New Insights into Plastid and Mitochondria Evolution in Wild Peas (Pisum L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Organellar DNA Extraction, High Throughput Sequencing, Assembly and Alignment of Organellar Genomes
2.3. Phylogenetic Analysis
3. Results
3.1. Mitochondrial and Plastid Genome Structure
3.2. Phylogenetic Reconstructions Based on the Plastid and Mitochondrial Genomes
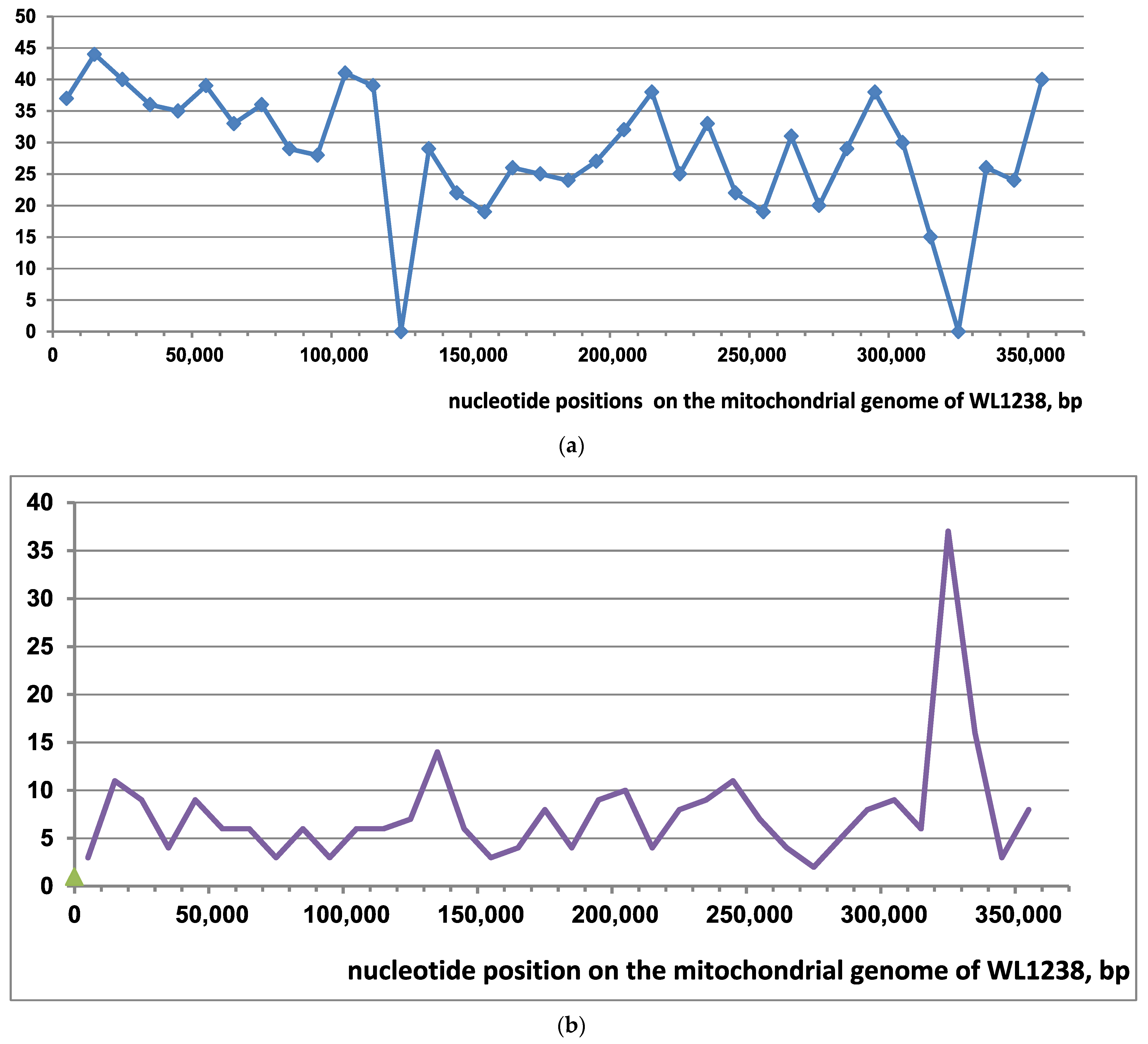
3.3. A Mitogenome Region with Unusual Variation in Accessions of M2 Branch
3.4. Phylogeography of European Wild Peas with Respect to Organellar Genomes
3.5. Organellar Genome Constitutions of Israeli Wild Representatives of Pisum Sativum
4. Discussion
4.1. Phylogenetic Reconstructions Based on the Updated Set of Organellar Genomes
4.2. A Mitochondrial Wild Relative of Pisum abyssinicum Found in Israel
4.3. A Probable Case of Ancestral Recombination between Mitogenomes of M1 and M2 Clades
4.4. Organellar Diversity of Israeli Wild Peas
4.5. Organellar Monophyly of Pisum fulvum and Putative Introgressions
4.6. Organellar Phylogeography of European Wild Peas
4.7. A Problem of Pea Crop Closest Wild Relatives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Camus, M.F.; Alexander-Lawrie, B.; Sharbrough, J.; Hurst, H.D.D. Inheritance through the cytoplasm. Heredity 2022, 129, 31–43. [Google Scholar] [CrossRef]
- Bogdanova, V.S.; Shatskaya, N.V.; Mglinets, A.V.; Kosterin, O.E.; Vasiliev, G.V. Discordant evolution of organellar genomes in peas (Pisum L.). Mol. Phylogenet. Evol. 2021, 160, 107136. [Google Scholar] [CrossRef]
- Maxted, N.; Kell, S.P. Establishment of A Global Network for the In Situ Conservation of Crop Wild Relatives: Status and Needs; FAO Commission on Genetic Resources for Food and Agriculture: Rome, Italy, 2009; p. 266. [Google Scholar]
- Kosterin, O.E. Prospects of the use of wild relatives for pea breeding. Russ. J. Genet. Appl. Res. 2016, 6, 233–243. [Google Scholar] [CrossRef]
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia, Europe and the Nile Valley, 3rd ed.; Clarendon Press: Oxford, MS, USA, 2000; p. 316. [Google Scholar]
- Bogdanova, V.S.; Kosterin, O.E.; Yadrikhinskiy, A.K. Wild peas vary in their cross-compatibility with cultivated pea (Pisum sativum subsp. sativum L.) depending on alleles of a nuclear-cytoplasmic incompatibility locus. Theor. Appl. Genet. 2014, 127, 1163–1172. [Google Scholar] [CrossRef] [PubMed]
- Bogdanova, V.S.; Zaytseva, O.O.; Mglinets, A.V.; Shatskaya, N.V.; Kosterin, O.E.; Vasiliev, G.V. Nuclear-cytoplasmic conflict in pea (Pisum sativum L.) is associated with nuclear and plastidic candidate genes encoding Acetyl-CoA carboxylase subunits. PLoS ONE 2015, 10, e0119835. [Google Scholar] [CrossRef]
