on the other hand, shares its two major ribotypes, As1 (6353 reads, 39% of the rDNA pool) and As2 (4571 reads, 28%), with tetra- and hexaploid oats. The main ribotype of the AB-genome species A. barbata Pott ex Link is As1 (4894 reads, 33% of the rDNA pool). The same main ribotype was found in A. vaviloviana (10,884 reads, 47%). The second major ribotype of A. barbata (AB-genome) is species-specific (B4, 1014 reads, 6%). The AB-genome oat A. agadiriana B.R.Baum & Fedak has a species-specific main ribotype B1 (7136 reads, 28%), and its ribotype As2 is only the fourth of most represented (1018 reads, 4%). The second of the major ribotypes in A. agadiriana is B2 (4919 reads, 41%); it is shared with the minor ribotype fractions of A. atlantica (15 reads) and A. magna (18 reads). The third major ribotype of A. agadiriana is species-specific (1275 reads, 5%). Two well-represented ribotypes of the AB-genome species A. vaviloviana, B5 (3255 reads, 14%) and B6 (1162 reads, 5%), are shared with A. barbata (as minor ribotypes of it).
All the AC-genome oats have at least one A-genome ribotype inherited from A. atlantica—the second variant, As2 (Figure 1a,b). This is the main ribotype in the AC-genome oats A. insularis and A. murphyi (8297 reads, 39% and 6900 reads, 44%, respectively). It is also the third of major ribotypes in A. magna (3847, 15%). The latter species has a species-specific A-related main ribotype (Am1, 6067 reads, 24%). In addition, the second major ribotype of A. insularis is species-specific (Ai, 3770 reads, 18%). Avena murphyi has the second major ribotype, Amp (4443 reads, 28%), that belongs to the A-related group (Figure 1, Table 2). It is common in a minor fraction of ribotypes of A. magna (467 reads), A. insularis (196 reads), and A. agadiriana (33 reads). In these species, the C-genome sequences remained only in a minor fraction of the rDNA pool (from 78 to 10 reads, Figure 1a,b). The ribotype tree obtained by Bayesian inference (Figure 2) shows the clear distinction of the Ac-genome sequences (A. canariensis) from other A-genome oats (PP = 0.86, BS = 93), except for two minor ribotypes.
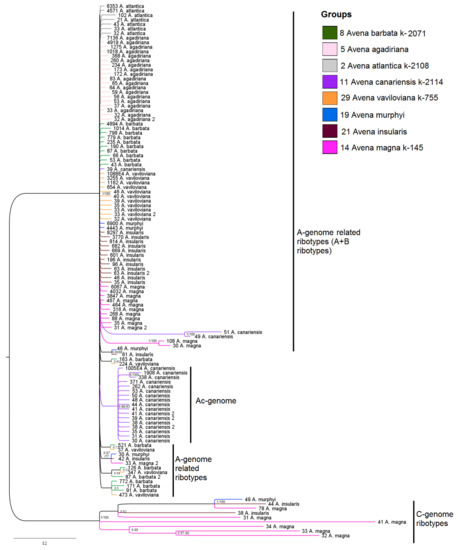
Figure 2.
Phylogenetic tree of the ribotypes of wild tetraploid Avena species and their putative parental species. The first index on the branch is the posterior probability in Bayesian inference, and the second is the bootstrap index obtained by the Maximum Likelihood algorithm. When only one index is shown on the branch, it is the posterior probability. Numbers before the species name indicate the number of reads per rDNA pool.
The C-genome-derived sequences of the AC-genome, in which the largest number of reads is 78 (PP = 1, BS = 100), form a separate cluster. The B-genome-related ribotypes in the tree do not separate from the A-related ones (Figure 2).
The second network (Figure 3a,b) depicts the relationships among hexaploids and their possible parental taxa. The main ribotypes of hexaploid oats A. fatua L., A. ludoviciana Durieu, and A. occidentalis (5203 reads, 42%; 4682 reads, 30%; and 4668 reads, 21%, respectively) are common and belong to a single family that does not occur in the tetraploids.
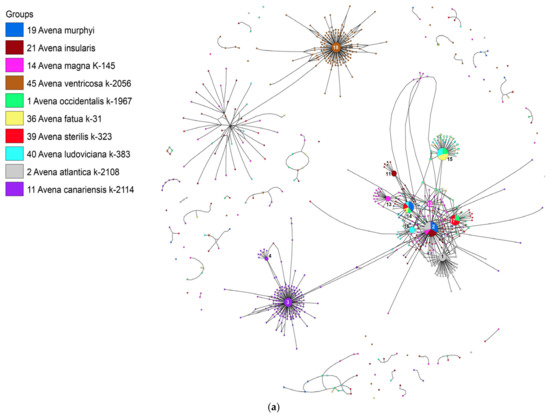

Figure 3.
(a) Ribotype network of wild hexaploid Avena species and their putative parental taxa. The radius of the circles on the ribotype network is proportional to the percentage of reads for each ribotype. Major ribotypes are larger than others (more than 1000 reads per rDNA pool) and marked with numbers. (b) Ribotype network of wild hexaploid Avena species and their putative parental taxa. More detailed picture of the major ribotypes.
These ribotypes can represent the D-genome. The A-related ribotypes of A. fatua (1608 reads, 13%), A. ludoviciana (1409 reads, 9%), and A. occidentalis (2694 reads, 12%) are homologous with the second major ribotype of A. murphyi (Amp). At the same time, A. sterilis and A. occidentalis have one common ribotype that can be named Ast. It is the main ribotype for A. sterilis (6138 reads, 47%) and the second for A. occidentalis (3763 reads, 17%). One of major ribotypes of A. ludoviciana (Ald, 3811 reads, 24%) is species-specific and closely related to the main ribotype of A. magna (Am1, 6067 reads, 24%) (Figure 3a,b). All major ribotypes of hexaploid Avena species are A-related, as are those of AC-genome species. The C-genome marker sequences remained only in a minor fraction (Figure 3a,b). They are related to the Cv-genome sequences of A. ventricosa but have accumulated many changes (Figure 3a,b). The ribotype tree shows the distinction of D-genome ribotypes (PP = 0.78, BS = 71), the separation of the main ribotypes of A. canariensis (PP = 0.88, BS = 98) and A. atlantica (PP = 0.86, BS = 73) (Figure 4).
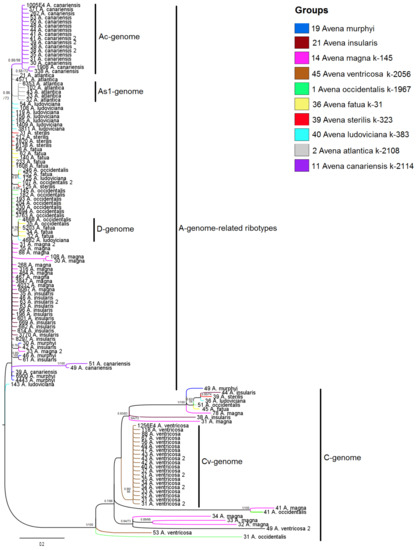
Figure 4.
