Alternaria muriformis sp. nov., a New Species in Section Chalastospora Isolated from Herbivore Dung in Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Isolation of Fungi
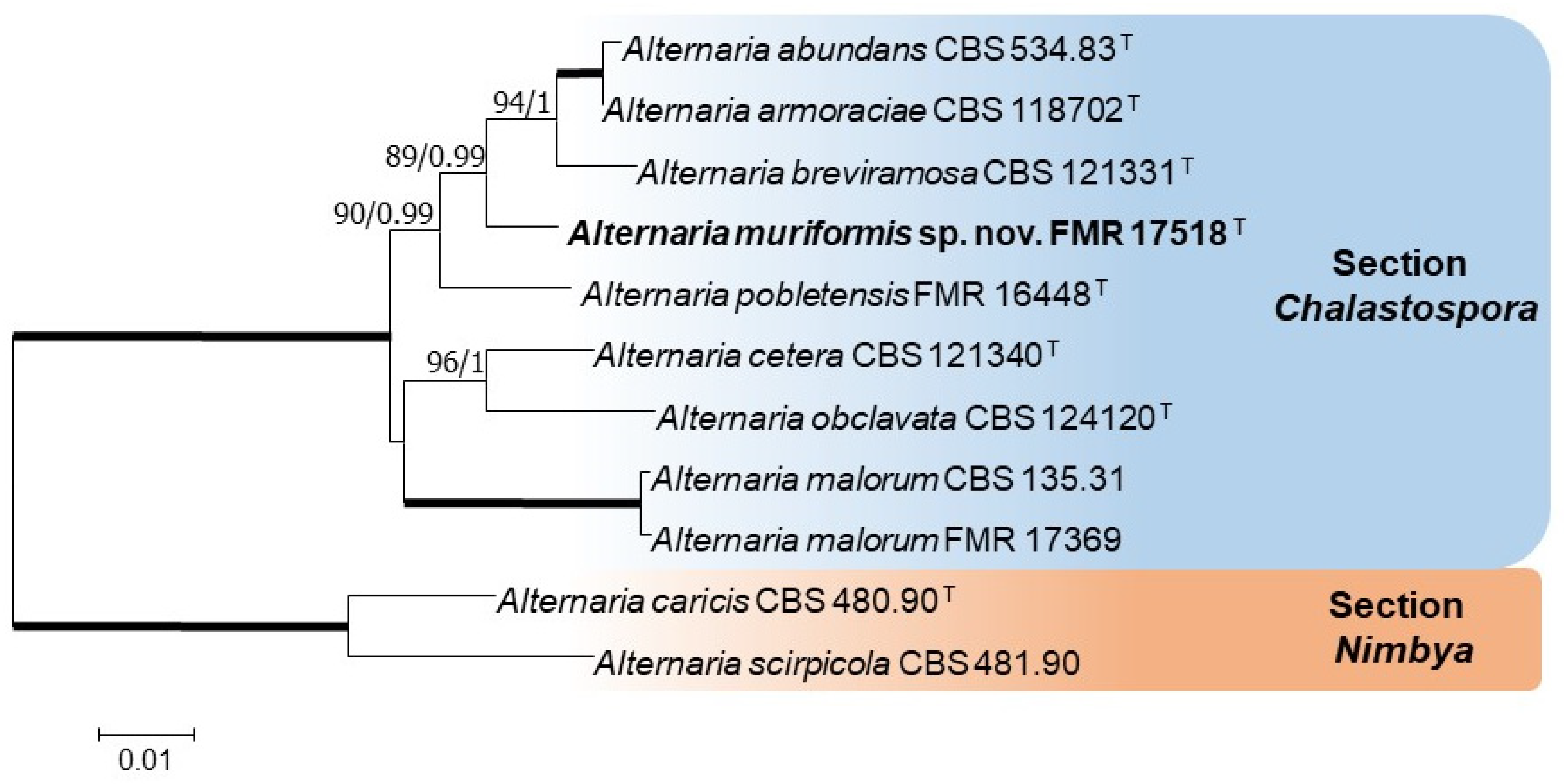
2.2. DNA Extraction, PCR, Sequencing, and Phylogenetic Analysis
| Species | Section | Isolates 1 | Sources | GenBank Accession Numbers 2 | References | ||
|---|---|---|---|---|---|---|---|
| ITS | gapdh | ATPase | |||||
| A. abundans | Chalastospora | CBS 534.83 T | Fragaria sp. and stolon | JN383485 | KC584154 | JQ671802 | [15,32] |
| A. armoraciae | Chalastospora | CBS 118702 T | Armoracia rusticana | KC584182 | KC584099 | LR134098 | [2,15] |
| A. breviramosa | Chalastospora | CBS 121331 T | Triticum sp. | FJ839608 | KC584148 | LR134099 | [2,15] |
| A. cetera | Chalastospora | CBS 121340 T | Elymus scabrus | JN383482 | AY562398 | LR134101 | [15,32] |
| A. malorum | Chalastospora | CBS 135.31 | Malus sylvestris and fruit | JQ693638 | JQ646278 | JQ671800 | [33] |
| Chalastospora | FMR 17369 | Rabbit dung | LR134074 | LR134077 | LR134029 | [2] | |
| A. obclavata | Chalastospora | CBS 124120 T | Air | KC584225 | KC584149 | LR134100 | [2,15] |
| A. pobletensis | Chalastospora | FMR 16448 T | Herbivore dung | LR133896 | LR133897 | LR133903 | [2] |
| A. muriformis | Chalastospora | FMR 17518 T | Herbivore dung | OQ421258 | OQ425406 | OQ425407 | Present study |
| A. caricis | Nimbya | CBS 480.90 T | Carex hoodii | AY278839 | AY278826 | JQ671780 | [15,32] |
| A. scirpicola | Nimbya | CBS 481.90 | Scirpus sp. | KC584237 | KC584163 | JQ671781 | [15,32] |
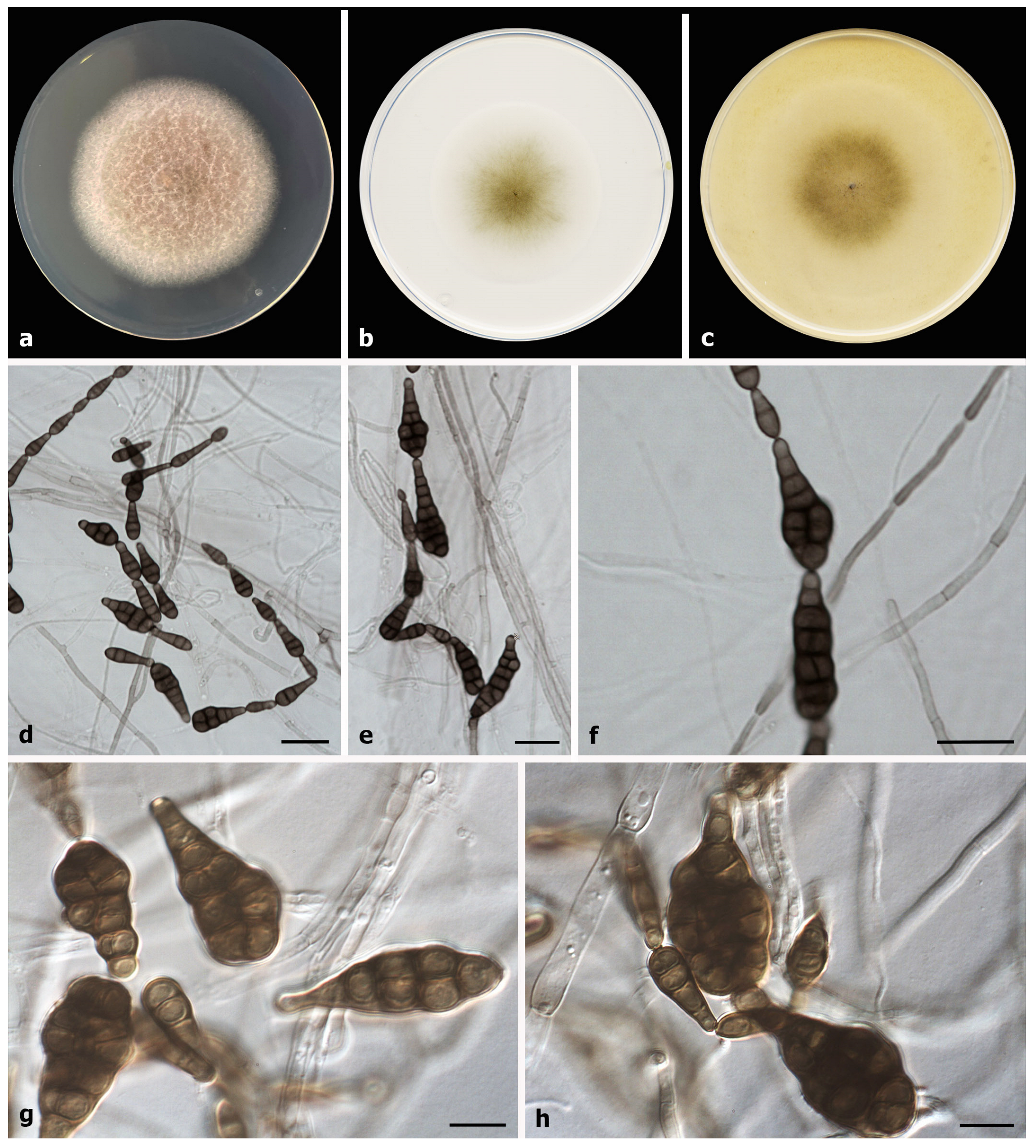
2.3. Phenotypic Study
3. Results
Taxonomy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Von Esenbeck, C.G.N. Das System der Pilze und Schwämme; Stahelschen Buchhandlung: Wurzburg, Germany, 1816. [Google Scholar] [CrossRef]
- Marin-Felix, Y.; Hernández-Restrepo, M.; Iturrieta-González, I.; García, D.; Gené, J.; Groenewald, J.Z.; Cai, L.; Chen, Q.; Quaedvlieg, W.; Schumacher, R.K.; et al. Genera of phytopathogenic fungi: GOPHY 3. Stud. Mycol. 2019, 94, 1–124. [Google Scholar] [CrossRef]