- Maxted, N.; Ambrose, M. Peas (Pisum L.). In Plant Genetic Resources of Legumes in the Mediterranean. Current Plant Science and Biotechnology in Agriculture 39; Maxted, N., Bennett, S.J., Eds.; Kluwer Academic Publishers: Dordrecht, Germany, 2001; Volume 3, pp. 181–190. [Google Scholar]
- Coulot, P.; Rabaute, P. Monographie de Leguminosae de France. Tome 4. Tribus des Fabeae, des Cicereae et des Genisteae. Bull. Société Bot. Cent.-Ouest 2016, 46, 1–902. [Google Scholar]
- Coulot, P.; Rabaute, P. Deuxièmes compléments à la Monographie des Leguminosae de France. Monde Plantes 2017, 516, 11–35. [Google Scholar]
- Schaefer, H.; Hechenleitner, P.; Santos-Guerra, A.; Menezes de Sequeira, M.; Pennington, R.T.; Kenicer, G.; Carine, M.A. Systematics, biogeography, and character evolution of the legume tribe Fabeae with special focus on the middle-Atlantic island lineages. BMC Evol. Biol. 2012, 12, 250. [Google Scholar] [CrossRef] [PubMed]
- Hellwig, T.; Abbo, S.; Ophir, R. Drivers of genetic differentiation and recent evolutionary history of an Eurasian wild pea. J. Biogeogr. 2021, 49, 794–808. [Google Scholar] [CrossRef]
- Weeden, N.F.; Coyne, C.J.; Lavin, M.; McPhee, K. Distinguishing among Pisum accessions using a hypervariable intron within Mendel’s green/yellow cotyledon gene. Genet. Resour. Crop Evol. 2021, 68, 2591–2609. [Google Scholar] [CrossRef]
- Lev-Yadun, S.; Gopher, A.; Abbo, S. The cradle of agriculture. Science 2000, 288, 1602–1603. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; de Pamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzym. 2005, 395, 348–384. [Google Scholar] [CrossRef]
- Chevreux, B.; Wetter, T.; Suhai, S. Genome sequence assembly using trace signals and additional sequence information. In Computer Science and Biology, Proceedings of the German Conference on Bioinformatics (GCB), Hannover, Germany, 4–6 October 1999; Hannover, Germany; pp. 45–56. Available online: https://dblp2.uni-trier.de/db/conf/gcb/gcb1999.html (accessed on 26 June 2017).
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.J.A.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using Tablet for visual exploration of second-generation sequence data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Kosterin, O.E. Abyssinian pea (Lathyrus schaeferi Kosterin nom. nov. pro Pisum abyssinicum A. Br.) is a problematic taxon. Vavilovskii Zhurnal Genet. Sel. Vavilov J. Genet. Breed. 2017, 21, 158–169, (In Russian, English Summary). [Google Scholar] [CrossRef]
- Kosterin, O.E. Abyssinian pea (Lathyrus schaeferi Kosterin nom. nov. pro Pisum abyssinicum A. Br.) is a problematic taxon. Acta Biol. Sib. 2017, 3, 97–110. [Google Scholar] [CrossRef]