Phylogenetic tree of the ribotypes of wild hexaploid Avena species and their putative parental species. The first index on the branch is the posterior probability in Bayesian inference, and the second is the bootstrap index obtained by the Maximum Likelihood algorithm. When only one index is shown on the branch, it is the posterior probability. Numbers before the species name indicate the number of reads per rDNA pool.
In addition, the strongly supported clade comprises the C-genome sequences of A. ventricosa and polyploids (PP = 1, BS = 100) (Figure 4). Avena ventricosa, in turn, occupies its own place in the C-genome clade (PP = 0.98, BS = 98), while C-genome-related sequences of other species form separate subclades (Figure 4). In the tree, the number of reads in the C-genome fraction varies from 53 to 31.
The third network shows the relationships between the ACD-genome hexaploids, A. clauda—a paleohomoploid species with the A- and C-genomes, and A. hirtula, the As-genome species.
The main ribotype of A. clauda (Cc1A, 5320 reads, 13%), together with one of the major ribotypes (Cc1B, 2695 reads, 7%), formed the C-genome ribotype network (26, b), but is not related to the C-genome-related sequences of the AC and ACD-genome species (Figure 5a,b). Meanwhile, the ribotypes of A. clauda CC2A–CC2C fell within the A-genome-related group [27], and some of them are shared with the A-genome ribotypes of polyploids (Figure 5a,b). Avena clauda ribotype CC2A (3195 reads, 8%) is common with A. ludoviciana, in which it is one of the major ribotypes (Ald, 3811 reads, 24%). The ribotype CC2B of A. clauda (1777 reads, 4%) is species-specific, and CC2C (1305 reads, 3%) is shared with the supposed the D-genome ribotype—the main ribotype of A. fatua, A. ludoviciana, and A. occidentalis (Figure 5a,b). We need to notice that possible the D-genome ribotype is common with a minor fraction of A. hirtula sequences (111 reads). The minor ribotype fraction of A. hirtula (579 reads) may be common with the As2 ribotype (Figure 5a,b). The ribotype tree shows the distinction of C-genome-related ribotypes (PP = 1, BS = 100, Figure 6).
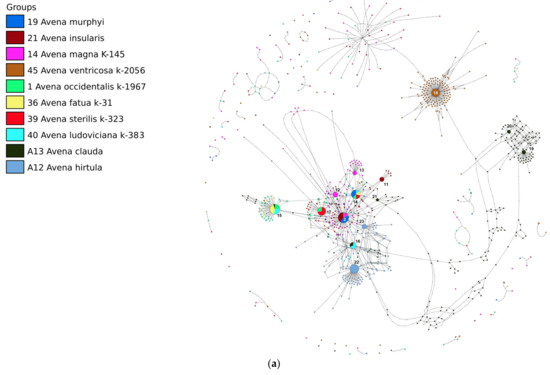
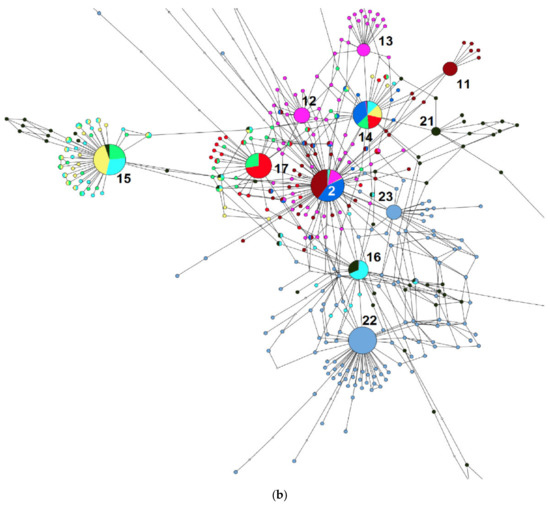
Figure 5.
(a) Ribotype network of wild hexaploid Avena species, the related tetraploids with AC-genome, and diploids A. clauda and A. hirtula. The radius of the circles on the ribotype network is proportional to the percentage of reads for each ribotype. Major ribotypes are larger than others (more than 1000 reads per rDNA pool) and marked with numbers. (b) Ribotype network of wild hexaploid Avena species, the related tetraploids with AC-genome, and diploids A. clauda and A. hirtula. More detailed picture of the major ribotypes.
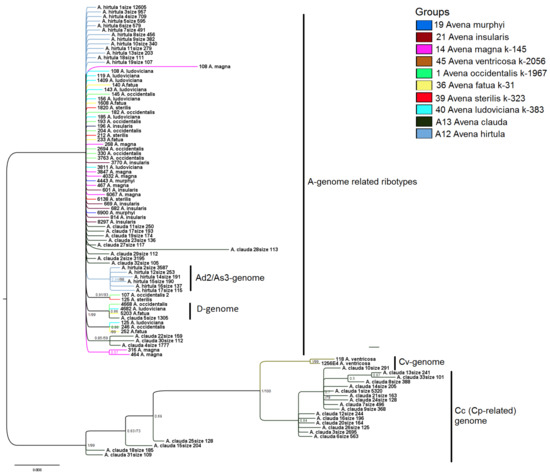
Figure 6.
Phylogenetic tree of the ribotypes of wild hexaploid Avena species, the related tetraploids with the AC-genome, and the diploids A. clauda and A. hirtula. The first index on the branch is the posterior probability in Bayesian inference, the second is the bootstrap index obtained by Maximum Likelihood algorithm. When only one index is shown on the branch it is the posterior probability. Numbers before the species name indicate number of the reads per rDNA pool. In the case of two species, A. clauda and A. hirtula number of reads per rDNA pool is placed after the word “size”.
The Cv-genome ribotypes on the tree form a clade within those of the C-genome group (PP = 1, BS = 99) that also contained the C-genome ribotypes of A. clauda (Figure 6). The A-genome-related ribotypes form separate clades that consist of the second major ribotype of A. hirtula with allies (PP = 0.94, BS = 0.98); the D-genome ribotypes with one of A. clauda (PP = 0.86, BS = 99); minor ribotypes of A. sterilis and A. occidentalis (PP = 0.91, BS = 93); minor ribotypes of A. ludoviciana, A. fatua, and A. occidentalis (PP = 0.86, BS = 89), and minor ribotypes of A. magna (PP = 0.57, BS unsupported).
3. Discussion
One of the main evolutionary pathways in the angiosperms and, in particular, in the grasses is allopolyploidization, which leads to new combinations of morphological features and adaptation to new environmental conditions. Then, after a series of massive chromosome rearrangements, a new chromosome set with the characteristics of a classic diploid may be established. Thus, for a complete understanding of the evolution in the active hybridizing group of some grass taxa, it is necessary to consider the chromosome set evolution against the background of the marker gene changes.