- Huang, D.M.; Liu, X.; Bai, L.; Zhang, S.J.; Zhang, Z.G.; Qin, Q.P. First Report of Alternaria alternata Causing Leaf Spot Disease on Daylily in China. Plant Dis. 2022, 106, 3200. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Phookamsak, R.; Jiang, H.; Bhat, D.J.; Camporesi, E.; Lumyong, S.; Kumla, J.; Hongsanan, S.; Mortimer, P.E.; Xu, J.; et al. Additions to the Inventory of the Genus Alternaria Section Alternaria (Pleosporaceae, Pleosporales) in Italy. J. Fungi 2022, 8, 898. [Google Scholar] [CrossRef]
- Thomma, B.P.H.J. Alternaria spp.: From general saprophyte to specific parasite. Mol. Plant. Pathol. 2003, 4, 225–236. [Google Scholar] [CrossRef] [PubMed]
- El-Alwany, A.M. Plant Pathogenic Alternaria Species in Libya. Open Access Libr. J. 2015, 2, e1662. [Google Scholar] [CrossRef]
- Lawrence, D.P.; Rotondo, F.; Gannibal, P.B. Biodiversity and taxonomy of the pleomorphic genus Alternaria. Mycol. Prog. 2016, 15, 3. [Google Scholar] [CrossRef]
- Meena, M.; Gupta, S.K.; Swapnil, P.; Zehra, A.; Dubey, M.K.; Upadhyay, R.S. Alternaria Toxins: Potential Virulence Factors and Genes Related to Pathogenesis. Front. Microbiol. 2017, 8, 1451. [Google Scholar] [CrossRef] [PubMed]
- Pastor, F.J.; Guarro, J. Alternaria infections: Laboratory diagnosis and relevant clinical features. Clin. Microbiol. Infect. 2008, 14, 734–746. [Google Scholar] [CrossRef]
- Cardona, S.; Yusef, S.; Silva, E.; Bustos, M.G.; Torres, M.I.; Leal, A.R.; Ceballos-Garzon, A.; Josa, D.F. Cerebral phaeohyphomycosis caused by Alternaria spp.: A case report. Med. Mycol. Case Rep. 2020, 27, 11–13. [Google Scholar] [CrossRef]
- De Hoog, G.S.; Guarro, J.; Gené, J.; Ahmed, S.A.; Al-Hatmi, A.M.S.; Figueras, M.J.; Vitale, R.G. Atlas of Clinical Fungi, 4th ed.; Foundation Atlas of Clinical Fungi: Hilversum, The Netherlands, 2020. [Google Scholar]
- Iturrieta-González, I.; Pujol, I.; Iftimie, S.; García, D.; Morente, V.; Queralt, R.; Guevara-Suarez, M.; Alastruey-Izquierdo, A.; Ballester, F.; Hernández-Restrepo, M.; et al. Polyphasic identification of three new species in Alternaria section Infectoriae causing human cutaneous infection. Mycoses 2019, 63, 212–224. [Google Scholar] [CrossRef]
- Abbas, H.K.; Riley, R. The presence and phytotoxicity of fumonisins and aal-toxin in Alternaria alternata. Toxicon. 1996, 34, 133–136. [Google Scholar] [CrossRef]