- Weeden, N. Domestication of pea (Pisum sativum L.): The case of the Abyssinian Pea. Front. Plant Sci. 2018, 9, 515. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Jorgensen, R.A.; Thompson, W.F. Chloroplast DNA variation and evolution in Pisum: Patterns of change and phylogenetic analysis. Genetics 1985, 109, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, O.O.; Bogdanova, V.S.; Mglinets, A.V.; Kosterin, O.E. Refinement of the collection of wild peas (Pisum L.) and search for the area of pea domestication with a deletion in the plastidic psbA-trnH spacer. Genet. Resour. Crop Evol. 2017, 64, 1427–1430. [Google Scholar] [CrossRef]
- Ross, M.G.; Russ, C.; Costello, M.; Hollinger, A.; Lennon, N.J.; Hegarty, R.; Nusbaum, C.; Jaffe, D.B. Characterizing and measuring bias in sequence data. Genome Biol. 2013, 14, R51. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ze’ev, N.; Zohary, D. Species relationship in the genus Pisum L. Israel J. Bot. 1973, 22, 73–91. [Google Scholar]
- Kosterin, O.E.; Zaytseva, O.O.; Bogdanova, V.S.; Ambrose, M. New data on three molecular markers from different cellular genomes in Mediterranean accessions reveal new insights into phylogeography of Pisum sativum L. subsp. elatuis (Beib.) Schmahl. Genet. Resour. Crop Evol. 2010, 57, 733–739. [Google Scholar] [CrossRef]
- Govorov, L.I. [Pea]. In Kul’turnaya Flora [Cultivated Flora of the USSR]; Vulf, E.V., Ed.; Gosudarstvennoe Izdatelstvo Sovkhoznoi i Kolkhoznoi Literatury: Moscow, Russia, 1937; Volume 4, Grain Legumes; pp. 229–336. (In Russian) [Google Scholar]
- Makasheva, R.K. Pea. In Kul’turnaya Flora SSSR [Cultivated Flora of the USSR], 2nd ed.; Brezhnev, D.D., Ed.; Kolos: Leningrad, Russia, 1979; Volume 4, Grain Legumes, Part 1; pp. 7–322. (In Russian) [Google Scholar]
- Kosterin, O.E.; Bogdanova, V.S. Reciprocal compatibility within the genus Pisum L. as studied in F1 hybrids: 3. Crosses involving P. abyssinicum A. Br. Genet. Resour. Crop Evol. 2020, 67, 967–983. [Google Scholar] [CrossRef]
- Kreplak, K.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1141–1422. [Google Scholar] [CrossRef] [PubMed]
- Vershinin, A.V.; Allnutt, T.R.; Knox, M.R.; Ambrose, M.J. Transposable elements reveal the impact of introgression, rather than transposition, in Pisum diversity, evolution, and domestication. Mol. Biol. Evol. 2003, 20, 2067–2075. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Vershinin, A.; Grzebota, J.; Shaw, P.; Smýkal, P.; Marshall, D.; Ambrose, M.J.; Ellis, T.H.N.; Flavell, A.J. The genetic diversity and evolution of field pea (Pisum) studied by high throughput retrotransposon based insertion polymorphism (RBIP) marker analysis. BMC Evol. Biol. 2010, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Trněný, O.; Brus, J.; Hradilova, I.; Rathore, A.; Das, R.R.; Kopecký, P.; Coyne, C.J.; Reeves, P.; Richards, C.; Smýkal, P. Molecular evidence for two domesticated events in the pea crop. Genes 2018, 19, 535. [Google Scholar] [CrossRef] [PubMed]