Previous research has found that two main groups of Avena chromosome sets—the A and C genomes—probably diverged from the common ancestor from 5 to 25 MYA, according to various estimates [32,33]. The A-genome also produced two variants, B and D, during its evolution [3,7,9,10,11]. Some recent studies placed the D-variant of the genome instead of the A-variant in previously defined AC-genome tetraploids [15,18,34], but our NGS analysis of marker sequences showed the presence of different variants of the A-genome not directly related to the putative D-genome in ACD-hexaploids. C-genome oats are characterized by serious differences in their ITS sequences [35], and C-genome tetraploid A. macrostachya is probably the most primitive species according to ITS sequences and karyological analysis [26,35]. Our NGS studies showed some other differences from previously obtained results, and in some cases, they clarified the hypotheses based on morphological and cytogenetic data.
Despite a significant susceptibility to homogenization, ITS sequences have been widely used in research for interspecific and, in some cases, intraspecific phylogenetic reconstructions for a rather long time, since the 1990s. “Due to their levels of variation, ITS frequently provide enough molecular markers suitable for evolutionary studies at the species level. These include topics such as the origin of polyploid taxa, hybridization, introgression, and, above all, phylogenetic inference (up to 40% sequence divergence in pair-wise comparisons between congeneric taxa reported in early studies)” [36].
Intraspecific rDNA polymorphism has long been constantly used to study introgression resulting from hybridization, including due to hypothetical horizontal transfer, as well as to build phylogenetic hypotheses [37,38,39], etc. Multiple nucleotide sequences of ITS (in this case, ITS2) obtained by the next-generation sequencing method were taken for the phylogenetic analysis of Abies [40], with a certain emphasis on the distinction of the related species. As one of the universal barcodes and very popular DNA markers, the ITS region is very popular for establishing the hybridization picture in the complicated subtribe Aveninae (Poeae, Poaceae) using chloroplast sequences (tpF–atpH, psbK–psbI, psbA–rps19–trnH, matK, trnL–trnF) as independent markers because of their mostly maternal inheritance [41,42]. Moreover, complex of these markers corroborates the division of hybrid complexes involving intersubtribal and even intertribal hybridizations [41]. In our analysis, we use the whole pool of rDNA marker sequences obtained via next-generation sequencing (NGS). The advantage of this method is that in the rDNA pool, the sequences of different parental taxa of the allopolyploid species are retained. These sequences usually cannot be amplified by direct sequencing because of nucleolar dominance Avena canariensis (a diploid with the Ac-genome) is endemic to the Canary Islands. It is thought to be the most ancient among the A-genome oats [15,43], having diverged 13–15 MYA. According to previous data, A. canariensis could be the A-genome donor to AC and ACD species [3], or only to A. agadiriana, which is supposed to be the AB-genome species [15]. Our results, on the contrary, indicate only a little possible participation of A. canariensis in the polyploid formation; the ribotype of the minor fraction of A. canariensis is shared with the As2-ribotype of A. atlantica [27]. The ribotype As2 of A. atlantica was probably inherited by AC-genome tetraploids.
The AB-genome tetraploids have the As1-ribotype in their genome (the main ribotype in A. atlantica). While As-genome-related ribotypes are also present in other diploids (A. wiestii, A. hirtula), only two major ribotypes of A. atlantica, As1 and As2, were detected in tetraploids and hexaploids. Avena barbata most often has two satellite chromosomes in its tetraploid karyotype; one of them is heterobrachial with a large satellite attached to the short arm of the chromosome, and the second chromosome is strongly heterobrachial with a small satellite [44]. Such a pattern of satellite chromosomes can determine the number of major ribotypes in A. barbata, the second major ribotype is species-specific, and it is only 6% of the rDNA pool. However, and it is interesting, sometimes the third satellite chromosome occurs [44], and thus theoretically a new ribotype pattern can be observed. Avena barbata belongs to the AB-genome tetraploids; species-specific ribotypes can be variants of the B-genome. Two short awns on the lemma apex of A. barbata can show us the relationship of this species with primary diploids. The second AB-genome species, A. vaviloviana, has three major ribotypes; two of them are shared with minor ribotypes of A. barbata. The chromosomes of A. vaviloviana are prone to translocations, including those in satellite regions [15]. Thus, the increasing number of major ribotypes can be connected with the translocations. The second and third major ribotypes (14% and 5%, respectively) are related to the A-genome. They can be regarded as the species-specific variants of the B-genome sequences. Avena vaviloviana was at first thought to be a member of A. strigosa Schreb. affinity, a subspecies of A. strigosa [45,46]. Their morphological resemblance may reflect the participation of the As-genome in this species.
The ribotype pattern in A. agadiriana differs from that of other AB-genome oats. Avena agadiriana has a special karyotype that is not found in any other tetraploids; in particular, it has two pairs of chromosomes with very small satellites and drastically asymmetrical centromeres [47]. In addition, early flowering forms of this species have three pairs of satellites on their chromosomes instead of two [48]. Nevertheless, its karyotype was designated as AB [8,18,34,49,50]. Avena wiestii and A. canariensis were supposed to be the ancestors of A. agadiriana by previous studies [7,8,34]. The results of FISH analysis of the karyotype of A. agadiriana, surprisingly, suggested determining its chromosome set as DD tetraploid instead of AB [15]. Our data, on the other hand, did not show a clear relationship of ribotypes of A. agadiriana and D-genome ribotypes of ACD-hexaploids. Instead, A. agadiriana has a species-specific main ribotype (Figure 1a,b) and two major ribotypes, one of which is shared with A. atlantica and A. magna (those among their minor ribotypes). Only the fourth of the ribotypes of A. agadiriana is shared with some other species as their major ribotype; it belongs to As2. The main ribotype of A. agadiriana, along with its different major ribotypes, can be derived from the A-genome and thus represent B-variants (specifically B1, B2, and B3). According to NGS data, A. agadiriana ribotype constitution may be described as BA instead of AB-variant as in A. vaviloviana and A. barbata because the main ribotype possibly represents B-variant, a derivative of ancestral A-genome. This special ribotype pattern may reflect the peculiar chromosome set of A. agadiriana. Results of the analysis of NGS data also confirmed the previous morphological observations: A. agadiriana resembles A. barbata in inflorescence characters [47]. Our data do not show the close relationship of A. agadiriana with A. canariensis (Ac-genome); it is related to As-genome of A. atlantica as well as to other AB-genome species.