- Lou, J.; Fu, L.; Peng, Y.; Zhou, L. Metabolites from Alternaria Fungi and Their Bioactivities. Molecules 2013, 18, 5891–5935. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.H.C.; Groenewald, J.Z.; Binder, M.; Crous, P.W. Alternaria redefined. Stud. Mycol. 2013, 75, 171–212. [Google Scholar] [CrossRef] [PubMed]
- Gannibal, P.B.; Orina, A.S.; Gasich, E.L. A new section for Alternaria helianthiinficiens found on sunflower and new asteraceous hosts in Russia. Mycol. Prog. 2022, 21, 34. [Google Scholar] [CrossRef]
- Al Ghafri, A.; Maharachchikumbura, S.S.N.; Hyde, K.D.; Al-Saady, N.A.; Al-Sadi, A.M. A new section and a new species of Alternaria encountered from Oman. Phytotaxa 2019, 405, 279–289. [Google Scholar] [CrossRef]
- Zhao, L.; Luo, H.; Cheng, H.; Gou, Y.N.; Yu, Z.H.; Deng, J.X. New Species of Large-Spored Alternaria in Section Porri Associated with Compositae Plants in China. J. Fungi 2022, 8, 607. [Google Scholar] [CrossRef] [PubMed]
- Simmons, E.G. Alternaria: An Identification Manual; CBS Biodiversity Series 6; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2007. [Google Scholar]
- Simmons, E.G. Helminthosporium allii as type of a new genus. Mycologia 1971, 63, 380–386. [Google Scholar] [CrossRef]
- Mirhendi, H.; Fatemi, M.J.; Bateni, H.; Hajabdolbaghi, M.; Geramishoar, M.; Ahmadi, B.; Badali, H. First case of disseminated phaeohyphomycosis in an immunocompetent individual due to Alternaria malorum. Med. Mycol. 2013, 51, 196–202. [Google Scholar] [CrossRef]
- Muller, F.M.C.; Werner, K.E.; Kasai, M.; Francesconi, A.; Chanock, S.J.; Walsh, T.J. Rapid extraction of genomic DNA from medically important yeasts and filamentous fungi by high-speed cell disruption. J. Clin. Microbiol. 1998, 36, 1625–1629. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing 794 of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and 796 Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- Lawrence, D.P.; Gannibal, P.B.; Peever, T.L.; Pryor, B.M. The sections of Alternaria: Formalizing species-group concepts. Mycologia 2013, 105, 530–546. [Google Scholar] [CrossRef]
- Berbee, M.L.; Pirseyedi, M.; Hubbard, S. Cochliobolus Phylogenetics and the Origin of Known, Highly Virulent Pathogens, Inferred from ITS and Glyceraldehyde-3-Phosphate Dehydrogenase Gene Sequences. Mycologia 1999, 91, 964–977. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Farris, J.S.; Kallersjo, M.; Kluge, A.G.; Bult, C. Testing significance of incongruence. Cladistics 1994, 10, 315–319. [Google Scholar] [CrossRef]