- Hellwing, T.; Abbo, S.; Ophir, R. Phylogeny and disparate selection signatures suggest two genetically independent domestication events in pea (Pisum L.). Plant J. 2022, 110, 419–439. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, O.O.; Bogdanova, V.S.; Kosterin, O.E. Phylogenetic reconstruction at the species and intraspecies levels in the genus Pisum L. (peas) using a histone H1 gene. Gene 2012, 504, 192–202. [Google Scholar] [CrossRef]
- Zaytseva, O.O.; Gunbin, K.V.; Mglinets, A.V.; Kosterin, O.E. Divergence and population traits in evolution of the genus Pisum L. as reconstructed using genes of two histone H1 subtypes showing different phylogenetic resolution. Gene 2015, 556, 235–244. [Google Scholar] [CrossRef]
- Stegemann, S.; Keuthe, M.; Greiner, S.; Bock, R. Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. USA 2012, 109, 2434–2438. [Google Scholar] [CrossRef]
- Gurdon, C.; Svab, Z.; Feng, Y.; Kumar, D.; Maliga, P. Cell-to-cell movement of mitochondria in plants. Proc. Natl. Acad. Sci. USA 2016, 113, 3395–3400. [Google Scholar] [CrossRef]
- Davis, C.C.; Wurdack, K.J. Host-to-parasite gene transfer in flowering plants: Phylogenetic evidence from Malpighiales. Science 2004, 305, 676–678. [Google Scholar] [CrossRef]
- Bergthorsson, U.; Adams, K.L.; Thomason, B.; Palmer, J.D. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 2003, 424, 197–201. [Google Scholar] [CrossRef]
- Gentry, H.S. Foreign Travel Report. Italy, Greece, Turkey, Germany, England. 8 April–28 June 1969; New Crops Research Branch: Beltsville, MD, USA, 1969; pp. 1–4, unpublished report. [Google Scholar]
- Abbo, S.; Gopher, A.; Bar-Gal, G.K. Plant Domestication and the Origin of Agriculture in the Ancient Near East; Cambridge University Press: Cambridge, UK, 2022; p. 288. [Google Scholar] [CrossRef]
- Schmidt, K. Sie Bauten Die Ersten Tempel. Das Rätselhafte Heiligtum der Steinzeitjäger; C.H. Beck: Munich, Germany, 2006; p. 288. [Google Scholar]
- Dietrich, O.; Dietrich, L.; Notroff, J. Cult as a driving force of human history: A view from Göbekli Tepe. Expedition 2017, 59, 657–663. [Google Scholar]
- Heun, O.; Schäffer-Pregl, R.; Kalavan, D.; Castagna, R.; Accerbi, M.; Borghi, B.; Salamini, F. Site of einkorn wheat domestication identified byDNA fingerprinting. Science 1997, 278, 1312–1314. [Google Scholar] [CrossRef]
- Kosterin, O.E. The lost ancestor or the broad bean (Vicia faba L.) and the origin of plant cultivation in the Near East. Vavilovskii Zhurnal Genet. Sel. Vavilov J. Genet. Breed. 2014, 18, 831–840. [Google Scholar] [CrossRef]