Avena magna, A. murphyi, and A. insularis probably have the AC-genome constitution, according to our analysis. Previously, the AC-karyotype in these species was determined by various cytogenetic methods [3,11,13,51]. FISH-analysis with probes on the different genomes and chromosome assignments, on the contrary, detected the CD-genome variant in the AC-genome oats [15,18,19]. Similar results were presented by chromosome assignment in the linkage map [52]. Our NGS data obtained by sequencing the whole rDNA pool showed different ribotype patterns in A. magna, A. murphyi, and A. insularis species, though all these oats have ribotypes inherited from the A-genome of diploids. Avena magna has two species-specific major ribotypes as well as one As2-ribotype common with A. atlantica (Figure 1a,b). Avena magna has three satellite chromosomes and also the A-genome chromosomes, which are more similar to than those of hexaploids than of oats with the Ac-genome [53,54]. Cytogenetic studies also did not support the presence of a B-genome in A. magna. Specific major ribotypes in A. magna are very closely related to the A-genome ribotype variant (Table 2) and are not shared with the hexaploids, which have specific ribotypes. Thus, the main ribotype of A. magna can represent its peculiar genome that can pair with D-related probes, as revealed in the cytogenetic study by Tomaszewska et al. [15], but also with the A. magna genomic set, which retained possible ancestral sequences. The C-genome ribotypes in all AC-genome species of oats remained only in minor fraction (Figure 1 and Figure 3a,b). There is a main network in the picture that contains the A-genome-related ribotypes (Figure 3a,b). The C-genome ribotypes, are related to the ribotypes of A. ventricosa as we see in Figure 3a,b, but have not any common ribotypes with it. Possibly, the C-genome-derived sequences in the allopolyploid genomes of tetra- and hexaploid species were drastically transformed through the post-hybridization processes. The elimination of the C-genome sequences in rDNA pool of allopolyploid species probably occurred because of significant differences between the sequences of two genome types [55].
From a morphological point of view, A. murphyi (AC) is close to A. magna (AC), and A. insularis (AC) is intermediate between A. murphyi and hexaploid A. sterilis (ACD) [11]. Avena murphyi and A. insularis are characterized by two denticles on the lemma apex [11]. The most represented ribotype in the rDNA pool of A. murphyi and A. insularis is As2 (Figure 1a,b and Figure 3a,b, Table 2). As in A. magna, this fact may point to the participation of A. atlantica (or some extinct closely related diploid species) in the formation of a tetraploid. We previously assumed that only A. atlantica has two specific ribotypes, As1 and As2 [27], that were retained in the polyploid genome set (Figure 1a,b and Figure 3a,b), whereas other species with the As-genome, A. wiestii and A. hirtula, have their own peculiar ribotypes that are partially common with the Ad-genome species and not further inherited by polyploid species. Both A. murphyi and A. insularis have two pairs of satellite chromosomes [13,56]. This probably caused the presence of two major ribotypes in these species (Figure 1a,b and Figure 3a,b). The second major ribotype of A. insularis is species-specific, and that of A. murphyi is common with minor ribotypes of A. magna, A. agadiriana, and A. insularis. While FISH-analysis with genome-specific probes detected the CD-genome structure of A. insularis and A. murphyi [15] our data showed a closer relationship of these species and the A-genome species and a drastic elimination of the C-genome sequences from their rDNA. Furthermore, the As-genome of A. insularis and A. murphyi (their main ribotype) is not present among the ribotypes of hexaploids that can be attributed to the D-genome (Figure 3a,b). The second most represented ribotype in A. murphyi (variant Amp) could be A-genome ribotype inherited by the ACD-genome hexaploids (Figure 3a,b).
The ACD-genome hexaploid group contains the most widespread oat species, A. fatua. The D-genome is thought to be a derivative of the A-genome [11,20,21]. In this group, the A-genome comprises two pairs of satellite chromosomes (one pair with a large satellite and the other with a small satellite), and the C and D-genomes each have one pair of satellite chromosomes with small satellite [57,58]. It is interesting that at first their karyotype constitution was determined as ABC [59]. Recent studies supported the ACD-genome variant in hexaploids [15] and suggested different hypotheses about their origin [19]. For example, SSR-FISH analysis of the hexaploid species A. byzantina and A. sativa showed A. insularis as a tetraploid species closest to hexaploid ones [60]. FISH-analysis with genome-specific probes revealed A. strigosa as the A-genome progenitor of the ACD-oats while A. insularis is thought to be the CD-genome donor of A. sterilis which is, in turn, the ancestral of all hexaploids [15]. Our data clearly supported the ACD-genome constitution for the studied hexaploid species (Figure 3a,b) although the pattern of relationships between tetra- and hexaploids differs from the assumptions based on FISH data [15]. The most frequent ribotype in the species A. occidentalis, A. fatua, and A. ludoviciana is specific variant that does not occur in any other diploid or polyploid species except paleopolyploid A. clauda (see below). This variant can be described as the D-subgenome. However, the main ribotype in hexaploid A. sterilis differs from the most represented ribotypes of other hexaploids. It is common with the second major ribotype of A. occidentalis. This ribotype is connected to the A-subgenome (Figure 3a,b) and can be named Ast or D’ (Table 2). Avena sterilis was thought to be an ancestral species for the hexaploids [45,61]. On the contrary, specific major ribotypes of A. sterilis and A. occidentalis may suggest the separate origin of these species, independent from other ACD-genome oats. The second of the major ribotypes of A. fatua is shared with A. murphyi, its second major ribotype, which may be named as Amp (Figure 3a,b, Table 2). This contradicts the previous hypotheses of A. canariensis as the Ac-genome progenitor of all hexaploids [3,11], and also of A. magna as the parental taxon for the hexaploids [62]. Instead, A. murphyi could form its specific ribotype in its polyploid evolution and then participate in the formation of A. fatua and other hexaploids. Avena ludoviciana is treated as a relative of A. sterilis by morphological criteria [11]. It has the species-specific second major ribotype along with possible D-genome-related and amp ribotypes (Figure 3a,b, Table 2). The specific ribotype Ald belongs to the A-genome group. According to the ribotype pattern, A. ludoviciana is not closely related to A. sterilis. Based on our NGS sequence analysis, A. ludoviciana, A. occidentalis, and A. fatua form the natural group of affinity, where A. fatua has fewer variants of ribotypes than others. Thus, A. fatua probably could have gone through the stages of hybrid stabilization or evolved independently from other hexaploid species. Regarding the results of the NGS analysis, the C-genome rDNA could derive from A. ventricosa or some extinct the Cv-genome species (Figure 3a,b and Figure 4), though its sequences have been probably greatly transformed by post-hybridization processes such as conversion. They were retained only in minor fraction being strongly different from the prevailing sequences of the A-genome. It supports the previous results of ITS analysis [63]. All sequences of hexaploid oats that were obtained by Sanger method clearly group with A-genome taxa [63]. This fact can indicate the elimination of the distantly related sequences from the pool rDNA of complex hybrid (see also [64], IGS data) and probably inactivating of some sequences for establishing the genome balance [55,65,66].