- Posada, D. jModelTest: Phylogenetic Model Averaging. Mol. Biol. Evol. 2008, 25, 1253–1256. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.X.; Li, M.J.; Paul, N.C.; Oo, M.M.; Lee, H.B.; Oh, S.K.; Yu, S.H. Alternaria brassicifolii sp. nov. isolated from Brassica rapa subsp. pekinensis in Korea. Mycobiology 2018, 46, 172–176. [Google Scholar] [CrossRef]
- Poursafar, A.; Ghosta, Y.; Orina, A.S.; Gannibal, P.B.; Javan-Nikkhah, M.; Lawrence, D.P. Taxonomic study on Alternaria sections Infectoriae and Pseudoalternaria associated with black (sooty) head mold of wheat and barley in Iran. Mycol. Prog. 2018, 17, 343–356. [Google Scholar] [CrossRef]
- Kornerup, A.; Wanscher, J.H. Methuen Handbook of Colour, 3rd ed.; Methuen: London, UK, 1978; pp. 1–256. [Google Scholar]
- Crous, P.W.; Braun, U.; Wingfield, M.J.; Wood, A.; Shin, H.D.; Summerell, B.A.; Alfenas, A.C.; Cumagun, C.J.; Groenewald, J.Z. Phylogeny and taxonomy of obscure genera of microfungi. Persoonia 2009, 22, 139–161. [Google Scholar] [CrossRef]
- Simmons, E.G. An aggregation of Embellisia species. Mycotaxon 1983, 17, 216–241. [Google Scholar]
- Simmons, E.G. Alternaria themes and variations (145–149). Mycotaxon 1996, 57, 391–409. [Google Scholar]
- Braun, U.; Crous, P.W.; Dugan, F.; Groenewald, J.Z.; De Hoog, G.S. Phylogeny and taxonomy of Cladosporium-like hyphomycetes, including Davidiella gen. nov., the teleomorph of Cladosporium s. str. Mycol. Prog. 2003, 2, 3–18. [Google Scholar] [CrossRef]
- Woudenberg, J.H.C.; Truter, M.; Groenewald, J.Z.; Crous, P.W. Large-spored Alternaria pathogens in section Porri disentangled. Stud. Mycol. 2014, 79, 1–47. [Google Scholar] [CrossRef] [PubMed]
- Woudenberg, J.H.C.; Seidl, M.F.; Groenewald, J.Z.; De Vries, M.; Stielow, J.B.; Thomma, B.P.H.J.; Crous, P.W. Alternaria section Alternaria: Species, formae speciales or pathotypes? Stud. Mycol. 2015, 82, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Romain, B.B.N.D.; Hassan, O.; Kim, J.S.; Chang, T. Alternaria koreana sp. nov., a new pathogen isolated from leaf spot of ovate-leaf Atractylodes in South Korea. Mol. Biol. Rep. 2022, 49, 413–420. [Google Scholar] [CrossRef]
- Ruehle, G.D. New apple-rot fungi from Washington. Phytopathology 1931, 21, 1141–1152. [Google Scholar]