- Smýkal, P.; Trněný, O.; Brus, J.; Hanáček, P.; Rathore, A.; Roma, R.D.; Pechanek, V.; Douchoslav, M.; Battacharrya, D.; Bariotakis, M.; et al. Genetic structure of wild pea (Pisum sativum subsp. elatius) populations in the northern part of the Fertile Crescent reflects moderate cross-pollination and strong effect of geographic but not environmental distance. PLoS ONE 2018, 13, e0194056. [Google Scholar] [CrossRef]
- Smýkal, P.; Hradilová, I.; Trněný, O.; Brus, J.; Rathore, A.; Bariotakis, M.; Das, R.R.; Battacharrya, D.; Richards, C.; Coyne, C.J.; et al. Genomic diversity and macroecology of the crop wild relatives of domesticated pea. Sci. Rep. 2017, 7, 17384. [Google Scholar] [CrossRef]
- Tanno, K.; Wilcox, G. How fast was wild wheat domesticated? Science 2005, 311, 1886. [Google Scholar] [CrossRef] [PubMed]
- Fuller, D.Q.; Willcox, G.; Allaby, R.G. Cultivation and domestication had multiple origins: Arguments against the core area hypothesis for the origins of agriculture in the Near East. World Archaeol. 2011, 43, 628–658. [Google Scholar] [CrossRef]
- Fuller, D.Q.; Willcox, G.; Allaby, R.G. Early agricultural pathways: Moving outside the ‘core area’ hypothesis in Southwest Asia. J. Exp. Bot. 2012, 63, 617–633. [Google Scholar] [CrossRef] [PubMed]
- Asouti, E.; Fuller, D.Q. From foraging to farming in the southern Levant: The development of the Epipaleolithic and Pre-pottery Neolithic plant managing strategies. Veg. Hist. Archeobot. 2012, 21, 149–162. [Google Scholar] [CrossRef]
- Berdnikov, V.A.; Bogdanova, V.S.; Rozov, S.M.; Kosterin, O.E. Geographic patterns of histne H1 allelic frequencies formed in the course of Pisum sativum L. (pea) cultivaion. Heredity 1993, 71, 199–209. [Google Scholar] [CrossRef]





| Accession Designation Used in This Paper | Taxonomic Attributions and Other Known Designations | Origin; Date of Collection (Where Relevant/Available), Collector | Lati- Tude (N) | Longi- Tude (E) | Organellar Constitution, Allele Combination of Three Markers | Plastid Spacer psbA-trnH. (Difference from Consensus) | Gene Bank IDs for Plastid and Mictochondrial Genomes |
|---|---|---|---|---|---|---|---|
| Pisum fulvum, wild | |||||||
| VIR_6071 | Pisum fulvum var. striatum Makasheva | Palestine, foothills approx. 30 km south-west of Jerusalem. Collected in 1960. | [31.6] | [35.0] | P1 M1, A | - | ON357683, ON186758, |
| Pisum sativum L. subsp. elatius (Bieb.) Aschers. et Graebn. s.l., wild | |||||||
| CE_23 | Morocco, Er-Rif Mts, Chefchaouen Province, Chefchaouen, Jbel Tissouka Mt western foot, a femce made of stones and dry branches between a tourist circuit trial and a small abandoned garden with sparse trees. Collected on 19 May 2021 by O. Kosterin and N. Solovyeva | 35.16837 | −5.25504 | P4 M3, C | - | OP928222, OQ078748 | |