We also need to mention that the As-genome diploid oat, A. hirtula (or its ancestor), could participate in the formation of polyploid species. Major ribotypes of A. hirtula are not shared with any of the polyploids, but some sequences of its minor fraction are shared with A. ludoviciana—its second major ribotype (Figure 5a,b). In addition, one ribotype of the minor fraction of A. hirtula, in addition, is identical with As2-ribotype that is common for A. murphyi, A. magna, and A. insularis. Previous data placed different marker sequences of A. hirtula near those of A. strigosa [67] or in a clade with A. atlantica and A. lusitanica (Tab.Morais) B.R.Baum (=A. barbata), sister to clade including the hexaploids [7]. As was supposed in previous studies, A. hirtula probably evolved from A. atlantica [11,15] and thus it could keep ancestral ribotype variants in its minor fraction of rDNA.
One of the most interesting questions is the possible participation of the paleopolyploid (now diploid) species A. clauda in the formation of hexaploids. Phylogenetic data obtained from low copy sequences showed that A. clauda was more closely related to the hexaploids than A. ventricosa [7]. However, our data indicated that C-genome rDNA in tetra- and hexaploid oats was obtained from A. ventricosa. Avena clauda, beinga putative ancient hybrid [26,27], in turn had A-genome-related ribotypes in its rDNA pool that could further form the D-genome rDNA in A. occidentalis, A. fatua, and A. ludoviciana. The possible belonging of the major ribotypes of A. occidentalis, A. fatua, and A. ludoviciana to the D-genome is proved by the fact that these sequences do not occur in any other studied oats except hexaploids. However, chromosome constitution of A. clauda characterizes this species as the C-genome bearer [46]. Four chromosomes Therefore, A. clauda could be the ancient hybrid between A- and C-genome oats from which the A-genome chromosomes were eliminated but some rDNA sequences have been retained after the post-hybridization transformation. In addition, one of the major A-genome-related ribotypes of A. clauda could be inherited by A. ludoviciana (Figure 5a,b). From the morphological point of view, A. clauda propagates by separate caryopses instead of the whole spikelets and has very unequal glumes; so, it may be one of the most primitive oats [3,56]. Probably, A. clauda or some extinct relative of this species participated in the formation of at least some hexaploids with the D-genomes.
4. Conclusions
NGS data, as we see, can be a convenient tool for phylogenetic reconstruction of the evolution of hybrid species. A rather small but stable quantity of different nucleotide positions, combined with the data obtained by cytogenetic methods, can provide us with detailed information about the allopolyploidization patterns in the genera of grasses. Our results supported some earlier hypotheses about the origin of polyploid Avena species. We assume that tetraploid oats that were previously defined as AC-genome species actually have this genomic constitution according to the marker ITS1 sequences. The D-genome sequences were not found in the AC-genome tetraploid oats that were related to those with the ACD-genome. The A-genome-related sequences were possibly inherited from the As-genome species A. atlantica, while the D-genome-related sequences were formed already in hexaploid oats or were taken from an unknown ancestor related to A. clauda. The A-genome sequences in the ACD-genome Avena species were probably inherited from A. murphyi (AC). Alternatively, the AC-genome-related ITS1 sequences could persist in the CD-genome oats because of translocation events detected by Yan et al. [59] when the A-genome chromosomes were transformed into the D-genome ones after a series of introgressive hybridizations.
5. Materials and Methods
We took into analysis 13 wild species of Avena belonging to three genome groups: AB (A. agadiriana, A. barbata, and A. vaviloviana), AC (A. insularis, A. magna, and A. murphyi), and ACD (A. fatua, A. ludoviciana, A. occidentalis, and A. sterilis). In addition, we studied putative diploid progenitors of the tetraploids—A. atlantica (the As-genome), A. canariensis (Ac), and A. ventricosa (Cv). All material was taken from the Federal Research Center N. I. Vavilov All-Russian Institute of Plant Genetic Resources (VIR). A summary of the studied species is presented in Table 1.
Genomic DNA was extracted from dried leaf and seed material using a Qiagen Plant Mini Kit (Qiagen Inc., Hilden, Germany). For NGS analysis, we took 18S–ITS1–5.8S rDNA region. The fragments were amplified and sequenced at the Center for Shared Use “Genomic Technologies, Proteomics, and Cell Biology” of the All-Russian Research Institute of Agricultural Microbiology on an Illumina Platform MiSeq. PCR was carried out in 15 µL of the reaction mixture containing 0.5–1 unit of activity of Q5® High-Fidelity DNA Polymerase (NEB, Ipswich, MA, USA), 5 pM of forward and reverse primers, 10 ng of DNA template, and 2 nM of each dNTP (Life Technologies, ThermoScientific, Waltham, MA, USA). It was amplified under the following conditions: initial denaturation at 94 °C for 1 min, followed by 25 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, and a final elongation 72 °C for 5 min using ITS 1P [68] and ITS 2 [69] primers. PCR products were purified according to the Illumina recommended method using AMPureXP (Beckman Coulter, Indianapolis, IN, USA). Then the libraries were prepared according to the manufacturer’s MiSeq Reagent Kit Preparation Guide (Illumina) (http://web.uri.edu/gsc/files/16s-metagenomic-library-prep-guide-15044223-b.pdf (accessed on 11 May 2020)). They were sequenced on an Illumina MiSeq instrument (Illumina, San Diego, CA, USA) using a MiSeq® ReagentKit v3 (600 cycles) with double-sided reading (2 × 300 n) following the manufacturer’s instructions. The sequences were trimmed with Trimmomatic [70], included in Unipro Ugene [71] using the following parameters: PE reads, sliding window trimming with size 4, quality threshold 12, and minimal read length 130. Further, paired marker sequences were combined, dereplicated, and sorted into the ribotypes with the aid of vsearch 2.7.1 [72]. Then the sequences that represent ribotypes with certain frequency in the whole genome pool were analyzed by TCS 1.21 [73] and visualized in TCSBU [74]. The resulting network was built according to the statistical parsimony algorithm and shows possible hybridization events in the species complex. The threshold for this analysis was 10 reads per the whole genome pool. Additionally, we built phylogenetic trees of the ribotypes by Bayesian inference and maximum likelihood method. The threshold for tree inference was 30 reads per the rDNA pool, except the dataset with hexaploid species and Avena clauda, here we took the threshold 100 reads per the rDNA pool. In the first case we used Mr. Bayes 3.2.2 [75] and in the second case, iqtree 1.6.12 (http://www.iqtree.org/, accessed on 1 March 2023) using the GTR+G model previously estimated by MEGA XI [76]. Bayesian analysis was conducted with 5–8 million of generations, sampling trees every 100 generations. and the first 25% trees were discarded as burn-in. The ML search was performed under the fast bootstrap option, 1000 generations. The obtained trees were edited in FigTree 1.4.3 (http://tree.bio.ed.ac.uk/, accessed on 1 March 2023). The resulting tree combines Bayesian and ML data. The first index is posterior probability, and the second is bootstrap index.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/d15060717/s1, Table S1. Aligned ribotype sequences of the tetraploid oats and their probable parental species obtained via NGS. Table S2. Aligned ribotype sequences of the hexaploid oats and their probable parental species obtained via NGS. Table S3. Aligned ribotype sequences of the hexaploid oats, their tetraploid relatives with AC-genome the and diploids A. clauda and the A. hirtula.