- Holter, P. Herbivore dung as food for dung beetles: Elementary coprology for entomologists. Ecol. Entomol. 2016, 41, 367–377. [Google Scholar] [CrossRef]
- Richardson, M. New and interesting records of coprophilous fungi. Bot. J. Scotl. 1998, 50, 161–175. [Google Scholar] [CrossRef]
- Ghosta, Y.; Poursafar, A.; Qarachal, J.F. Study on coprophilous fungi: New records for Iran mycobiota. Rostaniha 2016, 17, 115–126. [Google Scholar]
- Melo, R.F.R.; Monte, D.B.P.D.; Gondim, N.H.B.; Maia, L.C.; Miller, A.N. Coprophilous fungi from Brazil: New records for the Neotropics. Mycotaxon 2019, 134, 335–352. [Google Scholar] [CrossRef]
- Guevara-Suarez, M.; García, D.; Cano-Lira, J.F.; Guarro, J.; Gené, J. Species diversity in Penicillium and Talaromyces from herbivore dung, and the proposal of two new genera of penicillium-like fungi in Aspergillaceae. Fungal Syst. Evol. 2020, 5, 39–75. [Google Scholar] [CrossRef]
- Iturrieta-González, I.; García, D.; Gené, J. Novel species of Cladosporium from environmental sources in Spain. Mycokeys 2021, 77, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Li, J.; Liu, J.; Xiao, S.; Yang, S.; Mei, J.; Ren, M.; Wu, S.; Zhang, H.; Yang, X. Secondary metabolites of Alternaria: A comprehensive review of chemical diversity and pharmacological properties. Front. Microbiol. 2023, 13, 1085666. [Google Scholar] [CrossRef] [PubMed]
- Horiuchi, M.; Tokuda, H.; Ohnishi, K.; Yamashita, M.; Nishino, H.; Maoka, T. Porritoxins, metabolites of Alternaria porri, as anti-tumor-promoting active compounds. Nat. Prod. Res. 2006, 20, 161–166. [Google Scholar] [CrossRef]
- Shi, Y.N.; Pusch, S.; Shi, Y.M.; Richter, C.; Maciá-Vicente, J.G.; Schwalbe, H.; Kaiser, M.; Opatz, T.; Bode, H.B. (±)-Alternarlactones A and B, two antiparasitic alternariol-like dimers from the fungus Alternaria alternata P1210 isolated from the halophyte Salicornia sp. J. Org. Chem. 2019, 84, 11203–11209. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.D.; Yi, T.F.; Ma, Q.Y.; Xie, Q.Y.; Zhou, L.M.; Chen, J.P.; Dai, H.F.; Wu, Y.G.; Zhao, Y.X. Biphenyl metabolites from the patchouli endophytic fungus Alternaria sp. PfuH1. Fitoterapia 2020, 146, 104708. [Google Scholar] [CrossRef]
| Locus | Primer | Sequence (5′–3′) | References |
|---|---|---|---|