| PIS_2844 | España, Salamanca, 2 km de Puerto de Béjar, Camino de la Plata, 740 m a.s.l. | [40.35] | [−5.84] | P4 M3, C | - | MZ648183, MZ707507 | |
| CE_12 (-Dw, -d) | JI_3558, SE1, W6_56891 (Dw), W6_56893 (d), | España, Cataluña, comarca de Conca de Barberà, Muntanyes de Prades, Vall de Monestir de Poblet, Barranc de Castellfollit | 41.3517 | 01.0614 | P4 M3, C | - | MZ677459, ON165401 |
| CE_13 | JI_3553; W6_56891; FE1 | France, Région Sud Provence-Alpes-Côte d’Azur, département du Var, canton de Brignoles, commune de Rougiers, Massif de la Sainte-Baume (ca 50 km from Marseille), open stand of (Quercus pubescens Willd.). Collected by Michel Papazyan | 43.3839 | 05.8567 | P3 M3, C | 75: T→G | MZ677460, ON165402 |
| PIS_2850 | Italy, [Liguria,] Commune de Camogli, Mortola | [44.33] | [9.16] | P4 M3, C | - | MZ648184, MZ707508 | |
| JI_2055 | Italy, [Campania], Mt. Alburni | 40.55 | 15.30 | P4 M3, C | - | OP919340, OQ078752 | |
| PI_344539 | Italy, Sicily, Palermo [Prov.], Piana degli Albanesi. Collected by A. Di Martino before 1969 | [38.0] | [13.3] | P4 M4, C | - | ON259091, ON186755, | |
| JI_1092 | PI_344006, W6_8706, 22618 | Greece, Athos Peninsula, Xeropotamou Monastery, moist mountain slopes, rocky or well littered soil, 200 m a.s.l. Collected in June 1969 by H.S. Gentry | [40.23] | [24.22] | P4 M4, C | - | ON243975, ON165398 |
| JI_1093 | PI_344010, W6_8707, 22732, introgressed | Greece, Athos Peninsula, below Karyes, high macchia vegetation, 270 m a.s.l. Collected in June 1969 by H.S. Gentry | [40.26] | [24.25] | P4 M4, C | 75: T→G | ON243976, ON165399 |
| JI_1095 | PI_344012, W6_8709, 22734 | Greece, Athos Peninsula, above Ivyron Monastery, 180 m a.s.l. Collected in June 1969 by H.S. Gentry | [40.24] | [24.28] | P3 M4, C | - | ON243977, ON165400 |
| PI_344008 | W6_8710, 22735 | Greece, Athos Peninsula, 1 km S of Daphne. Collected in June 1969 by H.S. Gentry | [40.20] | [24.22] | P3 M4, C | 75: T→G | ON259089, ON186753, |
| PI_344009 | 22729 | Greece, Athos Peninsula, Panteleimonos Monestry, scrub oak macchia. Collected in June 1969 by H.S. Gentry | [40.28] | [24.20] | P3 M4, unusual (+—S) 1 | - | ON259090, ON186754, |
| PI_344001 | 22701 | Turkey, [Mersin Il], 17 km north of Mersin on road to Gonze, limestone rocks among macchia, 360 m a..l. Collected in May 1969 by H.S. Gentry | [37.0] | [34.6] | P2 M2 (no ins.), A | - | ON259088, ON186752, |
| Pe_6 | PI_639960, W6_2639 | Turkey, Mardin Il, 12 km on the road to Bozova from the main Shanlyurfa-Diyarbakyr road, edge of pistachio grove. Collected before 2005 by S. Abbo. | [37.29] | [38.67] | P5 M4, B | - | OP928224, OQ078750 |