Author Contributions
A.A.G. and N.N.N. carried out the experiments. A.A.G., N.N.N. and A.V.R. analyzed the data. I.G.L. and E.V.B. provided seed material. A.A.G., N.N.N. and A.V.R. wrote the manuscript. V.S.S. thoroughly corrected and edited the text. All authors have read and agreed to the published version of the manuscript.
Funding
Our research was supported by the Russian Science Foundation Grant No. 22-24-01085 (A.A.G., N.N.N., performing NGS, obtaining the material) and partially by No. 22-24-01117 (A.V.R., sample preparation, DNA isolation, data analysis).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
The Supplemetary data is available on https://susy.mdpi.com/user/manuscripts/review_info/6ea101d12ba7c1bdfd8fcf849f38d5c3#:~:text=manuscript%2Dsupplementary.7z (accessed on 23 March 2023).
Acknowledgments
The authors are grateful to A.G. Pinaev and all researchers of the Center for Shared Use “Genomic Technologies, Proteomics and Cell Biology” of the All-Russian Research Institute of Agricultural Microbiology for next-generation sequencing, to E.M. Machs for invaluable help in data processing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mitchell, C.C.; Parkinson, S.E.; Baker, T.J.; Jellen, E.N. C-banding and localization of 18S–5.8S S-26S rDNA in tall oatgrass species. Crop. Sci. 2003, 43, 32–36. [Google Scholar] [CrossRef]
- Leggett, J.M.; Markland, G.S. The genomic structure of Avena revealed by GISH. In Kew Chromosome Conference IV; Brandham, P.E., Bennett, M.D., Eds.; Royal Botanic Gardens: Kew, UK, 1995; pp. 133–139. [Google Scholar]
- Loskutov, I.G.; Rines, H.W. Avena. In Wild Crop Relatives: Genomic and Breeding Resources; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; Chapter 3; pp. 109–183. [Google Scholar]
- Stebbins, G.L. Chromosomal Evolution in Higher Plants; Edward Arnold: London, UK, 1971; 216p. [Google Scholar]
- Fabijanski, S.; Fedak, G.; Armstrong, K.; Altosaar, I. A repeated sequence probe for the C genome in Avena (oats). Theor. Appl. Genet. 1990, 79, 1–7. [Google Scholar] [CrossRef]
- Katsiotis, A.; Hagidimitriou, M.; Heslop-Harrison, J.S. The close relationship between the A and B genomes in Avena L. (Poaceae) determined by molecular cytogenetic analysis of total genomic, tandemly and dispersed repetitive DNA sequences. Ann. Bot. 1997, 79, 103–109. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, P.; Zhao, J.; Li, J.; Lai, S.; Tinker, N.A.; Liao, S.; Yan, H. Phylogenetic relationships in the genus Avena based on the nuclear Pgk1 gene. PLoS ONE 2018, 13, e0200047. [Google Scholar] [CrossRef]
- Chew, P.; Meade, K.; Hayes, A.; Harjes, C.; Bao, Y.; Beattie, A.D.; Puddephat, I.; Gusmini, G.; Tanksley, S.D. A study on the genetic relationships of Avena taxa and the origins of hexaploid oat. Theor. Appl. Gen. 2016, 129, 1405–1415. [Google Scholar] [CrossRef]
- Chen, Q.; Armstrong, K. Genomic in situ hybridization in Avena sativa. Genome 1994, 37, 607–612. [Google Scholar] [CrossRef]
- Linares, C.; González, J.; Ferrer, E.; Fominaya, A. The use of double fluorescence in situ hybridization to physically map the positions of 5S rDNA genes in relation to the chromosomal location of 18S-5.8S-26S rDNA and a C genome specific DNA sequence in the genus Avena. Genome 1996, 39, 535–542. [Google Scholar] [CrossRef]
- Loskutov, I.G. Oat (Avena L.). Distribution, Taxonomy, Evolution and Breeding Value; VIR: St. Petersburg, Russia, 2007; 336p. [Google Scholar]
- Leggett, J.M.; Thomas, H. Oat evolution and cytogenetics. In The Oat Crop Production and Utilization; Welch, R.W., Ed.; Chapman & Hall: London, UK, 1995; pp. 120–149. [Google Scholar]
- Ladizinsky, G. A new species of Avena from Sicily, possible the tetraploid progenitor of hexaploid oats. Genet. Resour. Crop Evol. 1998, 45, 263–269. [Google Scholar] [CrossRef]
- Li, C.D.; Rossnagel, B.G.; Scoles, G.J. The development of oar microsatellite markers and their use in identifying relationships among Avena species and oat cultivars. Theor. Appl. Genet. 2000, 101, 1259–1268. [Google Scholar] [CrossRef]
- Tomaszewska, P.; Schwarzacher, T.; Heslop-Harrison, J.S. Oat chromosome and genome evolution defined by widespread terminal intergenomic translocations in polyploids. Front. Plant Sci. 2022, 13, 1026364. [Google Scholar] [CrossRef]
- Yan, H.; Ren, Z.; Deng, D.; Yang, K. New evidence confirming the CD genomic constitutions of the tetraploid Avena species in the section Pachycarpa Baum. PLoS ONE 2021, 16, e0240703. [Google Scholar] [CrossRef]
- Morikawa, T. Isozyme and chromosome polymorphisms of the genus Avena and its geographic distribution in Morocco. Wheat Inform. Serv. 1991, 72, 104–105. [Google Scholar]
- Yan, H.-H.; Baum, B.R.; Zhou, P.-P.; Zhao, J.; Wei, Y.-M.; Ren, C.-Z.; Xiong, F.-Q.; Liu, G.; Zhong, L.; Zhao, G.; et al. Phylogenetic analysis of the genus Avena based on chloroplast intergenic spacer psbA-trnH and single-copy nuclear gene Acc1. Genome 2014, 57, 267–277. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, H.; Guo, L.; Deng, C.; Wang, C.; Wang, Y.; Kang, L.; Zhou, P.; Yu, K.; Dong, X.; et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 2022, 54, 1248–1258. [Google Scholar] [CrossRef]
- Jellen, E.N.; Phillips, R.L.; Rines, H.W. Chromosomal localization and polymorphisms of ribosomal DNA in oat (Avena spp.). Genome 1994, 37, 23–32. [Google Scholar] [CrossRef]