| Internal transcribed spacer (ITS) | ITS5 | GGAAGTAAAAGTCGTAACAAGG | [23] |
| ITS4 | TCCTCCGCTTATTGATATGC | ||
| Glyceraldehyde-3-phosphate dehydrogenase (gapdh) | gpd1 | CAACGGCTTCGGTCGCATTG | [25] |
| gpd2 | GCCAAGCAGTTGGTTGTGC | ||
| Plasma membrane ATPase (ATPase) | ATPDF1 | ATCGTCTCCATGACCGAGTTCG | [24] |
| ATPDR1 | TCCGATGGAGTTCATGATAGCC |
| Species | Conidia | References | ||||
|---|---|---|---|---|---|---|
| Shape | Size (µm) | Transverse Septa Numbers | Longitudinal or Oblique Septa Numbers (*) | Ornamentation | ||
| A. abundans | Ovoidal Obclavate | 20–30 × 10–12 40–50 × 8–12 | 3–6(–8) | 0–1 | Usually smooth | [36] |
| A. armoraciae | Ovoidal to ellipsoidal | 15–35 × 8–12 | 3–5 | 0–1 | Smooth | [19] |
| A. breviramosa | Ellipsoidal to fusiform | (8–)10–15(–17) × 3(–3.5) | 0–1(–2) | Absent | Smooth | [35] |
| A. cetera | Ellipsoidal to narrow-ovoid | 18–22 × 3–4(–5) | 1–3 | Absent | Smooth | [19,37] |
| A. malorum | Ellipsoidal-ovoidal, cylindrical, or fusiform | 6–14(17) × 2–4 | Absent | Absent | Smooth | [38] |
| A. obclavata | Obclavate | (23–)26–30(–35) × (3.5–)4 | 0–3 | Absent | Smooth | [35] |
| A. pobletensis | Obpyriform or obclavate, and some ellipsoidal or subcylindrical | 8–50 × 5–20 | (1–)3–7(–9) | 0–1(–2) | Smooth or verruculose | [2] |
| A. muriformis | Ellipsoidal or obclavate | 10–40 × 4–14 | (1–)3–5(–7) | 0–1(–2) | Smooth | Present study |
| Muriform | 37–45 × 16–33 | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iturrieta-González, I.; Gené, J. Alternaria muriformis sp. nov., a New Species in Section Chalastospora Isolated from Herbivore Dung in Spain. Diversity 2023, 15, 606. https://doi.org/10.3390/d15050606
Iturrieta-González I, Gené J. Alternaria muriformis sp. nov., a New Species in Section Chalastospora Isolated from Herbivore Dung in Spain. Diversity. 2023; 15(5):606. https://doi.org/10.3390/d15050606
Chicago/Turabian StyleIturrieta-González, Isabel, and Josepa Gené. 2023. "Alternaria muriformis sp. nov., a New Species in Section Chalastospora Isolated from Herbivore Dung in Spain" Diversity 15, no. 5: 606. https://doi.org/10.3390/d15050606
APA StyleIturrieta-González, I., & Gené, J. (2023). Alternaria muriformis sp. nov., a New Species in Section Chalastospora Isolated from Herbivore Dung in Spain. Diversity, 15(5), 606. https://doi.org/10.3390/d15050606