| W6_2107 | Psh 008, 120689-0302 | Turkey, Siirt Il, 6.3 km north of Batman (across from the airport), frequent as weed in lentil field on fine soil; 630 m a.s.l. Collected on 12 June 1989 by W.J. Kaiser, F.J. Muehlbauer, C.V. Sperling | 37.92 | 41.13 | P5 (no inv.) M2, unusual (- + S) 2 | - | ON357684, ON186759, |
| CE_15 | Phs 10–Phs 12, Phs 98–Phs 99, ‘southern humile’ | Israel, eastern Lower Galilee, 2.4 km north-east of Ginosar Kibbutz, near Khirbat al-Minya ruins and Atar Safir Pump Station, 420 m from Lake Tiberias north-western bank, in ruderal vegetation (including Lathyrus hierosolymnitanus and Vicia sp.) at a wheat field margin, 202 m below sea level. Collected on 15 April 2019 by S. Abbo and O. Kosterin | 32.86824 | 35.53594 | P4 M2, A | - | MZ677461, ON165403 |
| CE_16 | Pe 25–Pe 26, Pe 41–Pe 43, Pe 50–Pe 54, Pe 139–Pe 140 | Israel, Northern District, eastern Lower Galilee, 666 m north-east of Livnim Settlement, Vadi Amud, shrubbery, 141 m below sea levell. Collected on 15 April 2019 by S. Abbo and O. Kosterin | 32.86801 | 35.50259 | P4 M3, C | - | ON310561, ON186757, |
| 711 | JI_3272, PI_560068, L_99, ‘southern humile’ | Israel, 2 km west of Jerusalem, Jerusalem Forest, edges of the abandoned terrace field [31.0] | [~31.8] | [35.2] | P4 M1, A | - | ON310560, ON186756, |
| 714 | JI_3275, PI_560071, L_102, ‘southern humile’ | Israel, between Bet Shemesh and Bet Gurvin, field edges and roadsides [31.0] | [~31.7] | [~34.9] | P4 M1, A | - | OP919341, OQ078753 |
| CE_24 | Russia, Republic of Dagestan, Magaramkent District, Samur Forest, 2 km west-north-west of Primorskiy village, sparse oak stand in oak/hornbeam forest, 6 m below sea level. Collectedon 25 June 2021 by O.E. Kosterin | 41.85736 | 48.55370 | P5 M3/M4, B | 142–149 deleted | OP928223, OQ078749 | |
| YD-1 | Turkmenistan, Kopet-Dagh, Yol-Dere Valley. Collected on 1 June 2018 by S. Abbo | 38.50 | 56.38 | P5 M4, B | 142–149 deleted | OP919339, OQ078751 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shatskaya, N.V.; Bogdanova, V.S.; Kosterin, O.E.; Vasiliev, G.V. New Insights into Plastid and Mitochondria Evolution in Wild Peas (Pisum L.). Diversity 2023, 15, 216. https://doi.org/10.3390/d15020216
Shatskaya NV, Bogdanova VS, Kosterin OE, Vasiliev GV. New Insights into Plastid and Mitochondria Evolution in Wild Peas (Pisum L.). Diversity. 2023; 15(2):216. https://doi.org/10.3390/d15020216
Chicago/Turabian StyleShatskaya, Natalia V., Vera S. Bogdanova, Oleg E. Kosterin, and Gennadiy V. Vasiliev. 2023. "New Insights into Plastid and Mitochondria Evolution in Wild Peas (Pisum L.)" Diversity 15, no. 2: 216. https://doi.org/10.3390/d15020216
APA StyleShatskaya, N. V., Bogdanova, V. S., Kosterin, O. E., & Vasiliev, G. V. (2023). New Insights into Plastid and Mitochondria Evolution in Wild Peas (Pisum L.). Diversity, 15(2), 216. https://doi.org/10.3390/d15020216