- Linares, C.; Ferrer, E.; Fominaya, A. Discrimination of the closely related A and D genomes of the hexaploid oat Avena sativa L. PNAS 1998, 95, 12450–12455. [Google Scholar] [CrossRef] [PubMed]
- Ladizinsky, G. New evidence on the origin of hexaploidy oats. Evolution 1969, 23, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Murphy, H.C.; Sadanaga, K.; Zilinsky, F.J.; Terrell, E.; Smith, R.T. Avena magna: An important new tetraploid species in oats. Science 1968, 159, 103–104. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jellen, E.N.; Murphy, J.P. Progenitor germplasm of domesticated hexaploid oat. Crop. Sci. 1999, 39, 1208–1214. [Google Scholar] [CrossRef]
- Baum, B.R. Avena occidentalis, a hitherto overlooked species of oats. Can. J. Bot. 1971, 49, 1055–1057. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Machs, E.M.; Blinova, E.V.; Probatova, N.S.; Langdon, T.; Rodionov, A.V. New insights into the genomic structure of the oats (Avena L., Poaceae): Intragenomic polymorphism of ITS1 sequences of rare endemic species Avena bruhnsiana Gruner and its relationship to other species with C-genomes. Euphytica 2022, 218, 3. [Google Scholar] [CrossRef]
- Gnutikov, A.A.; Nosov, N.N.; Loskutov, I.G.; Blinova, E.V.; Shneyer, V.S.; Probatova, N.S.; Rodionov, A.V. New Insights into the Genomic Structure of Avena L.: Comparison of the Divergence of A-Genome and One C-Genome Oat Species. Plants 2022, 11, 1103. [Google Scholar] [CrossRef]
- Brassac, J.; Blattner, F.R. Species-Level Phylogeny and Polyploid Relationships in Hordeum (Poaceae) Inferred by Next-Generation Sequencing and In Silico Cloning of Multiple Nuclear Loci. Syst. Biol. 2015, 64, 792–808. [Google Scholar] [CrossRef]
- Schultz, J.; Maisel, S.; Gerlach, D.; Müller, T.; Wolf, M. A common core of secondary structure of 33 the internal transcribed spacer 2 (ITS2) throughout the Eukaryota. RNA 2005, 11, 361–364. [Google Scholar] [CrossRef]
- Coleman, A.W. Nuclear rRNA transcript processing versus internal transcribed spacer 29 secondary structure. Trends Genet. 2015, 31, 157–163. [Google Scholar] [CrossRef]
- Zhang, X.; Cao, Y.; Zhang, W.; Simmons, M.P. Adenine· cytosine substitutions are an alternative 13 pathway of compensatory mutation in angiosperm ITS2. RNA 2020, 26, 209–217. [Google Scholar] [CrossRef]
- Peng, Y.Y.; Wei, Y.M.; Baum, B.R.; Zheng, Y.L. Molecular diversity of the 5S rRNA gene and genomic relationships in the genus Avena (Poaceae: Aveneae). Genome 2008, 51, 137–154. [Google Scholar] [CrossRef]
- Maughan, P.J.; Lee, R.; Walstead, R.; Vickerstaff, R.J.; Fogarty, M.C.; Brouwer, C.R.; Reid, R.R.; Jay, J.J.; Bekele, W.A.; Jackson, E.W.; et al. Genomic insights from the first chromosome-scale assemblies of oat (Avena spp.) diploid species. BMC Biol. 2019, 17, 92. [Google Scholar] [CrossRef]
- Peng, Y.-Y.; Baum, B.R.; Re, C.-Z.; Jiang, Q.-T.; Chen, G.-Y.; Zheng, Y.-L.; Wei, Y.-M. The evolution pattern of rDNA ITS in Avena and phylogenetic relationship of the Avena species (Poaceae: Aveneae). Hereditas 2010, 147, 183–204. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Tiupa, N.B.; Kim, E.S.; Machs, E.M.; Loskutov, I.G. Genomic structure of the autotetraploid oat species Avena macrostachya inferred from comparative analysis of the ITS1 and ITS2 sequences: On the oat karyotype evolution during the early stages of the Avena species divergence. Genetika 2005, 41, 646–656. (In Russian) [Google Scholar]
- Baldwin, B.G.; Sanderson, M.J.; Porter, J.M.; Wojciechowski, M.F.; Campbell, C.S.; Donoghue, M.J. The ITS Region of Nuclear Ribosomal DNA: A Valuable Source of Evidence on Angiosperm Phylogeny. Ann. Mis. Bot. Gard. 1995, 82, 247–277. [Google Scholar] [CrossRef]
- Mahelka, V.; Krak, K.; Kopecký, D.; Fehrer, J.; Šafář, J.; Bartoš, J.; Hobza, R.; Blavet, N.; Blattner, F.R. Multiple horizontal transfers of nuclear ribosomal genes between phylogenetically distinct grass lineages. Biol. Sci. 2017, 114, 1726–1731. [Google Scholar] [CrossRef] [PubMed]
- Sochorová, J.; Garcia, S.; Gálvez, F.; Symonová, R.; Kovařík, A. Evolutionary trends in animal ribosomal DNA loci: Introduction to a new online database. Chromosoma 2018, 127, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.D.P.N.; Riina, R.; Valduga, E.; Caruzo, M.B.R. A new species of Croton (Euphorbiaceae) endemic to the Brazilian Pampa and its phylogenetic affinities. Pl. Syst. Evol. 2022, 308, 14. [Google Scholar] [CrossRef]
- Suyama, Y.; Hirota, S.K.; Matsuo, A.; Tsunamoto, Y.; Mitsuyuki, C.; Shimura, A.; Okano, K. Complementary combination of multiplex high-throughput DNA sequencing for molecular phylogeny. Ecol. Res. 2022, 37, 171–181. [Google Scholar] [CrossRef]
- Saarela, J.M.; Bull, R.D.; Paradis, M.J.; Ebata, S.N.; Peterson, P.M.; Soreng, R.J.; Paszko, B. Molecular phylogenetics of cool-season grasses in the subtribes Agrostidinae, Anthoxanthinae, Aveninae, Brizinae, Calothecinae, Koeleriinae and Phalaridinae (Poaceae, Pooideae, Poeae, Poeae chloroplast group 1). PhytoKeys 2017, 87, 1–139. [Google Scholar] [CrossRef]
- Tkach, N.; Schneider, J.; Döring, E.; Wölk, A.; Hochbach, A.; Nissen, J.; Winterfeld, G.; Meyer, S.; Gabriel, J.; Hoffmann, M.H.; et al. Phylogenetic lineages and the role of hybridization as driving force of evolution in grass supertribe Poodae. Taxon 2020, 69, 234–277. [Google Scholar] [CrossRef]
- Fu, Y.B. Oat evolution revealed in the maternal lineages of 25 Avena species. Sci. Rep. 2018, 8, 4252. [Google Scholar] [CrossRef]
- Ladizinsky, G. The cytogenetic position of Avena prostrata among the diploid oats. Can. J. Genet. Cytol. 1973, 15, 443–450. [Google Scholar] [CrossRef]
- Malzew, A.I. Wild and Cultivated Oat. Sectio Euavena Griseb; Publishers of the All-Union Institute of Applied Botany and New Cultures under the Council of People’s Commissars of the USSR: Leningrad, Russia, 1930; 522p. [Google Scholar]
- Emme, E.K. Karyosystematic research of oat of section Euavena Griseb. Works Appl. Bot. Plant Breed Seria II 1932, 1, 147–168. [Google Scholar]
- Baum, B.R.; Fedak, G. A new tetraploid species of Avena discovered in Morocco. Can. J. Bot. 1985, 63, 1379–1385. [Google Scholar] [CrossRef]
- Morikawa, T. Isozyme and chromosome variations of the Avena species in the Canary Islands and Morocco. In Proceedings of the 4th International Oat Conference, Adelaide, SA, Australia, 19–23 October 1992; Volume 3, pp. 138–140. [Google Scholar]
- Leggett, J.M. Interspecific diploid hybrids in Avena. Genome 1989, 32, 346–348. [Google Scholar] [CrossRef]
- Badaeva, E.D.; Shelukhina, O.; Diederichsen, A.; Loskutov, I.G.; Pukhalskiy, V.A. Comparative cytogenetic analysis of Avena macrostachya and diploid C-genome Avena species. Genome 2010, 53, 125–137. [Google Scholar] [CrossRef]
- Murray, B.E.; Craig, J.L.; Rajhathy. T. A protein electrophoretic study of three amphiploids and eight species in Avena. Can. J. Genet. Cytol. 1970, 12, 651–655. [Google Scholar] [CrossRef]
- Yan, H.; Bekele, W.A.; Wight, C.P.; Peng, Y.; Langdon, T.; Latta, R.G.; Fu, Y.-B.; Diederichsen, A.; Howarth, C.J.; Jellen, E.N.; et al. High-density marker profiling confirms ancestral genomes of Avena species and identifies D-genome chromosomes of hexaploid oat. Theor. Appl. Genet. 2016, 129, 2133–2149. [Google Scholar] [CrossRef]
- Sadasivaiah, R.S.; Rajhathy, T. Genome relationships in tetraploid Avena. Can. J. Genet. Cytol. 1968, 10, 655–669. [Google Scholar] [CrossRef]
- Rajhathy, T. Chromosome polymorphism in Avena ventricosa. Chromosoma 1971, 35, 206–216. [Google Scholar] [CrossRef]
- Rodionov, A.V.; Shneyer, V.S.; Gnutikov, A.A.; Nosov, N.N.; Punina, E.O.; Zhurbenko, P.M.; Loskutov, I.G.; Muravenko, O.V. Species dialectics: From initial uniformity, through the greatest possible diversity to ultimate uniformity. Bot. Zhurn. 2020, 105, 835–853. (In Russian) [Google Scholar] [CrossRef]
- Rajhathy, T.; Thomas, H. Cytogenetics of Oats (Avena L.) Miscellaneous Publications of the Genetics Society of Canada 2; Ontario (Canada) Genetics Society of Canada: Ottawa, ON, Canada, 1974; 90p. [Google Scholar]
- Rajhathy, T. A standard kariotype for Avena sativa. Can. J. Genet. Cytol. 1963, 5, 127–132. [Google Scholar] [CrossRef]
- Rajhathy, T.; Morrison, J.W. Chromosome morphology in the genus Avena. Can. J. Bot. 1959, 37, 331–337. [Google Scholar] [CrossRef]
- Nishiyama, I. The genetic and cytology of certain cereals. I. Morphological and cytological studies in triploid, pentaploid and hexaploid Avena hybrids. Jpn. J. Genet. 1929, 5, 1–48. [Google Scholar] [CrossRef]
- Fominaya, A.; Loarce, Y.; González, J.M.; Ferrer, E. Cytogenetic evidence supports Avena insularis being closely related to hexaploid oats. PLoS ONE 2021, 16, e0257100. [Google Scholar] [CrossRef] [PubMed]
- Ladizinsky, G.; Zohary, D. Notes on species delimination species relationships and poliploidy in Avena L. Euphytica 1971, 20, 380–395. [Google Scholar] [CrossRef]
- Sadanaga, K. Chromosome Homology of Avena magna Murphy and Terrell and the Hexaploid Species A. sativa L. and A. byzantina C. Koch. Crop Sci. 1976, 16, 425–428. [Google Scholar] [CrossRef]
- Nikoloudakis, N.; Skaracis, G.A.; Katsiotis, A. Evolutionary insights inferred by molecular analysis of the ITS1-5.8S-ITS2 and IGS Avena sp. sequences. Mol. Phyl. Evol. 2008, 46, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.; Viegas, W.; Silva, M. 45S rDNA external transcribed spacer organization reveals new phylogenetic relationships in Avena genus. PLoS ONE 2017, 12, e0176170. [Google Scholar] [CrossRef]
- Clarkson, P.M.; Hoffman, E.P.; Zambraski, E.; Gordish-Dressman, H.; Kearns, A.; Hubal, M.; Harmon, B.; Devan, J.M. ACTN3 and MLCK genotype associations with exertional muscle damage. J. Appl. Physiol. 2005, 99, 564–569. [Google Scholar] [CrossRef]
- Mandáková, T.; Pouch, M.; Harmanová, K.; Zhan, S.H.; Mayrose, I.; Lysak, M.A. Multispeed genome diploidization and diversification after an ancient allopolyploidization. Mol. Ecol. 2017, 26, 6445–6462. [Google Scholar] [CrossRef]
- Nikoloudakis, N.; Katsiotis, A. The origin of the C-genome and cytoplasm of Avena polyploids. Theor. Appl. Genet. 2008, 117, 273–281. [Google Scholar] [CrossRef]
- Ridgway, K.P.; Duck, J.M.; Young, J.P.W. Identification of roots from grass swards using PCR-RFLP and FFLP of the plastid trnL (UAA) intron. BMC Ecol. 2003, 3, 8. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; the UGENE team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahe, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Clement, M.; Posada, D.; Crandall, K.A. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1660. [Google Scholar] [CrossRef]
- Múrias dos Santos, A.; Cabezas, M.P.; Tavares, A.I.; Xavier, R.; Branco, M. tcsBU: A tool to extend TCS network layout and visualization. Bioinformatics 2016, 32, 627–628. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).